Abstract
PURPOSE
Genomewide linkage scans were performed in Caucasian (CAUC) and Old Order Amish (OOA) families to identify genomic regions containing genes responsible for refractive error control. We also performed a meta-analysis by combining these results with our previous linkage results from Ashkenazi Jewish (ASHK) and African American (AFRAM) families.
METHODS
Two hundred seventy-one CAUC and 411 OOA participants (36 and 61 families, respectively) were recruited to participate in the Myopia Family Study. Recruitment criteria were designed to enrich the sample for multiplex myopic families. Genomewide, model-free, multipoint linkage analyses were performed separately for each population by using >370 microsatellite markers. Empirical significance levels were determined via gene-dropping simulations. A meta-analysis was performed by combining linkage results from the CAUC, OOA, AFRAM, and ASHK samples, and results were compared to previously reported loci for myopia and refraction.
RESULTS
Suggestive evidence of linkage was found at 12q24 (LOD = 4.583, P = 0.00037) and 4q21 (LOD = 2.72, P = 0.0028) in the CAUC sample and at 5qter (LOD = 3.271, P = 0.0014) in the OOA. Meta-analysis linkage results were largely driven by population-specific signals from ASHK and AFRAM families. The meta-analysis showed suggestive evidence of linkage to 4q21-22 (meta-P = 0.00214) adjacent to the previously reported MYP9 and MYP11 loci.
CONCLUSIONS
The results showed suggestive evidence of linkage of ocular refraction to 12q24 and 4q21 in CAUC and to 5qter in OOA families. The meta-analysis supports the view that several genes play a role in refractive development across populations. In MFS families, four broad genomic regions (on 1p, 4q, 7p, and 12q) most likely contain genes that influence ocular refraction.
Ocular refraction is the quantitative basis behind the common optical defects of myopia and hyperopia. Although myopia and hyperopia can generally be corrected with glasses, contact lenses, or by surgical means, uncorrected refractive error is the most common cause of visual impairment worldwide. In addition, high degrees of myopia and hyperopia are associated with serious sequelae that can lead to permanent vision loss.
The current body of epidemiologic and experimental evidence suggests that, in addition to environmental and behavioral factors, genetic predispositions significantly influence refractive development. Genetic epidemiologic studies in a wide variety of populations have consistently produced high heritability estimates for refraction.1–6 Though refraction is highly heritable within populations, the prevalence of refractive errors varies among ethnic groups and geographical locations (see Morgan and Rose7 for an excellent review). For example, East Asian populations, particularly the Chinese ethnic groups in Singapore and Taiwan, have some of the highest reported prevalences of myopia in the world.8–13 These high rates are thought to be, in part, due to high levels of exposure to environmental and behavioral risk factors in these populations (myopia is associated with higher education,11,13,14 greater urbanization,15–17 intensive studying and reading habits,18–20 and a visually demanding work environment21,22). Exposure to intense near-work activities from an early age is also common in the Orthodox Jewish community, which suffers from a high prevalence of myopia. Zylbermann et al.20 has suggested that unique visual demands and intense study habits account for the high rates of myopia among Orthodox Jewish male students. Conversely, myopia tends to be less common in rural and agrarian societies in which access to formal education is limited.4,23–25 Recent work also suggests that outdoor activity during childhood has a protective effect against myopia26 which would partially explain the lower rates of myopia in rural areas. A recent study by Peet et al.4 reported lower crude prevalences of both myopia and hyperopia among the Old Order Amish compared with the overall population in the United States. The Old Order Amish live rural agrarian lifestyles that have remained relatively unchanged for centuries and do not educate their children past eighth grade.27
A number of genetic linkage studies have been performed to identify genetic loci implicated in myopization or refractive modulation.28–43 The Online Mendelian Inheritance in Man database (OMIM, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM/ provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD) currently lists 14 regions (MYP1 through MYP14) linked to refractive phenotypes in humans. Many of these loci were identified in small samples of families with severe forms of autosomal dominant myopia.31,33–35,40,41 Familial myopia also exists as part of the clinical picture of a multitude of ocular and systemic syndromes. For instance, two loci, MYP136 and MYP13,43 were mapped in families exhibiting myopia, amblyopia, and color vision abnormalities, and transmitted through an X-linked recessive mode of inheritance. Genes for severe and/or syndromic forms of myopia, however, are unlikely to account for a significant proportion of refractive errors in the population at large, and few loci linked to these more extreme traits have been replicated in independent samples.44–47
One approach to linkage analysis is to treat the phenotype as a quantitative rather than a qualitative (or binary) trait. These quantitative trait locus (QTL) methods offer the advantage of using the entire spectrum of the phenotypic distribution in the calculation of linkage statistics. QTL methods seem particularly well suited for studies of refraction because all binary classification schemes define myopia as an arbitrary cutoff of an underlying continuous trait (i.e., ocular refraction). Various QTL methods have been used to provide evidence for linkage to at least six loci that are putatively responsible for refractive variation in the populations studied.28–30,39
We previously identified linkage regions for two QTLs for refraction in two ethnically distinct populations.28,39 The first locus was mapped to 1p36 in a sample of 49 large Ashkenazi Jewish American pedigrees.39 A subsequent study of 94 predominantly nuclear African American families showed significant evidence of linkage to 7p13–25.28 In the present study, we report results of two separate QTL analyses of ocular refraction in Old Order Amish and Caucasian families. We further performed a meta-analysis by combining these linkage results with those from our earlier analyses of Ashkenazi Jewish39 and African American28 families. We found that the putative QTLs for refraction in our studies are largely population-specific, suggesting significant locus heterogeneity across studies and/or populations. We also showed that a locus of smaller effect on 4q21–22 may be common across populations. Moreover, our analyses provide some statistical support for refraction QTLs in previously reported areas of linkage to familial high myopia (at 12q,40 4q,42 and 10q33) and refraction (4q).29
METHODS
Family Recruitment and Evaluation
For the Myopia Family Study (MFS), cohorts of families were recruited from four American ethnic groups: African Americans (AFRAM) and Caucasians (CAUC) from the greater Philadelphia, Pennsylvania, area; Orthodox Ashkenazi Jewish (ASHK) families primarily from Lakewood Township, New Jersey; and Old Order Amish (OOA) families from Lancaster county, Pennsylvania. The MFS was designed to search for myopia susceptibility genes by using a systematic, consistent approach across study populations. Recruitment criteria were developed to enhance the samples for families aggregating myopia and were adapted to the cultural and demographic characteristics of each population. We describe recruitment methods for the OOA and CAUC populations. Recruitment methods and statistical analyses for the ASHK and AFRAM populations were similar and are described in detail elsewhere.28,37
For the OOA, myopic individuals were identified through liaisons in the community. These probands and their family members were then considered for recruitment in the study and received eye examinations at the Amish Eye Clinic in Strasburg, Pennsylvania. CAUC study participants were primarily recruited at the time of their vision examinations at The Eye Institute of the Pennsylvania College of Optometry (Philadelphia, PA). Recruitment of CAUC families was also accomplished through targeted mailings to established myopic patients at The Eye Institute as well as through referrals from local optometrists and ophthalmologists.
