Abstract
The TIP60 tumor suppressor is a histone acetyltransferase involved in transcriptional regulation, checkpoint activation, and p53-directed pro-apoptotic pathways. We report that Human Papilloma Virus (HPV) E6 destabilizes TIP60 both in vivo and in vitro. TIP60 binds to the HPV major early promoter and acetylates histone H4 to recruit Brd4, a cellular repressor of HPV E6 expression. Both low- and high-risk HPV E6 destabilize TIP60, thereby derepressing their own promoter. Destabilization of TIP60 by HPV E6 also relieves cellular promoters from TIP60-initiated repression and abrogates p53-dependent activation of apoptotic pathway. Degradation of TIP60 is therefore a new pathway by which low- and high-risk HPV promote cell proliferation, and cell survival.
Introduction
TIP60 (Tat-interacting protein 60 kDa) is implicated in multiple cellular pathways such as transcription, DNA damage induced checkpoint activation, and apoptosis (Jha and Dutta, 2009; Sapountzi et al., 2006; Squatrito et al., 2006). TIP60 is a catalytic subunit of the NuA4 (Nucleosomal Acetyltransferase of H4) complex and a member of evolutionarily related histone acetyltransferase enzymes (HAT) that belong to the MYST (Moz, Ybf2/Sas3, Sas2 and TIP60) family (Doyon and Cote, 2004; Ikura et al., 2000; Jha and Dutta, 2009; Smith et al., 1998). By acetylating histones, TIP60 modulates chromatin structure and influences interaction of sequence-specific DNA binding proteins and transcriptional machinery (Doyon and Cote, 2004; Robert et al., 2004; Ruthenburg et al., 2007; Sterner and Berger, 2000). Modification of histones by TIP60 is also important for the DNA damage response pathway (Bird et al., 2002; Jha and Dutta, 2009; Jha et al., 2008; Kusch et al., 2004). Upon DNA damage, ATM/ATR kinases are activated and phosphorylate the histone variant H2AX on Ser139, marking the site of damaged DNA and thereby recruiting repair proteins (Bonner et al., 2008; Sancar et al., 2004). The TIP60 complex is recruited to these sites and plays an important role in dephosphorylating phosphoH2AX for termination of the DNA damage response (Ikura et al., 2007; Jha et al., 2008; Kusch et al., 2004; Murr et al., 2006; Sapountzi et al., 2006; Squatrito et al., 2006).
TIP60 also acetylates non-histone proteins involved in transcription and activation of DNA damage checkpoint pathways (Gaughan et al., 2002; Sun et al., 2005; Sykes et al., 2006; Tang et al., 2006). Acetylation of p53 on K120 by TIP60 discriminates between cell-cycle arrest and apoptosis as acetylated p53 preferentially binds to the promoter of pro-apoptotic genes (Sykes et al., 2006; Tang et al., 2006). In addition, TIP60-dependent acetylation of ATM on K3016 is required for efficient activation of the ATM-dependent DNA damage signaling pathway (Sun et al., 2005; Sun et al., 2007). Consistent with a major role of TIP60 in gene expression and in checkpoint responses, mice that are haploid for TIP60 are predisposed to tumors, making TIP60 a bona fide haploinsufficient tumor suppressor (Gorrini et al., 2007).
Human papillomaviruses (HPVs) are non-enveloped DNA viruses that infect mucosal or cutaneous squamous epithelium and cause hyperproliferation (Howley and Livingston, 2009; zur Hausen, 2002). The high-risk HPV can cause cancers because the viral E7 and E6 oncoproteins destabilize pRB and p53, respectively (Munger and Howley, 2002; zur Hausen, 2002). High-risk HPV E6 interacts by its N-terminal residues with p53 and with a cellular ubiquitin ligase E6AP, forming a complex that results in ubiquitin-mediated degradation of p53 by the proteasome (Cooper et al., 2003; Huibregtse et al., 1991; Huibregtse et al., 1993; Scheffner et al., 1993). Additionally, the C-terminal region of high-risk E6 contain a PDZ ligand that interacts with PDZ domain-containing proteins and polyubiquitinates them in E6AP-dependent (Grm and Banks, 2004; Handa et al., 2007; Jing et al., 2007; Kuballa et al., 2007; Matsumoto et al., 2006; Nakagawa and Huibregtse, 2000) or -independent manner (Pim et al., 2000; Shai et al., 2007).
