Abstract
The etiology of Parkinson’s disease is unclear but appears to involve mitochondrial dysfunction, proteasome inhibition, and environmental toxins. It has been shown that pesticides, including the complex I inhibitor rotenone, cause proteasome inhibition but the mechanism of rotenone-induced proteasome dysfunction remains largely unknown. In this study, we examined the role of mitochondrial inhibition, oxidative stress, and microtubule dysfunction as potential mediators of rotenone-induced proteasome inhibition. Proteasome activity (26S) was measured in HEK and SK-N-MC cells expressing an EGFP-U degron fusion protein that is selectively degraded by the proteasome. We found that complex I and III inhibition led to the production of peroxides and decreased proteasome activity. We also found that rotenone increased nitric oxide production and nitric oxide and peroxynitrites led to proteasome inhibition. The effects of rotenone were attenuated by anti-oxidants and nitric oxide synthase inhibition. Since rotenone can also inhibit microtubule assembly, we tested a specific MT inhibitor and found it led to proteasome dysfunction. Rotenone also led to a decrease in 20S proteasome activity and 20S proteasome subunit immunoreactivity without a change in subunit mRNA. Together, these data suggest that rotenone-induced decreases in proteasome activity are due to increased degradation of proteasome components secondary to oxidative damage and possibly microtubule dysfunction.
Keywords: Parkinson’s disease, mitochondria, nitric oxide, microtubules
1: Introduction
The etiology of Parkinson’s disease (PD) remains unclear although it may involve mitochondria dysfunction and environmental toxins such as pesticides (Beal, 2003, Brown et al., 2006). The pesticide rotenone has been extensively studied because it is a mitochondria complex I inhibitor. Chronic rotenone infusion in rats results in nigrostriatal dopaminergic degeneration, signs of oxidative stress in the brain, and motor dysfunction (Betarbet et al., 2000, Fleming et al., 2004, Sherer et al., 2003). These animals also developed alpha-synuclein positive intraneuronal inclusions that resembled Lewy Bodies, the pathological hallmark of PD (Betarbet, Sherer, 2000).
The reason for the relatively selective toxicity to nigral neurons following systemic administration of rotenone is not clear. Complex I impairment can lead to compromised cell respiration, loss of mitochondrial membrane potential, and generation of free radicals such as superoxide anion (O2.−) and hydrogen peroxide (H2O2) (Barrientos and Moraes, 1999, Kudin et al., 2008). At low rotenone concentrations (5 nM), ATP production is not significantly altered (Betarbet et al., 2006). It is possible that rotenone generates oxidative stress, which is compounded by the increased production of reactive oxygen species (ROS) from dopamine metabolism making dopaminergic neurons particularly vulnerable. The exact pathophysiology of how oxidative stress leads to cell death is controversial. One potential downstream effector is the cellular ubiquitin proteasome system (UPS).
The UPS is responsible for degrading damaged proteins and has been implicated in the etiology of PD (Kitada et al., 1998, Leroy et al., 1998, McNaught et al., 2002). Studies by our group and others have demonstrated that treatment of cells with rotenone leads to proteasomal dysfunction (Betarbet, Canet-Aviles, 2006, Shamoto-Nagai et al., 2003, Wang et al., 2006). However, although several mechanisms have been proposed, including decreased ATP levels, acrolein-modification of proteasome subunits, and induction of oxidative stress (Hoglinger et al., 2003, Shamoto-Nagai, Maruyama, 2003), the exact mechanism by which rotenone causes inhibition of the proteasome remains unclear.
Since rotenone, a complex I inhibitor, can cause many of the pathological features of PD in rats and complex I dysfunction has been associated with PD in humans, it is important to determine the mechanisms by which rotenone acts. One known effect of rotenone-induced toxicity is reduced UPS activity. The aim of this study was to evaluate the mechanisms by which rotenone inhibits the proteasome. A better understanding of these mechanisms could provide important insights into the pathological processes underlying PD. We found that rotenone likely acts through multiple mechanisms including oxidative stress, nitric oxide (NO) production and possibly microtubule dysfunction. These processes, and possibly others, led to increased degradation of UPS 20S subunits.