All CAUC and OOA participants received comprehensive eye examinations including objective and subjective refraction, slit lamp biomicroscopy, and dilated fundus examination. Cycloplegia was used for subjects under 40 years of age. To be eligible for participation in the study, the families were required to have at least two myopic siblings (>5 years of age), a maximum of one myopic parent, and at least three participating family members. This ascertainment scheme was designed to enrich the study samples for myopic individuals while minimizing the likelihood of bilineal transmission of potential causative alleles. In the recruitment phase, myopia was defined as a refractive error of at least −1.0 D in each meridian of both eyes. This threshold was chosen to minimize false-positive misclassifications due to measurement error.48 Because of the natural history of changes in refractive error, individuals under age 21 with less than 1 D of myopia in either principal meridian were classified as unknown and were not included in the study. Also excluded from the study were individuals with significant age-related lens changes or previous cataract surgery, a history of premature birth, or a systemic or ocular disease that could have affected their refraction. For the purposes of analysis, ocular refraction was defined as the spherical equivalent (SE) spectacle refraction, averaged between the eyes. The sample characteristics of the ASHK, AFRAM, CAUC, and OOA participating families are summarized in Table 1.
TABLE 1.
Sample Characteristics of All Populations
| Genotyping and Analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Characteristics | ||||||||||||
| Genotyped |
Male |
Female |
Mean Age |
Myopic |
Statistical Model Parameters† |
|||||||
| Population | Families | n | n(%) | n(%) | n(%) | y (SD) | n(%) | MSE* (SD) | Markers (n) |
µ(D) | h2 | Sims‡ (n) |
| CAUC | 36 | 271 | 187 (69) | 125 (46) | 146 (54) | 36.8 (20) | 206 (76) | −2.90 (312) | 385 | 0 | 0.6 | 4,898 |
| OOA | 61 | 411 | 327 (80) | 183 (45) | 228 (55) | 37.7 (18) | 210 (51) | −1.68 (2.78) | 371 | +0.75 | 0.6 | 1,460 |
| AFRAM | 96 | 493 | 393 (80) | 206 (42) | 287 (58) | 40.4 (19) | 330 (67) | −2.86 (3.58) | 385 | 0 | 0.6 | 4,898 |
| ASHK | 49 | 542 | 401 (74) | 289 (53) | 253 (47) | 39.6 (19) | 417 (77) | −3.5 (3.34) | 371 | −1 | 0.6 | 943 |
| Total | 242 | 1717 | 1308 (76) | 803 (47) | 914 (53) | 39.0 | 1163 (68) | |||||
MSE, mean spherical equivalent refraction.
Putative parameters for the distribution of ocular refraction in the underlying populations used in MERLIN-REGRESS.
Whole-genome simulations.
Written informed consent was obtained from all subjects after explanation of the nature and requirements for participation in the study. The study protocol adhered to the tenets of the Declaration of Helsinki and was approved by the institutional review boards of the University of Pennsylvania, the Pennsylvania College of Optometry, and the National Human Genome Research Institute, National Institutes of Health.
DNA Extraction and Genotyping
Peripheral blood was collected from family members by a trained physician or phlebotomist. Genomic DNA was extracted with a commercial DNA extraction kit (Puregene; Gentra Systems, Inc; Minneapolis, MN) from leukocytes obtained from whole blood collected into EDTA-coated tubes (Vacutainer; BD Biosciences, Franklin Lakes, NJ). DNA samples were stored under a unique code and whole-genome genotyping was performed at the Center for Inherited Disease Research by use of automated fluorescent microsatellite analysis. Polymerase chain reaction products were sized on a sequencer (model 3700; Applied Biosystems, Inc. [ABI], Foster City, CA). The marker sets used were modifications of the Cooperative Human Linkage Center version 9 marker set (371 autosomal markers for OOA and ASHK, and 385 for CAUC and AFRAM; average spacing, 9 cM; average heterozygosity, 0.76). The error rate, based on paired genotypes from masked duplicate samples, was 0.06% and the overall missing data rate was 3.6%.
Statistical Analysis
Our genotyping data were subjected to several quality checks to ensure accuracy. Mendelian inconsistencies and potential relationship errors were evaluated and corrected before data analysis using the SIBPAIR49 and GAS50 software packages. The accuracy of putative relationships was checked using RelCheck.51,52 Mendelian inconsistencies from individuals at multiple markers that could not be resolved were coded as missing for the purpose of the analysis.
Before linkage analysis, refraction data were transformed with a logarithmic transformation to approximate a normal distribution and limit the influence of outliers. The following transformation was applied separately for each study population to the spherical equivalent refraction data: ln(—SE + 6.16) for CAUC; ln(—SE + 9.81) for OOA; ln(—SE + 5.78) for AFRAM; and ln(—SE + 20.26) for ASHK.
Marker allele frequencies were calculated independently for each population by using the maximum-likelihood estimation. The Marsh-field clinic (Marshfield, WI) database (http://research.marshfieldclinic.org/genetics/home/index.asp) was used to determine sex-averaged genetic map distances (in Kosambi centimorgans). All linkage analyses were conducted separately for the CAUC, OOA, AFRAM, and ASHK samples using population-specific allele frequencies. Multipoint QTL linkage statistics were calculated at 2-cM intervals across the autosomal genome using the regression-based method in the software package MERLIN, version 1.0.1.53 The MERLIN statistical package implements an extended Haseman-Elston regression-based linkage method through its MERLIN-REGRESS subroutine.54 This method is applicable to extended pedigrees and to samples ascertained on the basis of trait values, provided that appropriate values for population parameters of the trait distribution are specified. Consequently, this regression approach requires a priori specification of the underlying population-specific mean (µ), variance (V), and heritability (h2) of the trait.54 Although the true underlying population parameters of the refractive error distribution are unknown for the samples studied, numerous epidemiologic studies have reported these values in various ethnic groups. Heritability estimates for refraction have consistently been estimated to lie between 0.5 and 0.9 across populations.2–6 Hence, we set h2 to 0.6 in all population-specific analyses. The population means (µ) for the CAUC and AFRAM samples were set to 0 D, which roughly reflects values reported by previous population-based surveys in Caucasian American and African American populations.6,55,56 An underlying mean (µ) of +0.75 D was chosen for the OOA sample. This figure corresponds to the mean refractive error of an unselected sample of older OOA participants in a study of age-related macular degeneration.4 The prevalence of myopia is thought to be higher among Orthodox Ashkenazi Jews because of the unique visual demands experienced by individuals in the community.18,20 Consequently, we specified a mean refraction of −1.0 D for the ASHK sample. Variance (V) parameters were set to the population-specific sample variances, after transformation. Population parameters used in our analyses are summarized in Table 1.