HPV-18 E6 is essential for efficient viral DNA amplification in organotypic cultures of primary human keratinocytes (Wang et al., 2009). However, since the low-risk HPV E6 proteins do not destabilize any of the host proteins targeted by the high-risk HPV E6, the role of the E6 protein and the mechanism of its action in the viral life cycle are not understood. Infection by HPV results in a global changes in cellular gene expression that facilitate cellular proliferation or S phase re-entry.
In this study we report that the cellular tumor suppressor TIP60 is destabilized by HPV E6 both in vivo and in vitro through proteasomes. Deletion and mutational analysis of E6 reveals that this is through the N-terminal region of E6 and does not require interaction with E6AP. Interestingly, TIP60 binds to the HPV major early promoter in YY1 dependent manner and acetylates histone H4. TIP60-dependent acetylation of H4 is required for recruiting Brd4, resulting in the repression of E6. Both high-risk and low-risk HPV E6 target TIP60 for degradation and thereby de-repress the viral major early promoter, modulate the cellular transcriptional program, and attenuate DNA damage-induced activation of p53-dependent apoptotic pathways. The results suggest that TIP60 degradation is a major pathway by which both low- and high-risk HPV promote viral gene expression and deregulate cell proliferation.
Results
Inhibition of proteasome decreases levels of phosphorylated H2AX and stabilizes TIP60 in HPV positive cell lines
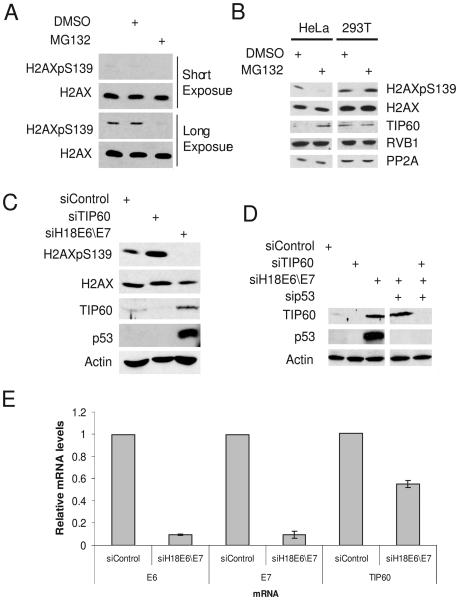
In HeLa cervical cancer cells treated with MG132, a proteasome inhibitor, the levels of phosphorylated H2AX was significantly reduced relative to those in the untreated cells, both in the presence or absence of a DNA damaging agent such as camptothecin and UV (Fig. 1A and S1). We reasoned that the decrease in phosphoH2AX could be due to stabilization of a phosphatase or of other proteins involved in down-modulation of phosphoH2AX. TIP60 and RVB1 reduce phosphorylated H2AX through their effects on chromatin remodeling whereas PP2A is the phosphatase which dephosphorylates H2AX (Chowdhury et al., 2005; Jha et al., 2008; Keogh et al., 2006; Kusch et al., 2004). Addition of MG132 stabilized TIP60, but not RVB1 or PP2A (Fig. 1B). Interestingly, this stabilization was only observed in HeLa, SiHa and CaSki cells, cervical cancer cell lines that harbor the high-risk HPV18 or HPV16, but not in other cell lines such as 293T (kidney epithelial transformed by adenovirus E1A and E1B that also expresses the SV40 T antigen), U2OS (osteosarcoma), or C33A (HPV-negative cervical cancer) cell lines (Fig. 1B, S2 and data not shown). These data suggest that inhibition of proteasomes by MG132 in HPV-infected cervical cancer cells increases TIP60.
Figure 1.
TIP60 is destabilized by human papillomaviral protein E6. (A) Inhibition of proteasome in HeLa cells by MG132 decreases basal phosphoH2AX levels. (B) Stabilization of TIP60 and decrease of basal phosphoH2AX after addition of MG132 are specific for HeLa cells. Cells were incubated with MG132 for 4 hrs and lysates were probed with indicated antibodies. (C) TIP60 is stabilized and phosphoH2AX decreased in E6\E7 depleted cells. HeLa cells were transfected with indicated siRNA and lysates prepared from cells 72 hrs after transfection were probed for indicated proteins. (D) Stabilization of TIP60 after depletion of HPV E6\E7 in HeLa cells is seen even when p53 is also knocked down, so the stabilization of TIP60 is not secondary to the increase in p53. Rest as in (C). (E) HPV18 E6 and E7 mRNA levels are decreased in HeLa cells after transfection of siRNA to HPV18 E6\E7. Also, TIP60 mRNA is not increased by E6/E7 knockdown. The level of a given mRNA was measured by Q-RT-PCR relative to that of actin mRNA: mean and standard deviation of three measurements are shown.