2: Methods
2.1 Chemicals
All compounds were purchased from Sigma-Aldrich (St. Louis, MO), ChemService (West Chester, PA), EMD Chemicals (Gibbstown, NJ), or Dojindo Molecular Technologies (Kumamoto, Japan).
2.2 Proteasome Activity Measurement
UPS activity was measured as previously described (Wang et al., 2006). Briefly, human embryonic kidney (HEK) and neuroblastoma SK-N-MC cells were grown in DMEM/F12 supplemented with 10% FBS and 1% Pen/Strep and passaged every 2–3 days. HEK and SK-N-MC cells transfected with the EGFP-degron fusion protein (GFP-U) were grown in the same media supplemented with 400 μg/mL of geneticin for selection. UPS activity was measured in HEK and SK-N-MC cells by quantitating the level of EGFP-degron fusion protein that is selectively degraded by the proteasome, using flow cytometry (Bence et al., 2001). We chose to use these cell lines since SK-N-MC cells are neuronal-like and HEK cells are non-neuronal. In some experiments, cells were also stained with 10 μg/mL of propidium iodide for 30 min at room temperature to detect dead cells. After PI staining, cells were trypsinized and triturated before analysis using a Beckman XL-MCL flow cytometer. Cells were gated for GFP-U measurement by forward and side scatter, and fluorescence was expressed relative to vehicle control. In experiments examining the effects of anti-oxidants on the toxicity of rotenone, fluorescence was expressed as a percent of the increase in fluorescence caused by rotenone (anti-oxidant GFP-U – control GFP-U)/(rotenone GFP-U – control GFP-U).
Proteasome chymotryptic activity (20S) was also measured in cell homogenates by the conversion of N-Suc-LLVY-AMC (100 uM, Sigma) into the fluorescent compound AMC by active protease cleavage. Epoxomicin (5 μM, Sigma) or Lactacystin (5 μM) were used to estimate total proteasome activity and fluorescence was measured using a Wallac multi-plate reader.
2.3 Oxidative Stress Measurement
Oxidative stress was measured by quantitating the oxidation of carboxy-DCF (Invitrogen) in untransfected HEK cells. Cells were loaded with 10 μM of carboxy-DCF for 1 h. The media containing the fluorescent probe was then removed and new media containing the treatment compound was added and incubated for 2 h before being analyzed by flow cytometry as previously described (Wang et al, 2006). Nitric oxide was quantitated using the NO-specific dye DAF-FM acetate (20 μM, Invitrogen) and assayed in a similar manner as carboxy-DCF.
2.4 Western Blot Analysis
Cells were harvested and lysed in PBS containing 0.1% (vol/vol) Triton, 5 mM EDTA and protease inhibitor cocktail (Complete-mini; Roche Applied Science, Indianapolis, IN, USA). Cells were then sonicated for 3–5 s while kept cold on ice, followed by centrifugation for 20 min at 10,000 rpm at 4°C. Protein concentration was determined by Bradford dye binding assay (Bio-rad, Hercules, CA). Equal volume of 2X SDS loading buffer (125 mM Tris, pH 6.8, 5% (wt/vol) SDS, 2.5 mg/ml Bromophenol Blue, 25% (vol/vol) glycerol) was added to samples and boiled for 5 min prior to loading. Proteins were separated by SDS-polyacrylamide gel using a 6% stacking phase and 12% separating phase. Proteins were then transferred to a Protran nylon membrane (Schleicher and Schuell, Dassel, Germany) using a wet transfer method. Membranes were blocked in PBS with 0.1% (vol/vol) Tween 20 (Sigma, St Louis, MO, USA) and 5% non-fat dry milk for at least 1 h. Membranes were washed and then probed with a mouse antibody to proteasome α-subunit 1,2,3,5,6 and 7 (Affiniti, UK) overnight at 4°C. Membranes were washed and then probed with anti-mouse secondary antibody (Amersham Biosciences, UK) for 1 h at room temperature. Membranes were washed a final time before detection by ECL plus (Amersham Biosciences, UK). Each wash consisted of 5 10-min washes in PBS plus 0.1% (vol/vol) Tween 20.