Simulations, Genomewide Significance Levels, and Power
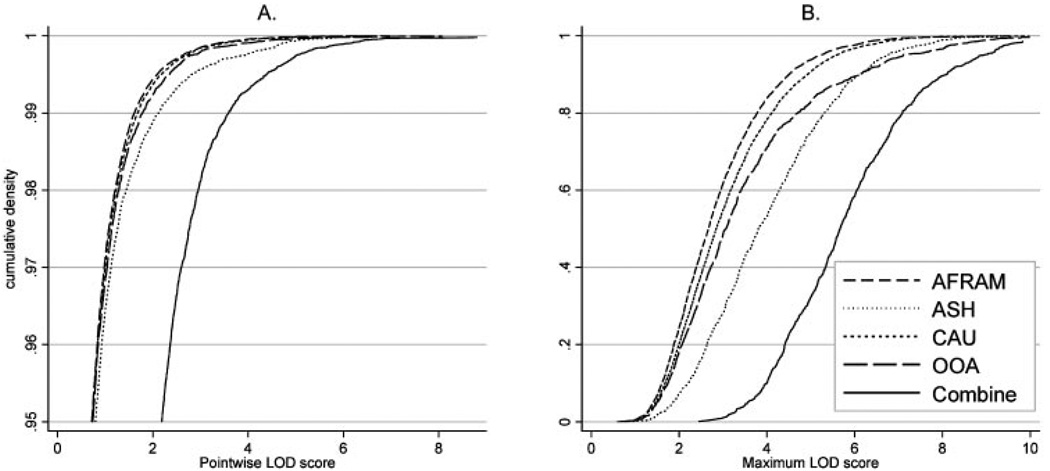
The MERLIN-REGRESS procedure provides locus-specific, nominal, LOD scores and significance levels based on the asymptotic χ2 distribution.54 We previously reported that LOD scores estimated by MERLIN-REGRESS can be inflated relative to expectations, yielding anti-conservative P-values and inflated type-1 error rates.39 Unbiased empirical significance levels can be determined, however, via a gene-dropping simulation procedure implemented in the software package53,57 (see the MERLIN tutorial for more information: http://www.sph.umich.edu/csg/abecasis/Merlin/reference/simulation.html). Briefly, MERLIN assigns random chromosomes to founders while preserving family structure, marker allele frequencies, marker spacing, the observed pattern of missing genotypes, and individual phenotypic information. Markers are then segregated through the pedigrees using the same relationships and genetic map as specified in the original dataset. Genomewide analyses are then repeated using these simulated marker datasets. Because the markers are generated and transmitted independently from trait values, these simulations yield empirical null distributions of linkage statistics. The simulation procedure offers a means of assessing both pointwise and genomewide significance levels (i.e., false-positive rates) in a given sample. Of importance, because these simulations preserve the family structures, phenotypes, and marker information of the original datasets, they can account for idiosyncrasies in particular samples and do not, in general, yield identical empirical null distributions across samples (Fig. 1).
FIGURE 1.
Cumulative densities of MERLIN-REGRESS linkage statistics for all samples. Cumulative densities of (A) pointwise and (B) maximum genomewide MERLIN-REGRESS LOD scores under no linkage for four populations, and their combined LOD scores. LOD scores under the hypothesis of no linkage were determined via gene-dropping simulations. Pointwise cumulative densities are shown only in the more relevant range ≥ 0.95, corresponding to PPW ≤ 0.05. The number of whole-genome replicates in our simulations was: 4898 for AFRAM, 906 for ASHK, 4898 for CAUC, and 1405 for OOA.
We performed whole-genome simulations independently for each population to compute sample-specific empirical P-values. The following number of genomewide simulations was performed: 4898 for AFRAM, 1460 for OOA, 943 for ASHK, and 4898 for the CAUC. All analyses and simulations were performed on a 120-node Linux cluster made up of dual 64-bit processors (Opteron; AMD, Sunnyvale, CA) with 4 Gb of random access memory and 60 Gb of “scratch” space per node. For the OOA and ASHK samples, the number of whole-genome simulations was limited by the computational burden required to compute multilocus allele-sharing statistics in large pedigrees.58
In each sample population, the maximum QTL LOD score was determined for each replicate, and the empirical genomewide P (PGW) was estimated as the proportion of simulated datasets in which the maximum LOD score was greater than or equal to the maximum LOD score obtained using the original data. Locus-specific (i.e., pointwise) empirical P-values (PPW) were obtained by comparing pointwise LOD scores from the original data to the distribution of all LOD scores under the null hypothesis of no linkage (i.e., from our simulations). Since linkage statistics were evaluated at 1750 locations in each simulated dataset, PPW was based on the distribution of: ~8.6 million LOD scores for AFRAM and CAUC; ~2.55 million for OOA; and ~1.65 million for ASHK. All pointwise LOD scores were converted to sample-specific empirical P to allow for comparisons between populations and metaanalysis. Cumulative distributions of MERLIN-REGRESS LOD score statistics are presented in Figure 1.
Though comprehensive power studies are not feasible given the extensive additional simulations required to obtain valid type II error estimates, sample-specific expected LOD scores (ELODs) can be estimated from the data, assuming complete marker informativeness. ELODs represent estimates of the conditional expectation of the LOD scores under the alternative hypothesis of linkage, given family structures, trait data, and a predefined genetic model. As expected, ELODs computed by the MERLIN-REGRESS program suggest that the power to detect quantitative trait loci of small effect may be limited in all our study populations. For example, for a locus-specific heritability of 20%, the ELODs were 1.41 (ASHK), 0.93 (AFRAM), 1.67 (OOA), and 0.84 (CAUC), whereas a locus-specific effect of 40% yield ELODs of 5.65 (ASHK), 3.72 (AFRAM), 6.68 (OOA), and 3.36 (CAUC). These relative power estimates show that, assuming identical underlying genetic mechanisms for refraction across populations, the statistical power to detect linkage is highest for OOA and ASHK compared to AFRM and CAUC.
Sensitivity Analyses
We have previously shown that the methodology implemented in MERLIN-REGRESS is relatively robust to alternate parameter specifications (within plausible parameter ranges), as long as statistical significance is determined empirically.39 To ensure that this was also the case across all samples, we conducted sensitivity analyses by individually varying µ and h2 in our population-specific statistical models. Our results were similar after varying µ by ±1 D, and h2 by ±0.2 in each of our populations and are not presented here.
Meta-analyses
Meta-analyses were performed by combining multipoint linkage evidence at 2-cM intervals across our samples from four populations, and determining the associated significance levels. Formally, the meta-analysis tests the null hypothesis of no linkage in any sample versus the alternative of linkage in at least one population sample. Sample-specific P-values were combined by using a variation of Fisher’s method59 by summing the negative natural logarithm of the P-values at each locus. We computed meta-analysis P (meta-PPW) empirically using our gene-dropping simulations. These were calculated by comparing the unweighted sums of locus-specific —ln(PPW) of the original data to the distribution of summed — ln(PPW) in 1 million random draws from our simulations.
