Abstract
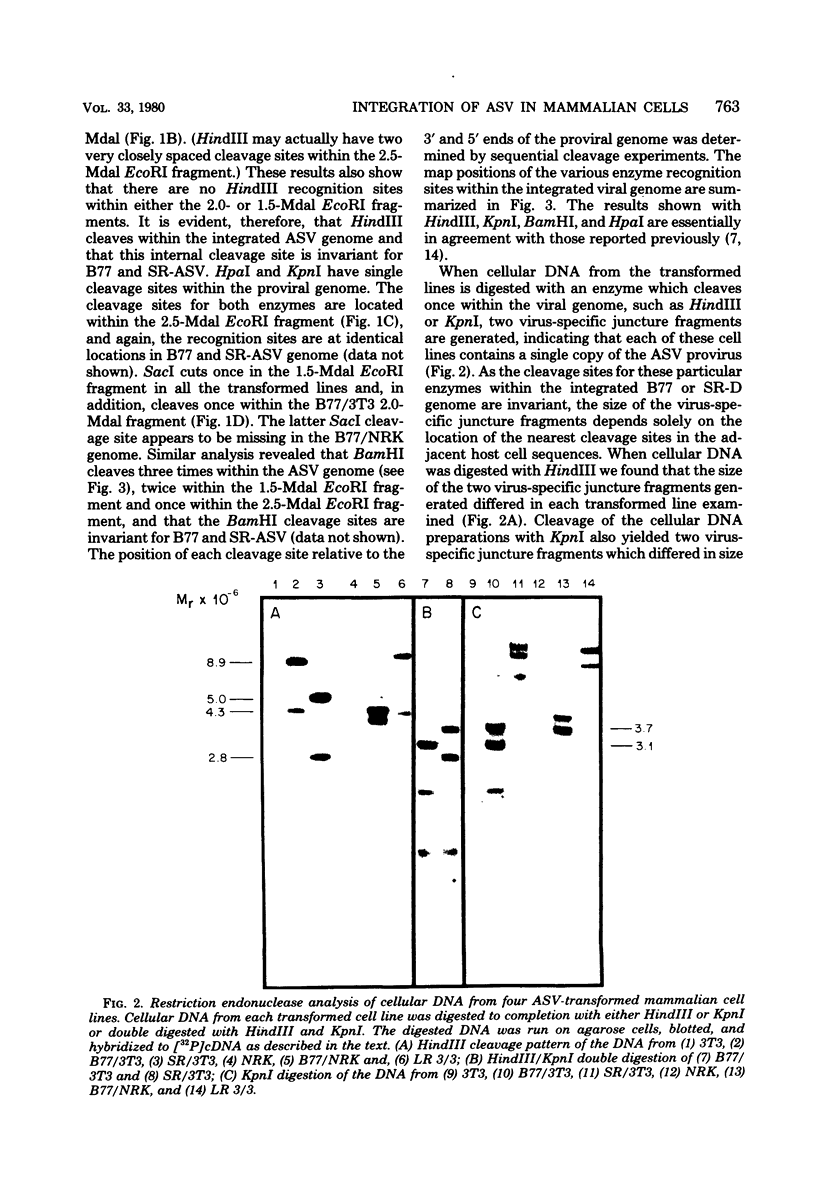
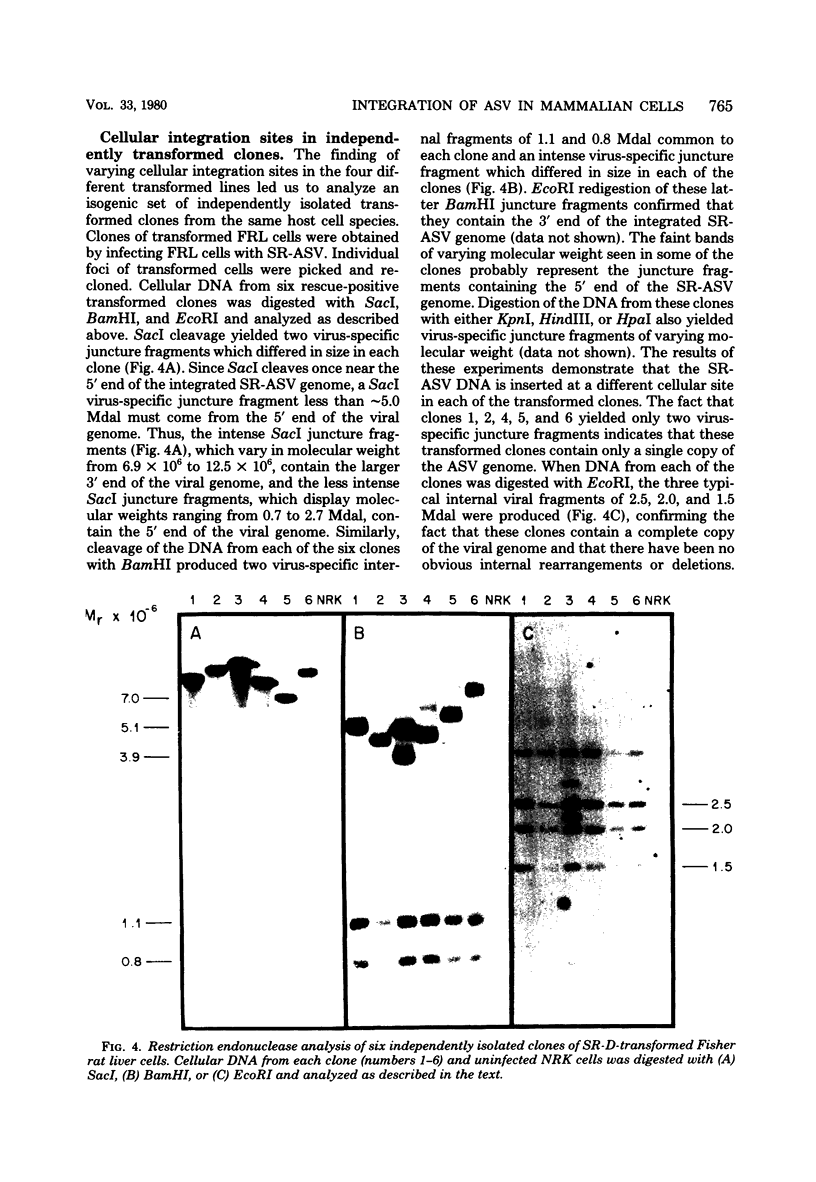
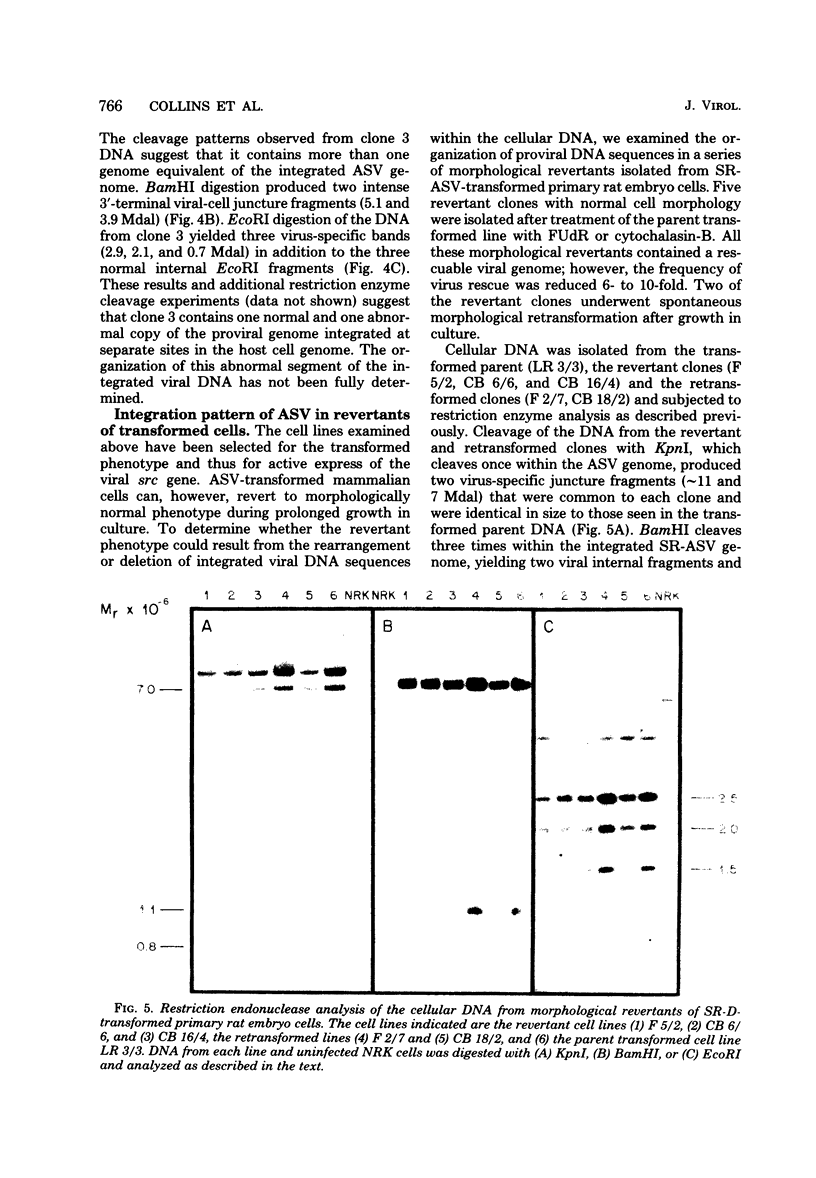
We have examined the arrangement of integrated avian sarcoma virus (ASV) DNA sequences in several different avian sarcoma virus transformed mammalian cell lines, in independently isolated clones of avian sarcoma virus transformed rat liver cells, and in morphologically normal revertants of avian sarcoma virus transformed rat embryo cells. By using restriction endonuclease digestion, agarose gel electrophoresis, Southern blotting, and hybridization with labeled avian sarcoma virus complementary DNA probes, we have compared the restriction enzyme cleavage maps of integrated viral DNA and adjacent cellular DNA sequences in four different mouse and rat cell lines transformed with either Bratislava 77 or Schmidt-Ruppin strains of avian sarcoma virus. The results of these experiments indicated that the integrated viral DNA resided at a different site within the host cell genome in each transformed cell line. A similar analysis of several independently derived clones of Schmidt-Ruppin transformed rat liver cells also revealed that each clone contained a unique cellular site for the integration of proviral DNA. Examination of several morphologically normal revertants and spontaneous retransformants of Schmidt-Ruppin transformed rat embryo cells revealed that the internal arrangement and