2.5 Quantitative real-time RT-PCR (qRT-PCR)
To measure proteasome mRNA levels, SK-N-MC cells were treated with 0, 10, or 100 nM of rotenone in fresh media for 48 h. Total RNA was isolated using RNAwiz (Ambion) according to the manufacturer’s instructions and was reverse-transcribed to cDNA with SuperScript First Strand Synthesis System for RT-PCR (Invitrogen) using oligo-dT primers. Relative cDNA levels were quantitated by real-time PCR using SYBR-Green (SYBR-green master mix, Applied Biosystems) as reporter dye on an ABI PRISM 7700 at the UCLA Sequencing and Genotyping Core. Proteasome cDNA levels were standardized to the levels of GAPDH and quantitated by the relative Ct method (2−DDCt). Vehicle treated cells were used as the calibrator. Primers were designed with Primer3 software (Table 1).
Table 1.
2.6 Statistical analysis
Statistical analysis was performed with GraphPad Prism software. Unless otherwise specified, values are expressed as Mean ± SEM. The level of significance is shown as * p<0.05 or ** p<0.01. Statistical significance was determined using One-way ANOVA with Dunnett’s or Bonferroni’s Multiple Comparison Post-Test.
3: Results
3.1 Proteasome inhibition by mitochondria inhibitors
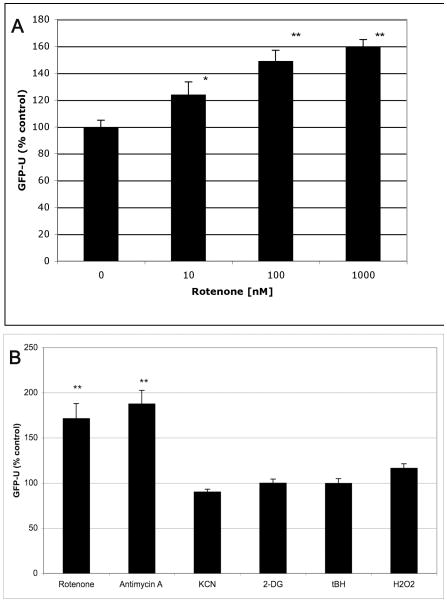
HEK and SK-N-MC cells expressing GFP-U were exposed to various concentrations of rotenone to determine its dose-response relationship with UPS inhibition (Figure 1a). Rotenone significantly lowered UPS activity at doses as low as 10 nM and lowered it further at higher concentrations. Similar results were obtained using other pesticides that inhibit complex I including pyridaben and fenazaquin (data not shown).
Figure 1.
Inhibition of mitochondrial respiration, oxidative stress and 26S UPS activity. HEK cells were exposed to rotenone (complex I), antimycin A (2μg/mL, complex III), KCN (2 mM, complex IV), 2-DG (10 mM, ATP depletion), and the oxidants tBH and H2O2 (100 μM) for 24 h and GFP-U fluorescence (26S UPS activity) was measured. Increased GFP-U indicates decreased UPS activity. In Figure 1B, 100 nM rotenone was used. N=8 per data point. Statistics by One-way ANOVA with Dunnett’s Multiple Comparison Post-Test (* p < 0.05, ** p < 0.01).