RESULTS
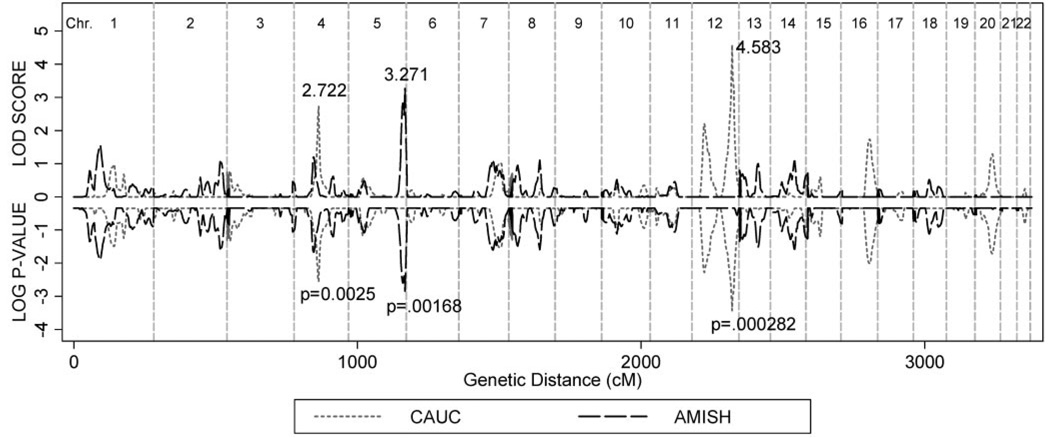
Multipoint MERLIN-REGRESS LOD scores and their corresponding PPW for the CAUC and OOA populations are shown in Figure 2. The maximum LOD score for the CAUC population was 4.583 at 142 cM on chromosome 12. This LOD score corresponded to an empirical PPW = 0.00037 and a genomewide significance (PGW) = 0.13. For the CAUC population, three additional regions had linkage peaks with associated PPW below 0.01: chromosome 4 at 98 cM (LOD = 2.72, PPW = 0.00279), 12 at 44 cM (LOD = 2.21, PPW = 0.0053), and 16 at 102 cM (LOD = 1.744, PPW = 0.0096).
FIGURE 2.
Multipoint linkage statistics for CAUC and OOA families. Multipoint MERLIN-REGRESS LOD scores and empirical PPW for CAUC and OOA populations. Empirical P-values were obtained through gene-dropping simulations. There were 4898 whole genome replicates in the CAUC sample and 1405 in the OOA population. PPW was based on the distribution of ~5.4 million LOD scores for CAUC and ~1.5 million for OOA under no linkage.
The maximum LOD score for the OOA families was 3.271 (PPW = 0.0014; PGW = 0.388) at 200 cM on chromosome 5. No linkage peak in either population reached the genomewide significance threshold of PGW < 0.05. There was no correspondence between the linkage profiles of the CAUC and OOA populations at their respective linkage peaks. However, faint linkage signals overlapped on chromosomes 4 and 7 (Fig. 2).
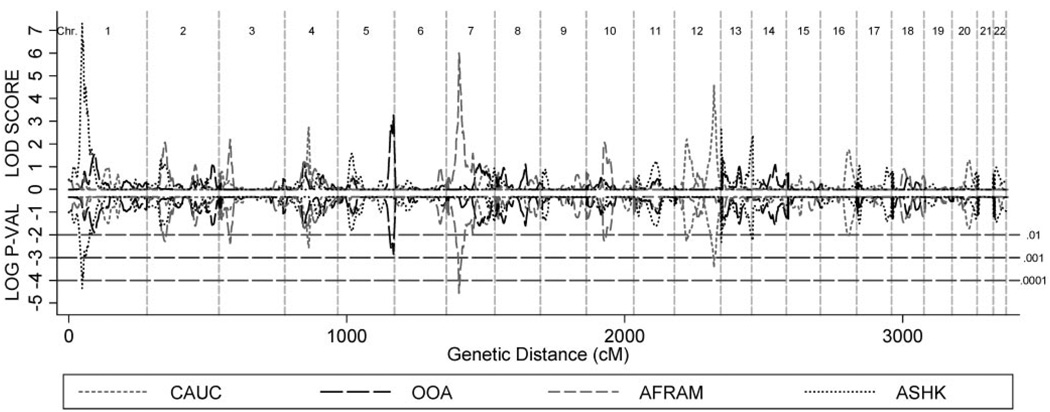
Multipoint MERLIN-REGRESS linkage scores and empirical PPW for the AFRAM, ASHK, CAUC, and OOA populations are presented in Figure 3. For additional details about QTL linkage analyses of refraction in ASHK and AFRAM families, refer to Wojciechowski et al.39 and Ciner et al.28 The previously identified linkage peaks on chromosomes 1 (ASHK)39 and 7 (AFRAM)28 were not replicated in the CAUC and OOA samples. Moreover, in the ASHK or AFRAM samples there was no evidence of linkage of ocular refraction to loci identified in OOA (chromosome 5) and CAUC (chromosome 12) families (Fig. 3).
FIGURE 3.
Multipoint linkage statistics for all populations. Empirical P-values were obtained through gene-dropping simulations. There were 4898 whole genome replicates in the CAUC and AFRAM samples, 943 in ASHK and 1405 in the OOA population. PPW are based on the null distribution of ~5.4 million LOD scores for CAUC and AFRAM, ~1 million for ASHK, and ~1.6 million for OOA.
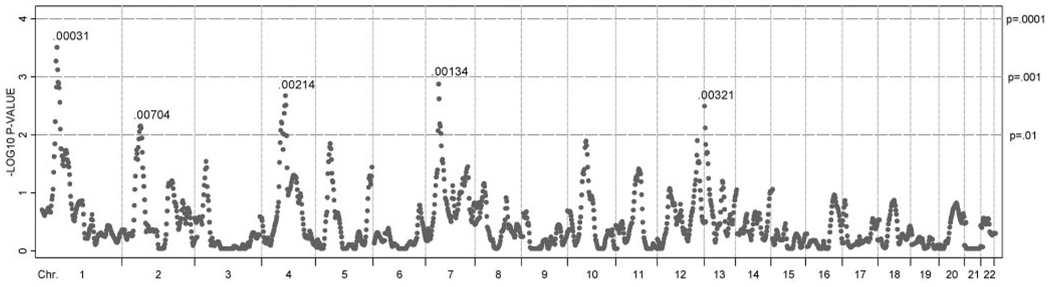
Meta-analysis empirical P-values at 2-cM intervals for all populations are shown in Figure 4. As expected, the population-specific linkage peaks on chromosomes 1 (ASHK)39 and 7 (AFRAM)28 also showed strong evidence of linkage in the meta-analysis. Hence, the two highest linkage peaks in our meta-analysis were located at 48 cM on chromosome 1 (meta-PPW = 0.00031), and at 46 cM on chromosome 7 (meta-PPW = 0.00134). Three additional areas showed meta-PPW = < 0.01: chromosome 2 at 64 cM (meta-PPW 0.0074), chromosome 4 at 96 cM (meta-PPW 0.00214), and chromosome 13 at 2 cM (meta-PPW = 0.00321).