Depletion of HPV E6\E7 stabilizes TIP60
HeLa cells constitutively express HPV-18 E6 and E7 genes. To test whether E6\E7 were involved in the degradation of TIP60, they were downregulated in HeLa cells by siRNA transfection. TIP60 protein and p53 were increased and phosphoH2AX levels were decreased (Fig. 1C). The increase in TIP60 protein was not due to an increase of TIP60 mRNA (Fig. 1E). As depletion of E6\E7 increases p53, the increase in TIP60 levels could to secondary to the p53 elevation. To rule this out, we co-depleted E6\E7 and p53. TIP60 was still stabilized, indicating that p53 elevation is not essential for this effect (Fig. 1D). Collectively, these data suggest that TIP60 is destabilized by HPV proteins E6 and/or E7, independent of p53.
HPV E6 destabilizes TIP60 in a proteasome dependent manner
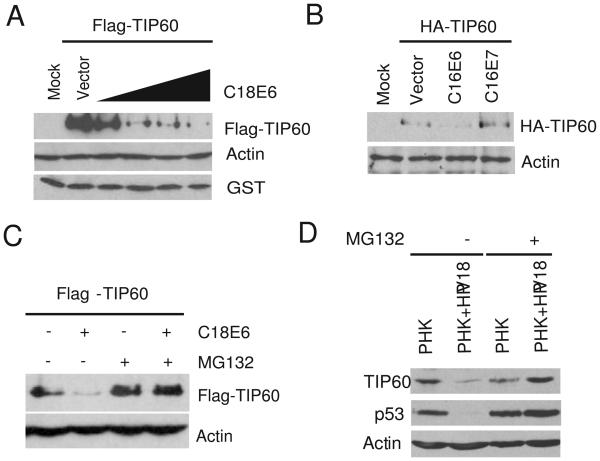
We next investigated whether E6 or E7 was responsible for TIP60's destabilization. 293T cells were co-transfected either with Flag-TIP60 or Myc-p53 expressing plasmids (both constructs driven by EF1α promoter) along with different amounts of HPV-18 E6 expressing plasmid. E6 alone destabilized both TIP60 and p53 (Fig. 2A and S3). This destabilization of TIP60 was not seen with E7 (Fig. 2B). E6-mediated destabilization of TIP60 was specific, as expression of E6 did not have any effect on RVB1, a subunit of TIP60\NuA4 complex or GST protein expressed from a cotransfected plasmid (Fig. 2A and S4). Additionally, the decrease in TIP60 level by E6 was promoter-independent as TIP60 expressed from two different promoters (Flag-TIP60 under EF1α promoter, HA-TIP60 under CMV promoter) was similarly decreased (Fig. 2A and 2B). MG132 restored the levels of TIP60 suggesting that the decrease was proteasome-dependent (Fig. 2C).
Figure 2.
TIP60 is destabilized by HPV18 E6 in proteasome dependent manner. (A) 293T cells were transfected with indicated plasmids and cells were harvested 48 hrs after transfection. Immunoblot with indicated antibodies are shown. 0.1, 0.25, 0.5 and 1.0 μg of C18E6 plasmid was transfected. A plasmid expressing GST was co-transfected to show that the degradation was specific to TIP60. (B) TIP60 is degraded by HPV16 E6 but not HPV16 E7. Rest as in (A). (C) Inhibition of proteasome restores TIP60 in HPV18 E6 transfected cells. 293T were transfected with indicated plasmids and MG132 was added for 4 hrs before cells were harvested. Lysates were probed with indicated antibodies. (D) TIP60 and p53 are decreased in primary human keratinocytes containing HPV18 genomic DNA. Addition of MG132 restores the levels of TIP60 and p53. Lysates from primary human keratinocytes (PHK) with or without HPV18 genome were resolved and immunoblotted with indicated antibodies.