cellular integration site of viral DNA sequences was identical with that of the transformed parent cell line. The loss of the transformed phenotype in these revertant cell lines, therefore, does not appear to be the result of rearrangement or deletions either within the viral genome or in adjacent cellular DNA sequences. The data presented support a model for ASV proviral DNA integration in which recombination can occur at multiple sites within the mammalian cell genome. The integration and maintenance of at least one complete copy of the viral genome appear to be required for continuous expression of the transformed phenotype in mammalian cells.
Full text
PDF

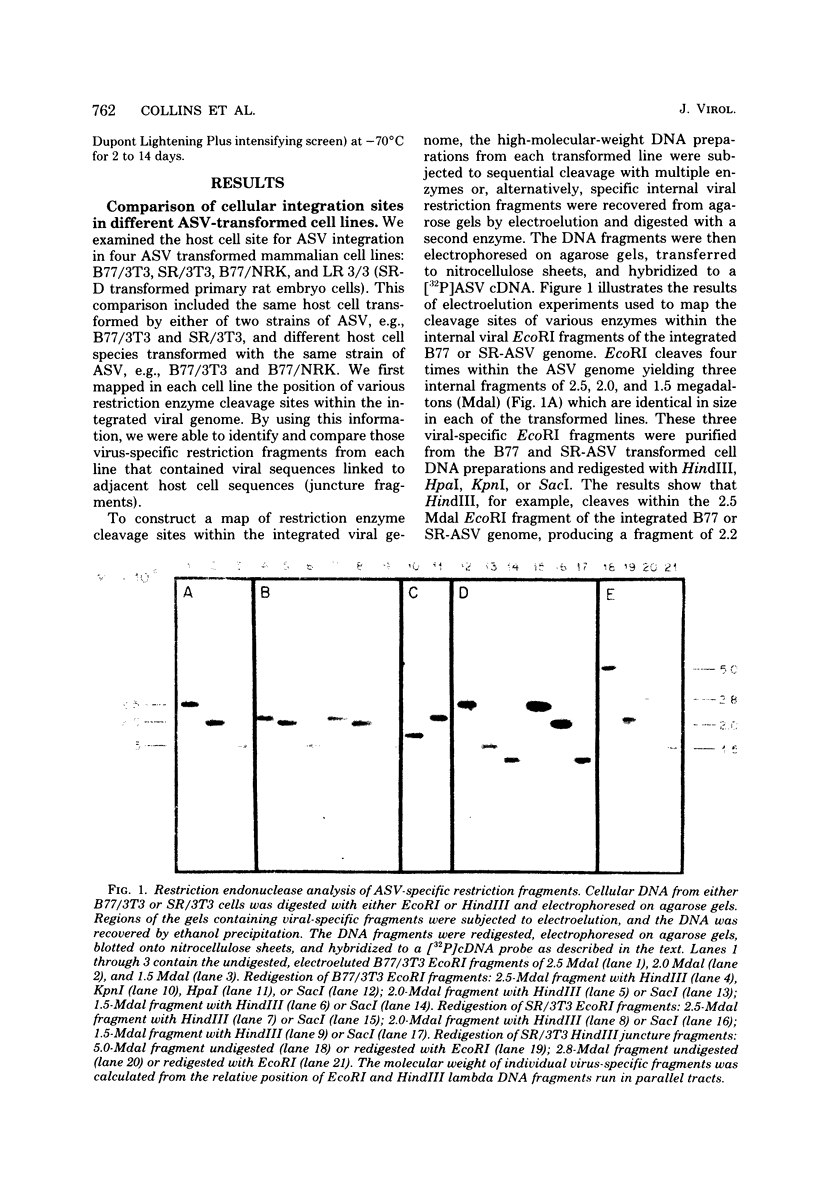



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama Y., Vogt P. K. Integration of different sarcoma virus genomes into host DNA: evidence against tandem arrangement and for shared integration sites. Proc Natl Acad Sci U S A. 1979 May;76(5):2465–2469. doi: 10.1073/pnas.76.5.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. M. Retroviruses. Annu Rev Biochem. 1978;47:35–88. doi: 10.1146/annurev.bi.47.070178.000343. [DOI] [PubMed] [Google Scholar]
- Boettiger D. Reversion and induction of Rous sarcoma virus expression in virus-transformed baby hamster kidney cells. Virology. 1974 Dec;62(2):522–529. doi: 10.1016/0042-6822(74)90412-7. [DOI] [PubMed] [Google Scholar]