FIGURE 4.
Meta-analysis linkage P-values for ocular refraction in four populations. Pointwise meta–P-values at 2-cM intervals. Dashed vertical line: chromosome boundaries; horizontal dashed lines: P-value thresholds of 0.01, 0.001, and 0.0001.
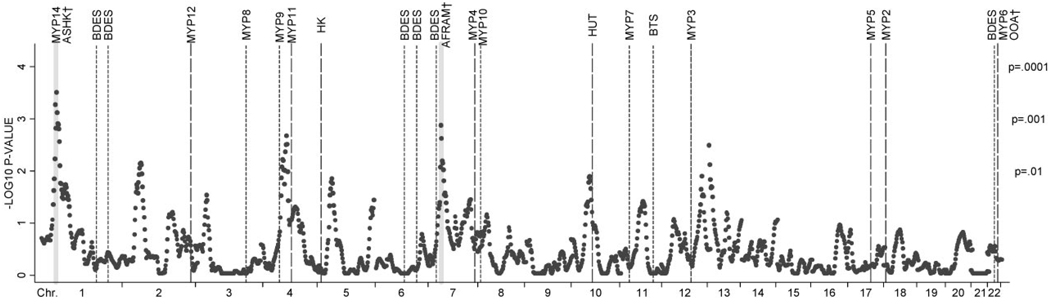
Figure 5 shows pointwise meta-analysis linkage results along with the locations of previously reported putative loci for myopia and ocular refraction, including (for completeness) loci found in ASHK37,39,60 and AFRAM28 families used in the current meta-analysis. Loci that showed suggestive evidence of linkage to refraction in the British Twin Study29 and the Beaver Dam Eye Study30 are also included. One region in our metaanalysis showed strong statistical evidence (P < 0.01) of replication of previously mapped regions in independent populations: Our chromosome 4 linkage peak at 96 cM lies between the MYP9 QTL (~70 cM) found in the British Twin Study29 and the MYP11 (~113 cM) locus for autosomal dominant high myopia which was recently localized in a large Chinese family.42 Of the individual populations in the present study, only the CAUC sample showed suggestive evidence of linkage (defined as PPW < 0.01) to this region (LOD = 2.722; PPW < 0.0028 at 98 cM). One additional region, with a peak at 66 cM on chromosome 10 in the meta-analysis, showed marginal evidence of replication (PPW < 0.05) of a high myopia locus (at ~75 cM) identified in a large Hutterite family.33
FIGURE 5.
Pointwise meta-analysis P-values for combined MERLIN-REGRESS linkage statistics. Vertical dashed lines: approximate locations of loci for myopia (long dashes) and putative QTLs for refraction (short dashes). Shaded areas show QTLs identified in the families used in the current meta-analysis. Ticks show chromosomal boundaries. Tags indicate either named loci (ex: MYP14) or the study in which currently unnamed loci were identified (BDES, Beaver Dam Eye Study30; BTS, British Twin Study29 HK, Hong Kong study31; HUT, Hutterite Family33). †Loci identified in Myopia Family Study populations used in the current meta-analysis. AFRAM,28 ASHK,37,39,60 and OOA.38
DISCUSSION
We performed QTL linkage analyses for ocular refraction in two culturally distinct European-derived populations: the Old Order Amish (OOA) and a more heterogeneous sample of Caucasian Americans (CAUC). We also performed a linkage meta-analysis by combining these results with our previous analyses of Orthodox Ashkenazi Jewish (ASHK) and African American (AFRAM) families.
In the CAUC sample, the two highest linkage signals mapped to 12q24.2–3 (LOD = 4.583; PPW = 0.000374 at 142 cM) and 4q21–22 (LOD = 2.72; PPW = 0.0028 at 98 cM). In the OOA group, the maximum multipoint linkage signal was seen at 5qter (LOD = 3.271; PPW = 0.0014 at 200 cM). Neither linkage signal in the CAUC or OOA analyses reached genome-wide statistical significance at PGW < 0.05. Specifically, an LOD ≥ 4.583 occurred in one of every 7.7 whole-genome simulations (PGW = 0.13) of the CAUC family data under the assumption of no linkage. An LOD ≥ 3.127 corresponded to PGW = 0.388 in the OOA sample (i.e., 1 of every 2.6 simulations in the OOA yielded a maximum LOD of at least 3.127). It should also be noted that the strongest linkage signal in OOA families occurred in a telomeric region (5qter), where linkage statistics are inherently unstable due to variations in genetic map precision and to lower marker information content for multipoint analysis. Although it is of some interest, this region should be considered tentative unless replicated in a different sample.
Although genetic epidemiologic studies have shown ocular refraction to be highly heritable in various European-derived populations,2–4,6 our linkage analyses in the CAUC and OOA samples do not provide conclusive evidence of linkage to any one genomic region. This finding may be the result of genetic heterogeneity or polygenic inheritance, wherein several genes of small effect contribute to the phenotypic distribution in the overall population. Such QTLs are difficult to detect in linkage analysis because each locus would account for only a small proportion of the total population variation of the phenotype. Moreover, our analyses may have been underpowered because of limited sample sizes combined with small locus effects or genetic heterogeneity. Though our ELODs show that our power to detect linkage was higher in the OOA sample than in the other populations, it should be noted that these power estimates assume identical genetic models of inheritance, and equivalent contributions of environmental factors, across populations.
It is generally acknowledged that the development of myopia, and hence the distribution of ocular refraction within populations, is influenced by genetic, behavioral, and environmental factors. Although the exact nature of the relationship between the visual environment and ocular refraction has not been worked out with certainty, excessive visual demand, and near-work activity do appear to affect the predisposition to myopia. It is therefore probable that susceptibility genes interact with environmental influences to determine refractive error. Hence, populations such as the OOA, in which the predisposing environmental factors for myopization are relatively scarce,27,61 may not express the myopic phenotype to the same extent as other ethnic groups.
To our knowledge, there has been no published population-based survey of ocular refraction in the OOA. However, data from a previous report suggest that myopia is less common in this population compared to other groups: Using data from a family study of age-related macular degeneration, we estimated the mean refraction in OOA to be 0.71 D (SD = 1.61).4 Moreover, the distribution of refraction was highly leptokurtotic (kurtosis = 14.9) and the prevalence of myopia (≤ —1.0 D) was only 8.7%.4 It is therefore possible that the genes mediating refractive error do not express fully in the OOA because of a relative paucity of interacting environmental influences. Nevertheless, our linkage sample was selected for multiplex myopic families which should have increased our power to detect these loci compared to a population-based study in the same sample.