We next investigated whether TIP60 is destabilized by human papillomavirus (HPV) infection of primary human keratinocytes (Wang et al., 2009). TIP60 was decreased in primary human keratinocytes (PHK) harboring the HPV18 genomic plasmids relative to naïve keratinocytes (Fig. 2D, lane 1 and 2). This decrease in TIP60 was proteasome dependent, as addition of MG132 restored its level (Fig. 2D, lane 3 and 4). In these experiments p53 behaved similarly as TIP60 and provided a positive control of a cellular protein known to be degraded by the high-risk HPV E6 via proteasomes (Scheffner et al., 1990).
HPV E6 accelerates degradation of TIP60 in vivo and in vitro
The half-life of TIP60 in HeLa cells was determined following cycloheximide treatment in cells depleted of E6\E7 (Fig. 3A). The half-life of TIP60 increased from 75 mins to >3 hr after depletion of HPV E6. Next, we used E6 protein produced by in vitro transcription and translation in rabbit reticulocyte lysates in an in vitro TIP60 degradation assay (Scheffner et al., 1990) (Fig. 3B). Addition of rabbit reticulocyte lysate programmed with E6 degraded TIP60 in a dose-dependent manner and addition of MG132 in the reactions stabilized TIP60 (Fig. 3C). These data demonstrate that TIP60 is degraded in an E6-dependent manner by proteasomes both in vivo and in vitro.
Figure 3.
HPV E6 destabilizes TIP60 in vivo and in vitro. (A) Depletion of HPV E6 increases the half-life of TIP60. HeLa cells were transfected with indicated siRNA. 72 hrs post-transfection cells were harvested (0 hr) or at indicated times after cycloheximide addition. More protein was loaded in the siControl lanes so that the TIP60 signal was comparable in the two 0 hr lanes. Graph: Levels of TIP60 in each lane were quantitated using Gene Tool software (Syngene). The level of TIP60 in each 0 hr lane is held at 100%. (B) In vitro transcribed and translated HPV18 E6 protein. Rabbit reticulocyte lysate was incubated with either empty vector or plasmid expressing HPV18 E6 in presence of S35 methionine. Fluorograph for the presence of HPV18 E6 proteins is shown. (C) TIP60 is degraded in vitro by HPV18 E6. Flag-TIP60 complex was purified from cells and used as a substrate in an in vitro degradation assay with increasing quantities of in vitro translated HPV18 E6. Immunoblot for levels of TIP60 is shown. * indicates a cross reacting band that serves as a loading control. MG132 blocks the in vitro degradation of TIP60 by HPV18 E6.
TIP60 is destabilized by HPV E6 defective in p53, E6AP and PDZ protein binding
High-risk HPV E6 degrades p53 by forming a complex with E6AP, a cellular ubiquitin ligase (Huibregtse et al., 1993; Scheffner et al., 1993). The domain within high-risk HPV E6 required for binding of p53 has been mapped to its N-terminal portion (Cooper et al., 2003). We tested mutant forms of E6 which cannot bind and degrade p53 but can still destabilize TIP60 (Δ5, F2V, F2VP5R). In co-transfection experiments TIP60 was destabilized by both wild type and p53-binding mutant forms of E6 (Fig. 4A, 4B and 4C). Interestingly, mutant form of HPV16 E6 that is unable to bind to E6AP (Y79N) was still capable of destabilizing TIP60 but not p53, suggesting E6 mediated destabilization of TIP60 is E6AP independent (Fig. 4A, 4B and 4C). The C-terminal region of high-risk E6 has the PDZ binding activity and E6 mutated in PDZ binding (Δ149-150) was still able to destabilize TIP60 (Fig. 4A and 4B). These data suggest that TIP60 is degraded by E6 via a different mechanism than p53 and PDZ family of proteins.
Figure 4.
TIP60 is destabilized by HPV E6 that is defective in binding to p53, E6AP and PDZ domain proteins. (A) Schematic representation of various mutant forms of HPV16 E6 tested to delineate the region of E6 responsible for destabilizing TIP60. ZnF indicates zinc finger domain. “A” indicates alanine substitution. Summary for data presented in Fig. 4B, 4C and 4D. (−) represents failure to degrade and (+) represents ability to degrade the indicated protein. The parenthetical signs represent expected results based on the published literature. (B-C) Screening of HPV E6 mutants for TIP60 destabilization. Mycp53 or HA-TIP60 was transfected along with expression plasmids of GST and various mutant forms of HPV E6 in 293T cells. Cells were harvested after 48 hrs of transfection and lysates were resolved and immunoblotted for indicated proteins. (D) N-terminal region of HPV E6 is sufficient for destabilization of TIP60. Rest as in (B). (E) HPV18 E6*I destabilizes TIP60 but not p53 in primary human keratinocytes. Lysates from primary human keratinocytes (PHK) with or without mutant HPV18 genomic plasmid expressing E6*I, but not full-length E6, were resolved and immunoblotted with indicated antibodies. (F) TIP60 interacts with HPV E6 in vitro. GST-pull down experiments was performed using bacterial lysate co-expressing GST or GST-16E6 along with TIP60. Proteins bound to glutatione beads were resolved on SDS-PAGE and blotted for indicated antibodies. (G) Charge-less mutant of 16E6 (1-43) disrupts this interaction. GST or GST-16E6 (1-43) or GST-16E6 (1-43)-A was expressed together with TIP60. Rest as in (F).
