Abstract
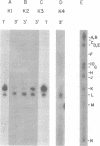
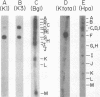
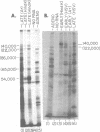
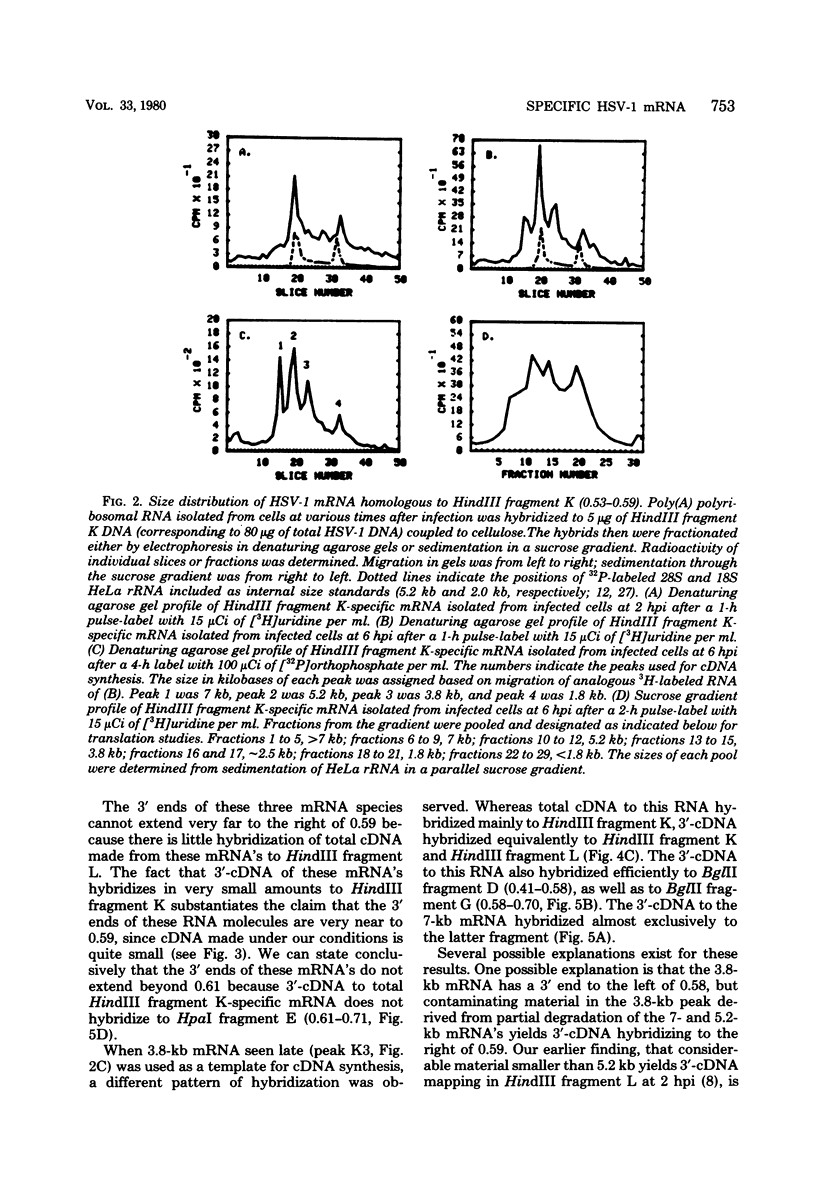
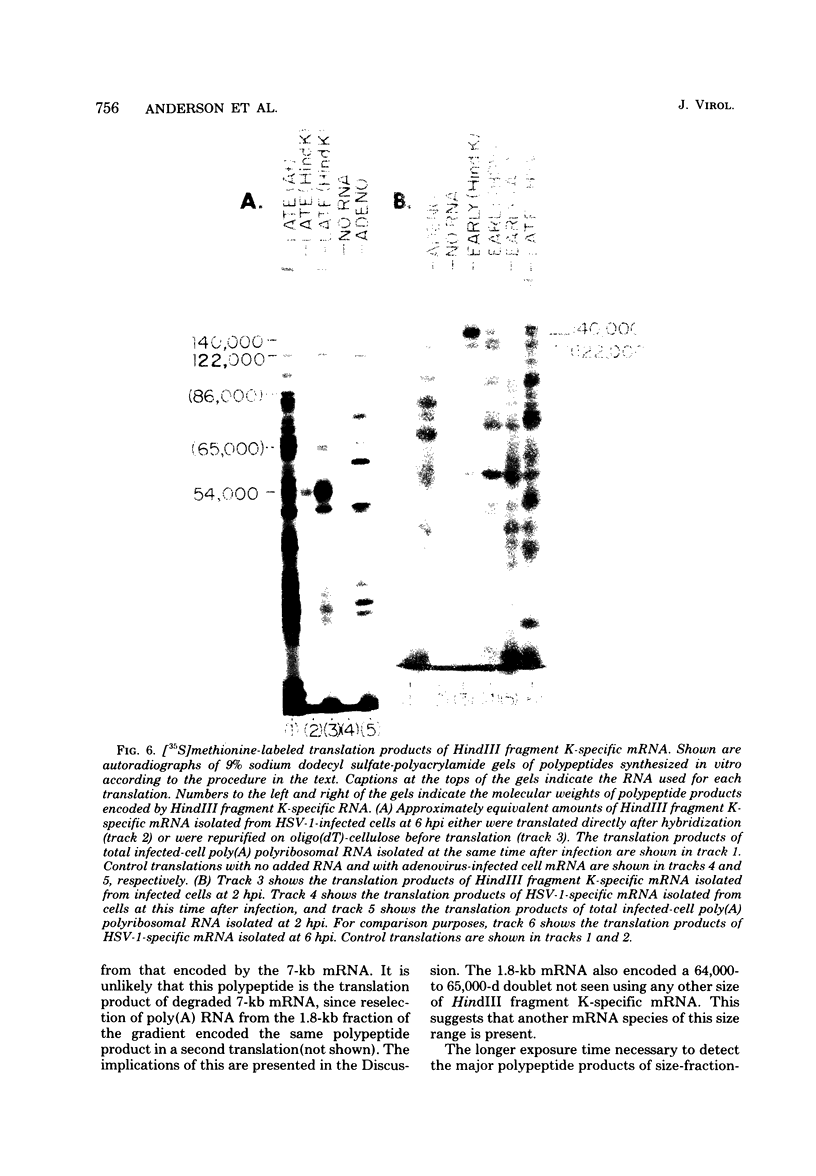
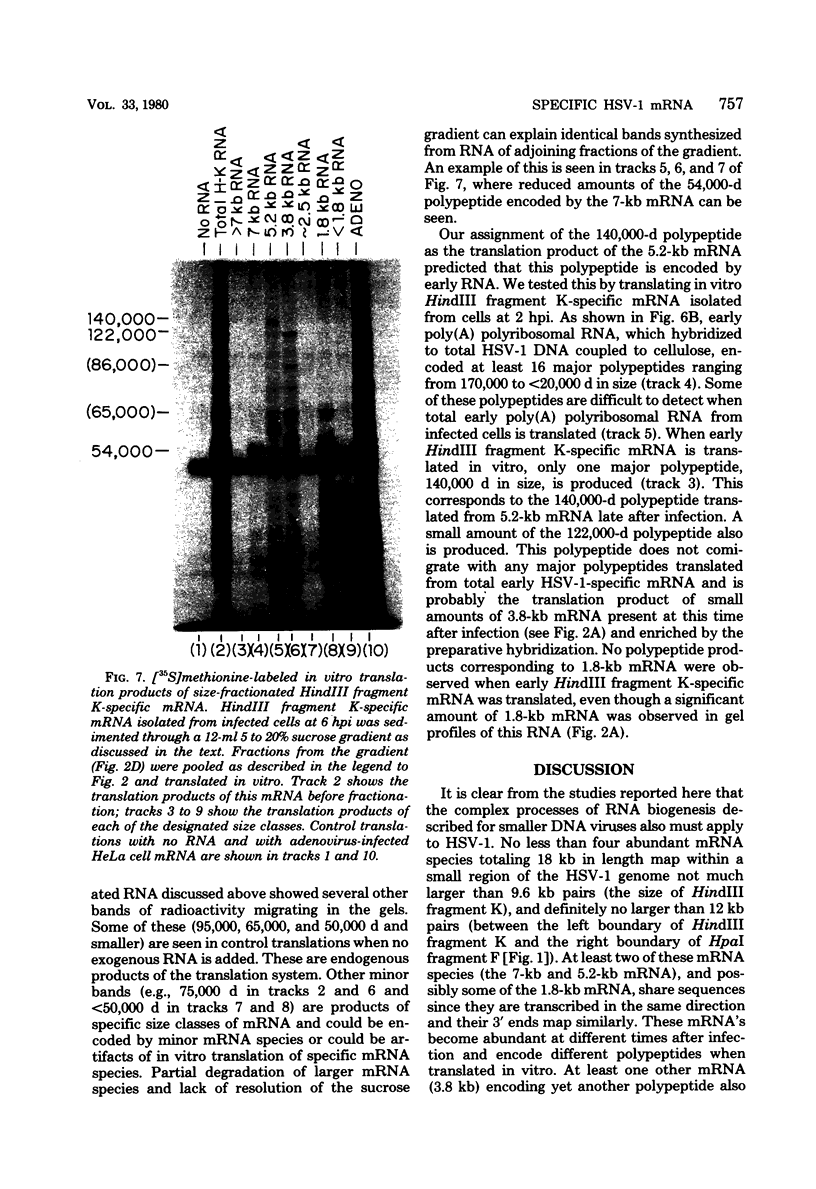
We have examined in detail the major mRNA species encoded by the region of the herpes simplex virus type 1 genome encoded by HindIII fragment K (0.53-0.59 from the left end of the prototype arrangement of the genome) by using this restriction fragment bound to cellulose as a reagent for isolation of this mRNA. Before viral DNA replication in infected cells (early), a major species of viral mRNA 5.2 kilobases (kb) in length is abundant. After the onset of viral DNA replication (late), four mRNA species are abundant: 7, 5.2, 3.8, and 1.8 kb in size. We have used reverse transcriptase from avian myeloblastosis virus to make DNA complementary to these RNA species and their 3′ ends. We have shown by hybridization of this complementary DNA to Southern blots of herpes simplex virus type 1 DNA that the 7-, 5.2-, and 1.8-kb mRNA species have their 3′ ends to the right of 0.59 and are at least partially colinear. The 3.8-kb mRNA has a 3′ end mapping to the left of the 3′ ends of these other species. In vitro translation of HindIII fragment K-specific mRNA in a reticulocyte lysate system yielded three major polypeptide products: 140,000, 122,000, and 54,000 daltons (d). Less prominent species of 86,000 and 65,000 d also were produced. Translation of size-fractionated HindIII fragment K-specific mRNA showed that the 7-, 5.2-, and 3.8-kb mRNA's encoded the 54,000-, 140,000-, and 122,000-d polypeptides, respectively. The 140,000-d polypeptide was the major polypeptide translated using early HindIII fragment