Our meta-analysis results were largely driven by individual-population linkage peaks. The two strongest linkage signals in the combined analysis (meta-PPW = 0.00031 at 46 cM on 1p36.1; meta-PPW = 0.00134 at 46 cM on 7p14) mirrored refraction QTLs previously identified in the ASHK39 and AFRAM28 subsamples, respectively. As would be expected when there is locus heterogeneity among populations, the pointwise significance levels of these linkage peaks were lower in the meta-analysis than their respective population-specific values. This phenomenon may also have been due to discrepancies between the locations of linkage signals among populations. For instance, two linkage peaks separated by a few centimorgans would not sum together to produce a stronger combined linkage signal, even though they may both be due to the same underlying trait locus. This failure can be a problem in meta-analyses of linkage studies because estimating the location of disease-predisposing loci is inherently prone to variation. To account for these uncertainties in gene location estimates, we repeated our meta-analyses after grouping the linkage data into 10- and 20-cM bins (data not shown). The results were consistent with the unbinned analyses, implying that poor alignment of estimated locus locations between populations did not significantly influence our meta-analysis results.
We compared the locations of linkage signals in our analyses to the positions of previously reported regions for high myopia and putative QTLs for refraction (Fig. 5). The chromosome 12 linkage peak in the CAUC sample was roughly 38 cM from the MYP3 locus for autosomal dominant high myopia first identified by Young et al.40 in a large American family of Italian and German descent. Mutti et al.45 provided support for linkage to this locus (nonparametric linkage P = 0.003) in a study of mild myopia in an ethnically diverse sample of American school children. The MYP3 region contains genes coding for the proteoglycans lumican (LUM), decorin, the dermatan sulfate proteoglycan 3 precursor (DSPG3), and fibromodulin (FMOD). These proteoglycans regulate collagen fibril assembly and interaction and are expressed in ocular tissues. In a case–control study of Taiwanese individuals, Wang et al.62 found that a single nucleotide polymorphism in the 5′ promoter region of the lumican gene was significantly associated with high myopia. Paluru et al.63 screened the same family used to identify the MYP3 locus for sequence alterations in the LUM and FMOD genes and found no relationship between affection status and polymorphisms within these genes. However, their study did not query sequence alterations in the 5′ promoter region of the lumican gene, which may explain their negative findings. Our linkage signal in the CAUC sample may correspond to the MYP3 locus and/or to the chromosome 12 proteoglycan gene complex. Certainly, given previous work and the biological plausibility of a role for proteoglycan genes in human refractive variation, this region should be investigated further using fine-mapping techniques.
The 4q21–22 (96 cM) linkage peak from our meta-analysis is ~20 cM telomeric to the center of the MYP929 locus and ~15 cM centromeric to MYP11.42 MYP9 was mapped to 4q12 by Hammond et al.29 in a QTL analysis of ocular refraction in 221 British twins. Zhang et al.42 localized MYP11 to 4q22–27 in a parametric linkage analysis of a large Chinese family with autosomal dominant high myopia. There was significant overlap between our meta-analysis linkage profile and the MYP9 and MYP11 loci: The interval within which our meta-analysis P-values were below 0.01 extended from 80 to 100 cM. Hence, our results also provide evidence supporting the presence of a gene controlling refraction in this region. However, it is not clear whether the chromosome 4q linkage signals from these three independent studies represent distinct QTLs or whether they correspond to a common underlying locus for refraction control.
Another local linkage peak (chromosome 10: P = 0.014 at 52 cM) from our meta-analysis of ocular refraction was adjacent to a locus (centered at ~75 cM) mapped in a large Hutterite family segregating high myopia.33 This signal, which mainly originated from the AFRAM population in our study, should be explored further for candidate genes for refractive regulation.
Our meta-analysis linkage peaks on 1p36.1 and 7p14 were previously reported for our ASHK39 and AFRAM28 samples, respectively. To our knowledge, the chromosome 1 (MYP14)39 QTL has not been replicated in other independent studies; and we saw no evidence of linkage to this region in analyses of the AFRAM, CAUC, and OOA pedigrees. This lack of replication is not unexpected given the heterogeneous and complex nature of the phenotype. It is also possible that the linkage signal to chromosome 1 in the ASHK families may have been a spurious (i.e., false-positive) result. Although false-positive errors cannot be entirely eliminated, our statistical methodology and extensive use of simulations resulted in a strict control of type 1 errors in all analyses.
The chromosome 7 locus found in AFRAM families,28 however, is in close proximity to an area of suggestive linkage (centered at 29 cM) reported by Klein et al.30 in the Beaver Dam Eye Study cohort. Further, though still preliminary, support for linkage to this region was provided by Paget et al. (IOVS 2005;46:ARVO E-Abstract 3816), who published an abstract that reported significant linkage of high myopia to 7p15.3 in 12 of 50 French families. Given that a linkage peak at 7p14–15 was identified in a least two ethnically and genetically distinct groups, it is possible that this QTL may be common to a variety of populations.
CONCLUSIONS
Refractive development is mediated by a complex interplay between biological factors (including several genes) and environmental influences. Linkage studies have proven successful in identifying genomic regions that merit scrutiny in future investigations, but they lack the resolution to refine these areas of interest with high precision. Specific causative polymorphisms and/or structural variations within these candidate regions can potentially be identified with more powerful analytical tools such as genetic association (i.e., linkage disequilibrium) studies. Indeed, association studies of high myopia have recently yielded interesting findings that will help unravel the complex genetics of common refractive errors in human populations.62,64 Future work on the genetic epidemiology of refractive error in human populations should not only use state-of-the-art genomic technologies and analytical methods (genomewide association studies, copy-number polymorphism assays, functional genomics and systems biology, to name a few) but build on a wealth of data provided by traditional laboratory science and epidemiology.
Our QTL linkage analyses in four distinct ethnic groups suggest that considerable genetic heterogeneity exists in the control of refractive development. Because we used composite P-values, we were unable to formally test for genetic heterogeneity in our meta-analyses. However, evidence of genetic heterogeneity in refractive control is reinforced by comparisons with previous linkage studies in independent populations, which have identified more than a dozen putative loci for nonsyndromic myopia or refractive error. In our meta-analysis, three of these genomic regions (on chromosomes 4, 7, and 12), corresponding to four previously mapped loci, showed some evidence of linkage to ocular refraction. In addition, although not replicated in other samples, the 1p36 region (MYP14) showed a highly significant linkage peak in the ASHK population.39 These regions should be further investigated with fine-mapping methodologies to identify the genes and genetic polymorphisms responsible for refractive control. Table 2 summarizes the characteristics of these broad linkage regions. Cumulatively, they contain ~1000 genes listed in the RefSeq database (http://www.ncbi.nlm.nih.gov/RefSeq/ provided in the public domain by the National Center for Bioinformatic Information, Bethesda, MD), of which more than 500 have been shown to be expressed in the human retina or retinal pigment epithelium65 (the likely sites of refractive regulation). In addition, more than 400 genes (or loci) in these regions are referenced in the OMIM database as related to human disease or quantitative phenotypes. Fine-mapping genotyping of these regions is currently being undertaken in our sample populations.