N-terminal region of HPV E6 is required to destabilize TIP60
To identify the region within E6 that is required for destabilizing TIP60 we generated various deletion and point mutations of HPV16 E6. E6 proteins deleted of the N-terminal but not C-terminal region ablated TIP60 (Fig. 4A and 4D). High-risk HPVs express an isoform of E6, a prematurely terminated E6*I peptide (Shirasawa et al., 1994), similar to the N-terminal region of the full-length E6 protein. Interestingly, expression of the N-terminal HPV16 E6 (1-43) (that is similar to E6*I) destabilized TIP60 but not p53 (Fig. 4A and 4D and data not shown). A C33G mutation was engineered into E6 (1-43) to disrupt the zinc finger in this peptide; it was still able to destabilize TIP60 (Fig. 4A and 4D). Thus, the zinc finger motif is dispensable. We hypothesized that the 43 amino acid-polypeptide from the N terminal region of HPV16 E6 has to interact and promote the degradation of TIP60 and thus decided to mutate all charged residues within this N-terminal domain of E6. Mutant HPV16E6 (1-43)-A (12 charged residues changed to alanine) failed to destabilize TIP60 suggesting that charged residues within this domain are indeed important for the degradation (Fig. 4A, 4D and S5).
E6*I is encoded by a predominant spliced mRNA (Schneider-Gadicke and Schwarz, 1986; Shirasawa et al., 1994) during natural infection of keratinocytes by HPV and its function is still being elucidated. To test whether E6*I in primary human keritinocytes destabilizes TIP60, we examined lysates from primary human keritinocytes containing a mutant HPV18 genome in which the E6*I intron coding sequence was deleted. The level of TIP60 but not p53 was decreased in the primary human keritinocytes with HPV18 E6*I being expressed at near physiological levels (Fig. 4E). Taken together these data suggest that N-terminal region of E6 encoded in E6*I is sufficient to destabilize TIP60.
Next, we tested if TIP60 interacted with E6. GST-E6 fusion protein was co-expressed with TIP60 in bacteria and GST-pull down were performed using the bacterial lysates. As shown in Fig. 4F, TIP60 specifically and directly interacted with E6. As the N-terminal region of E6 was sufficient to destabilize TIP60 in vivo we tested whether the interaction was mediated through the N-terminal 1-43 residues (Fig. 4G). E6 (1-43) interacted with TIP60 and this interaction was dependent on the charged residues, as the chargeless mutant, E6 (1-43)-A failed to interact with TIP60 (Fig. 4G). The close correlation of the effect of the mutations on interaction with and degradation of TIP60 suggests that E6 has to interact with TIP60 to promote TIP60 degradation.