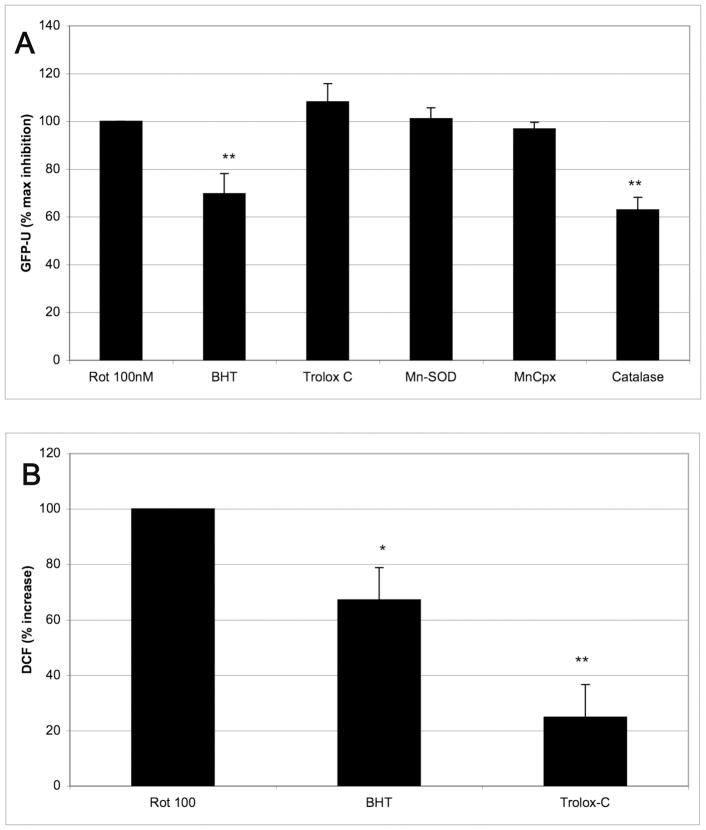
Oxidative stress generated by complex I inhibition has been postulated to be the main determinant of the ability of rotenone to lower UPS activity. In order to further dissect the mechanisms of rotenone, we tested other inhibitors of the mitochondrial electron transport chain (ETC). Complex III inhibition (antimycin A, 2 μg/mL) resulted in a 232% increase in GFP-U (Figure 1b). Rotenone at 100 nM and antimycin A at 2 μg/mL would be expected to inhibit 100% their respective mitochondrial respiratory complexes. On the other hand, complete inhibition of complex IV by 2 mM potassium cyanide (KCN), or ATP depletion by 10 mM 2-deoxyglucose (2-DG), did not result in proteasome inhibition (Figure 1b). Interestingly, complex IV inhibitors and 2-DG have been shown not to increase ROS in contrast to complex I and III inhibitors (McLennan and Degli Esposti, 2000). These data support the hypothesis that rotenone induces oxidative stress which leads to UPS inhibition. We further tested the contribution of ROS to proteasome inhibiton by exposing cells directly to oxidants that do not directly alter mitochondrial function and to anti-oxidants. Cells were exposed to H2O2, oxidant tert-butyl hydroperoxide (tBH), and iron chloride (FeCl2, not shown) for 24 h but all failed to inhibit UPS activity (Figure 1b). Some but not all anti-oxidants attenuated rotentone-induced (100 nM) UPS inhibition (Figure 2a) but this did not correlate with their ability to reduce ROS (measured by DCF fluorescence, Figure 2b) adding further doubt that oxidative stress is the primary mechanism for rotenone’s UPS inhibitory activity.
Figure 2.
(A) Beta-hydroxy toulene (BHT) and catalase, but not other antioxidants, attenuated rotenone’s UPS inhibitory effects. HEK cells were treated with 100 nM rotenone in combination with the various anti-oxidants for 24 h before being analyzed by flow cytometry. GFP-U is expressed as a percent of the increase in GFP-U caused by rotenone alone. A value of 100% would indicate no effect on rotenone, and 0% would indicate a complete reversal of rotenone’s effects (N=5–9). B: BHT and Trolox-C attenuated the formation of peroxides caused by 100 nM rotenone as measured by DCF flouresence (N=4–11 per data point). Note that although both reduced ROS produced by rotenone, only BHT attenuated UPS inhibiton.