TABLE 2.
Characteristics of Linkage Regions 1p, 4q, 7p, and 12q
| Chr. | Population | Linkage Evidence |
Evidence from Other Studies |
Cytogenetic Location |
Linkage Peak (cM) |
Approximate Boundaries of Linkage Area |
Genes in Linkage Area (n) |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic Map (cM) |
Physical Map (Mb) |
|||||||||
| Total | OMIM* | EyeSAGE† | ||||||||
| 1 | ASHK | GW significant | No | 1p36.13–1p34 | 49 | 37–72 | 17.5–41.4 | 390 | 180 | 220 |
| 4 | Meta-analysis AFRAM |
Suggestive | Yes29,42 | 4q12–4q25 | 96 | 69–106 | 57.5–108.8 | 328 | 141 | 157 |
| 7 | American | GW significant | Yes30‡ | 7p15.3-p14.1 | 47 | 33–62 | 20.5–38 | 146 | 63 | 74 |
| 12 | CAUC | Suggestive | Possible40§ | 12q24.22-q24.32 | 142 | 132–150 | 115–127 | 136 | 60 | 81 |
Referenced in the Online Mendelian Inheritance in Man database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM).
Expressed in human retina or retinal pigment epithelium in the EyeSAGE database (Bowes Rickman et al.65).
Paget S, et al. IOVS 2005;46:ARVO EAbstract 3816.
The center of the MYP3 locus for autosomal dominant high myopia is ≈38 cM from our suggestive linkage signal on 12q.
Acknowledgments
The authors thank the families for their participation in the study; Reuvain Shanik, MD, and Rabbi Yitzchok Rozsansky for their enthusiastic support of the Myopia Family Study; and the staff of The Eye Institute of the Pennsylvania College of Optometry and the Amish Eye Clinic in Strasburg, Pennsylvania, and local optometrists and ophthalmologists for their aid in participant recruitment.
Supported in part by U.S. Public Health National Eye Institute Grant EY12226 (DS, EC) and funds from the intramural program of the National Human Genome Research Institute, National Institutes of Health (JEB-W, GI, TNH, RW). RW received a William C. Ezell-CIBA Vision Fellowship from the American Optometric Foundation. Geno-typing services were provided by the Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the National Institutes of Health to Johns Hopkins University (contract number N01-HG-65403).
Footnotes
Disclosure: R. Wojciechowski, None; D. Stambolian, None; E. Ciner, None; G. Ibay, None; T.N. Holmes, None; J.E. Bailey-Wilson, None
References
- 1.Alsbirk PH. Refraction in adult West Greenland Eskimos. A population study of spherical refractive errors, including oculometric and familial correlations. Acta Ophthalmol (Copenh) 1979;57:84–95. doi: 10.1111/j.1755-3768.1979.tb06663.x. [DOI] [PubMed] [Google Scholar]
- 2.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–1236. [PubMed] [Google Scholar]
- 3.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peet JA, Cotch MF, Wojciechowski R, Bailey-Wilson JE, Stambolian D. Heritability and familial aggregation of refractive error in the Old Order Amish. Invest Ophthalmol Vis Sci. 2007;48:4002–4006. doi: 10.1167/iovs.06-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teikari JM, Kaprio J, Koskenvuo MK, Vannas A. Heritability estimate for refractive errors: a population-based sample of adult twins. Genet Epidemiol. 1988;5:171–181. doi: 10.1002/gepi.1370050304. [DOI] [PubMed] [Google Scholar]
- 6.Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, West SK. Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci. 2005;46:1588–1592. doi: 10.1167/iovs.04-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan I, Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24:1–38. doi: 10.1016/j.preteyeres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Lin LL, Shih YF, Tsai CB, et al. Epidemiologic study of ocular refraction among schoolchildren in Taiwan in 1995. Optom Vis Sci. 1999;76:275–281. doi: 10.1097/00006324-199905000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Lin LL, Chen CJ, Hung PT, Ko LS. Nation-wide survey of myopia among schoolchildren in Taiwan 1986. Acta Ophthalmol Suppl. 1988;185:29–33. doi: 10.1111/j.1755-3768.1988.tb02657.x. [DOI] [PubMed] [Google Scholar]
- 10.Matsumura H, Hirai H. Prevalence of myopia and refractive changes in students from 3 to 17 years of age. Surv Ophthalmol. 1999;44 suppl 1:S109–S115. doi: 10.1016/s0039-6257(99)00094-6. [DOI] [PubMed] [Google Scholar]
- 11.Saw SM, Wu HM, Seet B, et al. Academic achievement, close up work parameters, and myopia in Singapore military conscripts. Br J Ophthalmol. 2001;85:855–860. doi: 10.1136/bjo.85.7.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saw SM, Goh PP, Cheng A, Shankar A, Tan DT, Ellwein LB. Ethnicity-specific prevalences of refractive errors vary in Asian children in neighbouring Malaysia and Singapore. Br J Ophthalmol. 2006;90:1230–1235. doi: 10.1136/bjo.2006.093450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu N, Nomura H, Ando F, Niino N, Miyake Y, Shimokata H. Refractive errors and factors associated with myopia in an adult Japanese population. Jpn J Ophthalmol. 2003;47:6–12. doi: 10.1016/s0021-5155(02)00620-2. [DOI] [PubMed] [Google Scholar]
- 14.Tay MT, Au Eong KG, Ng CY, Lim MK. Myopia and educational attainment in 421,116 young Singaporean males. Ann Acad Med Singapore. 1992;21:785–791. [PubMed] [Google Scholar]
- 15.Dandona R, Dandona L, Srinivas M, et al. Refractive error in children in a rural population in India. Invest Ophthalmol Vis Sci. 2002;43:615–622. [PubMed] [Google Scholar]
- 16.Dandona R, Dandona L, Naduvilath TJ, Srinivas M, McCarty CA, Rao GN. Refractive errors in an urban population in Southern India: the Andhra Pradesh Eye Disease Study. Invest Ophthalmol Vis Sci. 1999;40:2810–2818. [PubMed] [Google Scholar]
- 17.Zhan MZ, Saw SM, Hong RZ, et al. Refractive errors in Singapore and Xiamen, China: a comparative study in school children aged 6 to 7 years. Optom Vis Sci. 2000;77:302–308. doi: 10.1097/00006324-200006000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Ben Simon GJ, Peiss M, Anis E, Nakra T, Luski A, Spierer A. Spectacle use and reduced unaided vision in third grade students: a comparative study in different educational settings. Clin Exp Optom. 2004;87:175–179. doi: 10.1111/j.1444-0938.2004.tb03171.x. [DOI] [PubMed] [Google Scholar]
- 19.Kinge B, Midelfart A, Jacobsen G, Rystad J. The influence of near-work on development of myopia among university students: a three-year longitudinal study among engineering students in Norway. Acta Ophthalmol Scand. 2000;78:26–29. doi: 10.1034/j.1600-0420.2000.078001026.x. [DOI] [PubMed] [Google Scholar]
- 20.Zylbermann R, Landau D, Berson D. The influence of study habits on myopia in Jewish teenagers. J Pediatr Ophthalmol Strabismus. 1993;30:319–322. doi: 10.3928/0191-3913-19930901-12. [DOI] [PubMed] [Google Scholar]