K-specific mRNA as a template. The 3.8-kb mRNA also encoded the 86,000-d polypeptide, whereas the 1.8-kb mRNA encoded a polypeptide that was indistinguishable from the 54,000-d polypeptide encoded by the 7-kb mRNA, in addition to the 65,000-d polypeptide. The implications of the data are discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson K. P., Stringer J. R., Holland L. E., Wagner E. K. Isolation and localization of herpes simplex virus type 1 mRNA. J Virol. 1979 Jun;30(3):805–820. doi: 10.1128/jvi.30.3.805-820.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Courtney R. J., Schaffer P. A., Powell K. L. Synthesis of virus-specific polypaptides by temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1976 Dec;75(2):306–318. doi: 10.1016/0042-6822(76)90030-1. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fenwick M., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XI. Apparent clustering of functions effecting rapid inhibition of host DNA and protein synthesis. J Virol. 1979 Feb;29(2):825–827. doi: 10.1128/jvi.29.2.825-827.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W., Hyman R. W. The terminal repetition of herpes simplex virus DNA. Virology. 1975 Sep;67(1):144–157. doi: 10.1016/0042-6822(75)90412-2. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. E., Anderson K. P., Stringer J. R., Wagner E. K. Isolation and localization of herpes simplex virus type 1 mRNA abundant before viral DNA synthesis. J Virol. 1979 Aug;31(2):447–462. doi: 10.1128/jvi.31.2.447-462.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C. A., Oliver S. G., Newman A. M., Holland L. E., McLaughlin C. S., Wagner E. K., Warner R. C. The molecular weight of yeast P1 double-stranded RNA. J Biol Chem. 1978 Nov 25;253(22):8332–8336. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marsden H. S., Stow N. D., Preston V. G., Timbury M. C., Wilkie N. M. Physical mapping of herpes simplex virus-induced polypeptides. J Virol. 1978 Nov;28(2):624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes B. E., Stark G. R. Nucleic acid hybridization using DNA covalently coupled to cellulose. Cell. 1975 Jul;5(3):301–310. doi: 10.1016/0092-8674(75)90105-1. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undergraded polysomes and messenger ribonucleic acid. Biochemistry. 1974 Aug 13;13(17):3606–3615. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- Preston C. M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979 Jan;29(1):275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R., Holland L. E., Swanstrom R. I., Pivo K., Wagner E. K. Quantitation of herpes simplex virus type 1 RNA in infected HeLa cells. J Virol. 1977 Mar;21(3):889–901. doi: 10.1128/jvi.21.3.889-901.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R., Holland L. E., Wagner E. K. Mapping early transcripts of herpes simplex virus type 1 by electron microscopy. J Virol. 1978 Jul;27(1):56–73. doi: 10.1128/jvi.27.1.56-73.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Shank P. R. X-Ray Intensifying Screens Greatly Enhance the Detection by Autoradiography of the Radioactive Isotopes 32P and 125I. Anal Biochem. 1978 May;86(1):184–192. doi: 10.1016/0003-2697(78)90333-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Swanstrom R. I., Rice M., Howell L., Lane J. Variation in the molecular size of the DNA from closely related strains of type I herpes simplex virus. Biochim Biophys Acta. 1976 Jun 18;435(2):192–205. doi: 10.1016/0005-2787(76)90250-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]