3.2 Reactive Nitrogen Species (RNS) and Proteasome Activity
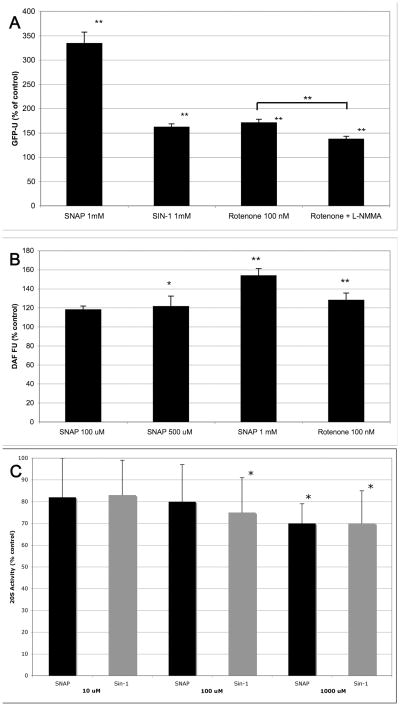
Given that ROS did not appear to account for the ability of rotenone to inhibit the proteasome, we examined the possibility that rotenone induces the generation of RNS and these lead to UPS inhibition. We first tested the effect of the NO donor SNAP and the peroxynitrite donor SIN-1 on UPS activity and found that both lead to 26S proteasome inhibition (Figure 3a). Furthermore, the NO synthase inhibitor L-NMMA attenuated the ability of rotenone to inhibit UPS activity by 20% (Figure 3a). We confirmed that rotenone induces NO formation by measuring DAF fluorescence and found that rotenone (100 nM) induced an increase in RNS in a similar manner as the NO donor SNAP (Figure 3b). Proteasome inhibition by SNAP was very potent, causing a 334% increase in GFP-U fluorescence at 1 mM. We also found that RNS can directly affect protease activity by using a 20S proteasome assay. NO and peroxynitrite donors added directly to cell homogenates led to a decrease in 20S activity (Figure 3c). Taken together, these observations suggest that NO and peroxynitrite can contribute to rotenone-induced UPS inhibition possibly by nitration of proteasome subunits.
Figure 3.
(A). NO donor SNAP and peroxynitrite donor SIN-1 cause 26S proteasome inhibition in SK-N-MC cells. Inhibiting NO synthase with L-NMMA attenuated rotenone’s ability toe inhibit the UPS (N=6). (B). Rotenone induced NO formation as measured by DAF staining (N=6). (C) The NO donor SNAP and peroxynitrite donor SIN-1 inhibited 20S proteasome activity in cell homogenates (N=8).
3.3 Microtubule dysfunction and proteasome activity
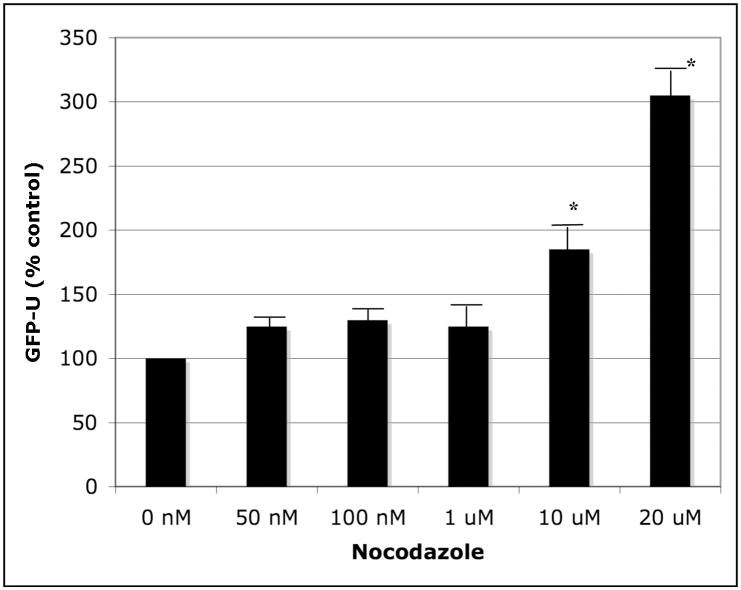
In addition to the direct effects of rotenone on mitochondrial respiration, it has also been shown to interfere with microtubule assembly at concentrations as low as 20 nM (Ren et al., 2005). We hypothesized that rotenone may inhibit UPS activity via its ability to alter microtubule assembly. To test this, we exposed GFP-U expressing SK-N-MC cells to the specific microtubule assembly inhibitor nocodazole for 48 h and found that it caused proteasome inhibition at 10 μM, the concentration needed to cause complete depolymerization of tubulin (Liao et al., 1995) (Figure 4). We have also found a similar effect of carbendazem (another microtubule inhibitor) on proteasome activity providing evidence that altering microtubules may be a mechanism by which rotenone alters proteasome function.