- Collins C. J., Parsons J. T. Integration of avian sarcoma virus DNA sequences in transformed mammalian cells. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4301–4305. doi: 10.1073/pnas.74.10.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dierks P. M., Highfield P. E., Parsons J. T. Deletion mutant of the Bratislava-77 strain of Rous sarcoma virus containing a fusion of the group-specific antigen and envelope genes. J Virol. 1979 Nov;32(2):567–582. doi: 10.1128/jvi.32.2.567-582.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Keshet E., Temin H. M. Sites of integration of reticuloendotheliosis virus DNA in chicken DNA. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3372–3376. doi: 10.1073/pnas.75.7.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury A. T., Hanafusa H. Synethesis and integration of viral DNA in chicken cells at different time after infection with various multiplicities of avian oncornavirus. J Virol. 1976 May;18(2):383–400. doi: 10.1128/jvi.18.2.383-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. T., Lewis P., Dierks P. Purification of virus-specific RNA from chicken cells infected with avian sarcoma virus: identification of genome-length and subgenome-leghth viral RNAs. J Virol. 1978 Jul;27(1):227–238. doi: 10.1128/jvi.27.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack R. E., Green H., Todaro G. J. Growth control in cultured cells: selection of sublines with increased sensitivity to contact inhibition and decreased tumor-producing ability. Proc Natl Acad Sci U S A. 1968 May;60(1):126–133. doi: 10.1073/pnas.60.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Steimer K. S., Boettiger D. Complementation rescue of Rous sarcoma virus from transformed mammalian cells by polyethylene glycol-mediated cell fusion. J Virol. 1977 Jul;23(1):133–141. doi: 10.1128/jvi.23.1.133-141.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J. Physical map of polyoma viral DNA fragments produced by cleavage with a restriction enzyme from Haemophilus aegyptius, endonuclease R-HaeIII. J Virol. 1975 Apr;15(4):946–953. doi: 10.1128/jvi.15.4.946-953.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Mechanism of cell transformation by RNA tumor viruses. Annu Rev Microbiol. 1971;25:609–648. doi: 10.1146/annurev.mi.25.100171.003141. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Bishop J. M., Vogt P. K. Appearance of virus-specific DNA in mammalian cells following transformation by Rous sarcoma virus. J Mol Biol. 1973 Mar 15;74(4):613–626. doi: 10.1016/0022-2836(73)90052-1. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Vogt P. K., Bishop J. M. Integration of deoxyribonucleic acid specific for Rous sarcoma virus after infection of permissive and nonpermissive hosts. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3067–3071. doi: 10.1073/pnas.70.11.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]