- 21.McBrien NA, Adams DW. A longitudinal investigation of adult-onset and adult-progression of myopia in an occupational group: refractive and biometric findings. Invest Ophthalmol Vis Sci. 1997;38:321–333. [PubMed] [Google Scholar]
- 22.Saw SM, Chia SE, Chew SJ. Relation between work and myopia in Singapore women. Optom Vis Sci. 1999;76:393–396. doi: 10.1097/00006324-199906000-00019. [DOI] [PubMed] [Google Scholar]
- 23.Naidoo KS, Raghunandan A, Mashige KP, et al. Refractive error and visual impairment in African children in South Africa. Invest Ophthalmol Vis Sci. 2003;44:3764–3770. doi: 10.1167/iovs.03-0283. [DOI] [PubMed] [Google Scholar]
- 24.Pokharel GP, Negrel AD, Munoz SR, Ellwein LB. Refractive Error Study in Children: results from Mechi Zone, Nepal. Am J Ophthalmol. 2000;129:436–444. doi: 10.1016/s0002-9394(99)00453-5. [DOI] [PubMed] [Google Scholar]
- 25.Saw SM, Hong RZ, Zhang MZ, et al. Near-work activity and myopia in rural and urban schoolchildren in China. J Pediatr Ophthalmol Strabismus. 2001;38:149–155. doi: 10.3928/0191-3913-20010501-08. [DOI] [PubMed] [Google Scholar]
- 26.Onal S, Toker E, Akingol Z, et al. Refractive errors of medical students in Turkey: one year follow-up of refraction and biometry. Optom Vis Sci. 2007;84:175–180. doi: 10.1097/OPX.0b013e3180335c52. [DOI] [PubMed] [Google Scholar]
- 27.Dewalt MW. Amish Education in the United States and Canada. Lanham, MD: Rowman and Littlefield Publishers, Inc; 2006. [Google Scholar]
- 28.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32(5):454–463. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond CJ, Andrew T, Tat MY, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–85. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 31.Lam CY, Tam PO, Fan DS, et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49(9):3768–3778. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 32.Naiglin L, Gazagne C, Dallongeville F, et al. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–124. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–236. [PMC free article] [PubMed] [Google Scholar]
- 34.Paluru P, Ronan SM, Heon E, et al. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–1836. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 35.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–2307. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease: linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–286. [PubMed] [Google Scholar]
- 37.Stambolian D, Ibay G, Reider L, et al. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004;75:448–459. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stambolian D, Ciner EB, Reider LC, et al. Genome-wide scan for myopia in the Old Order Amish. Am J Ophthalmol. 2005;140:469–476. doi: 10.1016/j.ajo.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Wojciechowski R, Moy C, Ciner E, et al. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–399. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young TL, Ronan SM, Alvear AB, et al. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–1424. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young TL, Ronan SM, Drahozal LA, et al. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–119. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22–q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–560. [PubMed] [Google Scholar]
- 43.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23–q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 UK families. Invest Ophthalmol Vis Sci. 2004;45:2879–2885. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- 45.Mutti DO, Semina E, Marazita M, Cooper M, Murray JC, Zadnik K. Genetic loci for pathological myopia are not associated with juvenile myopia. Am J Med Genet. 2002;112:355–360. doi: 10.1002/ajmg.10683. [DOI] [PubMed] [Google Scholar]
- 46.Young TL, Deeb SS, Ronan SM, et al. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004;122:897–908. doi: 10.1001/archopht.122.6.897. [DOI] [PubMed] [Google Scholar]
- 47.Ibay G, Doan B, Reider L, et al. Candidate high myopia loci on chromosomes 18p and 12q do not play a major role in susceptibility to common myopia. BMC Med Genet. 2004;5:20. doi: 10.1186/1471-2350-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goss DA, Grosvenor T. Reliability of refraction: a literature review. J Am Optom Assoc. 1996;67:619–630. [PubMed] [Google Scholar]
- 49.Duffy DL. Sibpair: a program for nonparametric linkage/association analysis. Am J Hum Genet. 1997;61:A197. [Google Scholar]
- 50.Young A. GAS (Genetic Analysis System) Oxford, UK: Oxford University; 1995. [Google Scholar]
- 51.Boehnke M, Cox NJ. Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63:1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 54.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz J, Tielsch JM, Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997;38:334–340. [PubMed] [Google Scholar]
- 56.Kempen JH, Mitchell P, Lee KE, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 57.Terwilliger JD, Speer M, Ott J. Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol. 1993;10:217–224. doi: 10.1002/gepi.1370100402. [DOI] [PubMed] [Google Scholar]
- 58.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisher RA. Statistical Methods for Research Workers. 13 ed. Edinburgh, UK: Oliver & Boyd; 1925. [Google Scholar]
- 60.Stambolian D, Ibay G, Reider L, et al. Genome-wide scan of additional Jewish families confirms linkage of a myopia susceptibility locus to chromosome 22q12. Mol Vis. 2006;12:1499–1505. [PubMed] [Google Scholar]
- 61.Hostetler JA. Amish Society. 4th ed. Baltimore: Johns Hopkins University Press; 1993. [Google Scholar]
- 62.Wang IJ, Chiang TH, Shih YF, et al. The association of single nucleotide polymorphisms in the 5′-regulatory region of the lumican gene with susceptibility to high myopia in Taiwan. Mol Vis. 2006;12:852–857. [PubMed] [Google Scholar]
- 63.Paluru PC, Scavello GS, Ganter WR, Young TL. Exclusion of lumican and fibromodulin as candidate genes in MYP3 linked high grade myopia. Mol Vis. 2004;10:917–922. [PubMed] [Google Scholar]
- 64.Mutti DO, Cooper ME, O’Brien S, et al. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–1019. [PMC free article] [PubMed] [Google Scholar]
- 65.Bowes Rickman C, Ebright JN, Zavodni ZJ, et al. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–2316. doi: 10.1167/iovs.05-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]