Figure 4.
Inhibition of microtubule assembly by nocodazole resulted in 26S UPS activity as measured by GFP-U flouresence (N=4, * p < 0.05).
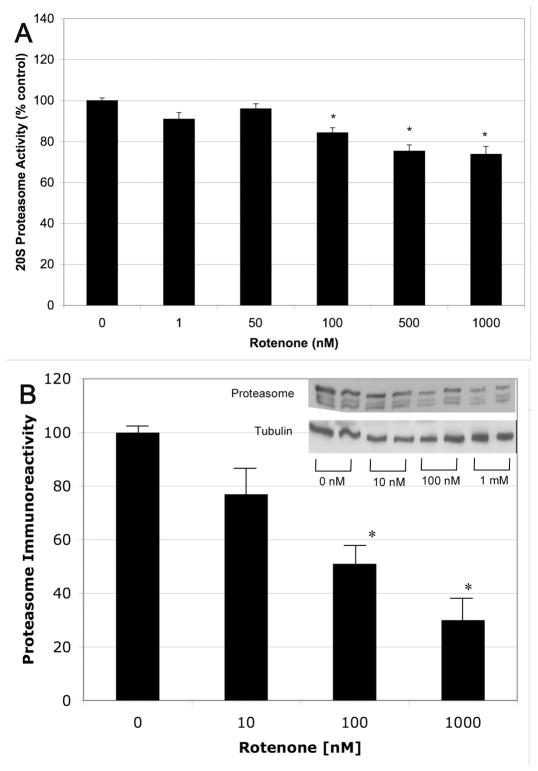
3.4 Rotenone lowers proteasome subunit levels
Our data suggest that ROS, RNS and MT dysfunction contribute to the UPS inhibition induced by rotenone but the mechanism through which these processes lower protease activity is unclear. One possibility is that the proteasome components are damaged by oxidation and/or nitration leading to their degradation. To test this possibility, we measured 20S proteasome activity and immunoreactivity in rotenone-treated lysates. We found that rotenone caused a reduction in the 20S proteasome activity and a dose-dependent decrease in proteasome α-subunit immunoreactivity (Figures 5a and b). To assess whether the observed decrease in proteasome levels were due to decreased synthesis, we performed qRT-PCR to measure proteasome mRNA levels. RT-PCR of α- and β-subunits all showed the same level of expression regardless of rotenone treatment (data not shown). These data suggest that rotenone increases proteasome subunit degradation but does not alter synthesis.
Figure 5.
Rotenone decreased the 20S proteasome activity and immunoreactivity. SK-N-MC cells were treated with rotenone for 48 h and the cell homogenate collected and assayed for the 20S proteasome activity (A, N=9–11) and immunoreactivity (B, N=4). Both 20S proteasome activity and immunoreactivity measured by Western Blot (α-subunits) were significantly lower at 100 nM or more rotenone.
4: Discussion
In this study, we examined the mechanisms by which rotenone inhibits proteasome activitiy and found that it likely involves multiple processes that appear to include mitochondrial inhibition, oxidative stress, RNS, microtubule assembly, and increased degradation of proteasome subunits. We propose that the unique toxicity of rotenone is due to a combination of these pathways, most of which have been implicated in the pathogenesis of PD.
Oxidative stress in part appears to contribute to the actions of rotenone. Compounds that inhibited mitochondrial respiration and also induced the generation of ROS (complex I and III inhibitors) led to proteasome inhibition. Blocking mitochondrial complex IV did not cause the generation of ROS nor did it inhibit the proteasome (Liu et al., 2002, Turrens and Boveris, 1980). Furthermore, the involvement of ROS in the actions of rotenone is supported by the fact that some anti-oxidants attenuated rotenone-induced proteasome inhibition. This is consistent with a previous report that demonstrated that α-tocopheral attenuated rotenone-induced (5 nM rotenone) proteasome inhibition (Betarbet, Canet-Aviles, 2006). In our study, some anti-oxidants could only protect rotenone-induced inhibition (100 nM) by approximately a third suggesting that other mechanisms are at play at this concentration. Furthermore, we did not find a good correlation with the ability of anti-oxidants to lower ROS and protect against rotenone-inuced UPS inhibition (Figure 2). In contrast to the study of Berarbet et al, we did not see any protective qualities of α-tocopheral but these differences may be due to the differences in the concentration of and the length of exposure to rotenone.
Induction of RNS is also likely to contribute to the toxicity of rotenone. RNS inhibited the proteasome both in cells and directly in cellular homogenates using NO and perinitirate donors. Based on DAF staining, rotenone stimulated NO production and the NOS inhibitor L-NMMA attenuated the effects of rotenone on proteasome activity. Although DAF staining may not be specific for NO, taken together with experiments using an NO and perinitrate donor and a NOS inhibitor, it is possible that RNS induced by rotenone play at least a partial role in inhibiting proteasome activity. Osna et al reported that nitrative stress in the form of NO or peroxynitrite can inhibit the proteasome in vitro (Osna et al., 2004) but nitrated proteasome subunits following rotenone treatment could not be detected (Shamoto-Nagai, Maruyama, 2003) although others have found that oxidation or nitration of UPS subunits can alter protease activity (Szweda et al., 2002) for review.
A potential third mechanism for the ability of rotenone to cause proteasome inhibition is via the disruption of microtubules as demonstrated by the ability of nocodazole to also inhibit the UPS. We have also found that carbendazim, another MT inhibitor, also leads to UPS dysfunction (data not shown). Nocodazole significantly inhibited the UPS at concentrations that causes almost complete MT depolymerization. The ability of rotenone to inhibit microtubule assembly is well established even at 10 nM (Ren, Liu, 2005, but it is likely that rotenone concentrations need to be closer to 0.2–1 (M to cause MT depolymerization similar to that of 10 (M nocodazole [Srivastava, 2007 #1516). Interestingly, MT dysfunction has been shown to induce selective dopaminergic cell death in primary cultures (Ren, Liu, 2005). The association of MT and the UPS has not been well studied but it has been shown that Parkin, an E3 ligase linked to PD, binds to tubulin and alters its degradation (Ren et al., 2003). It is possible that UPS components are associated with MT and disassembly of MTs leads to impaired proteasome activity but more work is needed to to establish causality between the ability of rotenone to alter MT assembly and decrease UPS activity.
We found that ROS, RNS and MT assembly are involved in rotenone’s proteasome inhibitory activity but the molecular events that lead to reduced protease activity remains unclear. We do know that the decrease in UPS activity is not simply reflecting a decrease in cell viability since some toxins kill cells but do not lead to decreased UPS activity (Wang et al., 2006). Importantly, we did find that proteasome subunit immunoreactivity was decreased following rotenone treatment. Changes in proteasome immunoreactive protein were not caused by decreased transcription of the subunits and therefore it is likely that the decreased protein level was caused by increased degradation of proteasome subunits. Considering the likely involvement of ROS and RNS in rotenone’s actions, it is possible that rotenone causes increased degradation of proteasome subunits by oxidation or nitration of the proteasome. This observation is in contrast to that by Shamoto-Nagai and coworkers who found no changes in the amount of proteasome protein (Shamoto-Nagai, Maruyama, 2003) and instead suggests that acrolein modification of the proteasome subunits is the cause for lowered proteasome activity. Additional studies need to be performed to directly test the effects of UPS subunit oxidation and nitration on its degradation.
In summary, we have found several pathological processes that can account for rotenone’s effects on the UPS. Synergistic action of these processes is an attractive hypothesis for the toxicity of rotenone and the pathogenesis of PD.
Acknowledgments
This study was supported by grants from the NIEHS (5 U54 ESO12078 and 1P01ES016732-01) and the Veterans Administration SW PADRREC. We would also like to thank Drs. Erik Schweitzer and Xue-Feng Wang, for their technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6: References
- Barrientos A, Moraes CT. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J Biol Chem. 1999;274:16188–97. doi: 10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria, oxidative damage, and inflammation in Parkinson’s disease. Ann N Y Acad Sci. 2003;991:120–31. doi: 10.1111/j.1749-6632.2003.tb07470.x. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Canet-Aviles RM, Sherer TB, Mastroberardino PG, McLendon C, Kim JH, et al. Intersecting pathways to neurodegeneration in Parkinson’s disease: Effects of the pesticide rotenone on DJ-1, alpha-synuclein, and the ubiquitin-proteasome system. Neurobiol Dis. 2006;22:404–20. doi: 10.1016/j.nbd.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–6. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Brown TP, Rumsby PC, Capleton AC, Rushton L, Levy LS. Pesticides and Parkinson’s disease--is there a link? Environ Health Perspect. 2006;114:156–64. doi: 10.1289/ehp.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SM, Salcedo J, Fernagut PO, Rockenstein E, Masliah E, Levine MS, et al. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J Neurosci. 2004;24:9434–40. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglinger GU, Carrard G, Michel PP, Medja F, Lombes A, Ruberg M, et al. Dysfunction of mitochondrial complex I and the proteasome: interactions between two biochemical deficits in a cellular model of Parkinson’s disease. J Neurochem. 2003;86:1297–307. doi: 10.1046/j.1471-4159.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kudin AP, Malinska D, Kunz WS. Sites of generation of reactive oxygen species in homogenates of brain tissue determined with the use of respiratory substrates and inhibitors. Biochim Biophys Acta. 2008;1777:689–95. doi: 10.1016/j.bbabio.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–2. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Liao G, Nagasaki T, Gundersen GG. Low concentrations of nocodazole interfere with fibroblast locomotion without significantly affecting microtubule level: implications for the role of dynamic microtubules in cell locomotion. J Cell Sci. 1995;108 ( Pt 11):3473–83. doi: 10.1242/jcs.108.11.3473. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem. 2002;80:780–7. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- McLennan HR, Degli Esposti M. The contribution of mitochondrial respiratory complexes to the production of reactive oxygen species. J Bioenerg Biomembr. 2000;32:153–62. doi: 10.1023/a:1005507913372. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, et al. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81:301–6. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- Osna NA, Haorah J, Krutik VM, Donohue TM., Jr Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–82. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- Ren Y, Liu W, Jiang H, Jiang Q, Feng J. Selective vulnerability of dopaminergic neurons to microtubule depolymerization. J Biol Chem. 2005;280:34105–12. doi: 10.1074/jbc.M503483200. [DOI] [PubMed] [Google Scholar]
- Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci. 2003;23:3316–24. doi: 10.1523/JNEUROSCI.23-08-03316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoto-Nagai M, Maruyama W, Kato Y, Isobe K, Tanaka M, Naoi M, et al. An inhibitor of mitochondrial complex I, rotenone, inactivates proteasome by oxidative modification and induces aggregation of oxidized proteins in SH-SY5Y cells. J Neurosci Res. 2003;74:589–97. doi: 10.1002/jnr.10777. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, et al. Mechanism of toxicity in rotenone models of Parkinson’s disease. J Neurosci. 2003;23:10756–64. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweda PA, Friguet B, Szweda LI. Proteolysis, free radicals, and aging. Free Radic Biol Med. 2002;33:29–36. doi: 10.1016/s0891-5849(02)00837-7. [DOI] [PubMed] [Google Scholar]
- Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–7. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Li S, Chou AP, Bronstein JM. Inhibitory effects of pesticides on proteasome activity: implication in Parkinson’s disease. Neurobiol Dis. 2006;23:198–205. doi: 10.1016/j.nbd.2006.02.012. [DOI] [PubMed] [Google Scholar]