TIP60 represses the HPV major early promoter
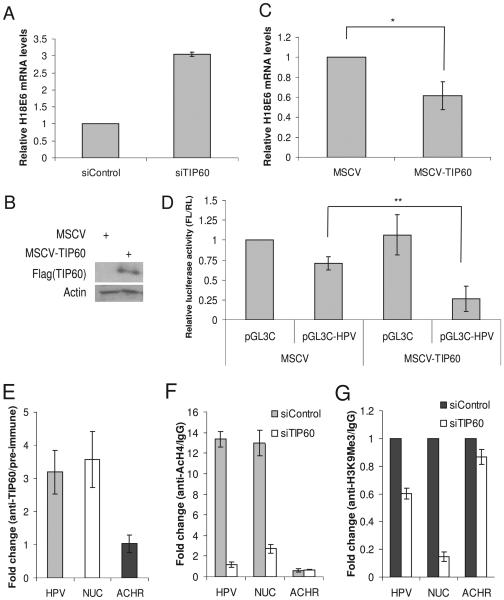
TIP60 acetylates histones, especially histone H4 (Bird et al., 2002; Jha et al., 2008; Sapountzi et al., 2006; Squatrito et al., 2006) and thereby usually activates transcription, though occasionally it acts as a co-repressor of transcription (Sapountzi et al., 2006). We wondered whether the degradation of TIP60 by E6 might regulate the activity of the HPV promoter during the varied conditions encountered in the HPV life cycle. Depletion of TIP60 by siRNA in HeLa cells significantly increases E6 mRNA (Fig. 5A) suggesting that TIP60 acts as a repressor at the E6 promoter. To further establish TIP60's repressive function on HPV E6 expression, we over-expressed epitope-tagged TIP60 by infecting HeLa cells with a retrovirus (Fig. 5B). Over-expression of TIP60 in HeLa cells decreased expression of the viral E6 mRNA (Fig. 5C). Acute over-expression of TIP60 decreased growth of HeLa cells and the level of the exogenous TIP60 decreased upon continuous passage of the cells (data not shown). TIP60's effect on the HPV promoter activity was additionally investigated by transiently transfecting a luciferase reporter driven by the HPV major early promoter in control cells and those over-expressing TIP60. TIP60 significantly repressed HPV promoter-driven luciferase activity (Fig. 5D). Collectively, these data suggest that TIP60 acts as a repressor for the major early promoter that drives E6 expression and E6 stimulates its own expression by targeting TIP60 for degradation.
Figure 5.
TIP60 represses HPV18 E6 mRNA. (A) Increase in HPV18 E6 mRNA after depletion of TIP60. HeLa cells were transfected with indicated siRNA and RNA was prepared from cells harvested 72 hrs after transfection. Q-RT-PCR was performed and normalized to actin and relative levels of HPV18 E6 mRNA is shown. Mean ± S.D. of three experiments. (B) Over-expression of TIP60 in HeLa cells. Flag-TIP60 was over-expressed in HeLa cells after infecting with a retrovirus and selecting the cells on puromycin. (C) HPV18 E6 mRNA is repressed in TIP60 over-expressing cells. Relative change in HPV18 E6 mRNA shown in the cells expressing either vector alone or Flag-TIP60. Mean ± S.D. of two experiments, (*p≤0.05). Rest as in (A) and (B). (D) HPV18 promoter is repressed by TIP60. Control firefly luciferase plasmid (pGL3C) or plasmid expressing firefly luciferase driven by the HPV18 major early promoter (pGL3C-HPV) were transfected into mock or TIP60 over-expressing HeLa cells. The ratio of the firefly luciferase to co-transfected renilla luciferase is calculated and normalized values are shown. Mean ± S.D. of two experiments, (**p≤0.05). (E) TIP60 binds to HPV18 promoter. ChIP assay was performed in HeLa cells to examine binding of TIP60 on HPV18 promoter. Ratio of Q-PCR signal in TIP60 immunoprecipitate relative to pre-immune (negative control) immunoprecipitate is plotted: mean ± S.D. of three experiments. (F) TIP60 acetylates histone H4 on HPV18 promoter. Level of acetylated histone H4 determined by ChIP assay and comparison between the control and TIP60 depleted cells is shown. Rest as in (E). (G) Depletion of TIP60 decreases tri-methylation on lysine 9 of histone H3. Rest as in (E)
TIP60 binds to HPV promoter and is required for acetylation of histone H4
To investigate whether this repression is a direct or an indirect effect of TIP60 we examined the occupancy of residual endogenous TIP60 and exogenous transfected TIP60 on the HPV promoter in HeLa cells. In chromatin immunoprecipitation (ChIP) endogenous and exogenous TIP60 were present on the HPV promoter (Fig. 5E and S6). Cellular Nucleolin [NUC, a known target of TIP60 (Frank et al., 2003)] and acetylcholine receptor (ACHR) were used as positive and negative controls of TIP60 binding (Fig. 5E). As TIP60 acetylates histones, we probed for acetylation of histone H4 on the HPV promoter. ChIP assay showed that histone H4 was acetylated on the HPV promoter (Fig. 5F). Acetylation of histone H4 was dependent on the residual TIP60 in the HeLa cells because depletion of TIP60 significantly decreased levels of acetylated histone H4 on the HPV promoter (Fig. 5F). Additionally, tri-methylation on lysine 9 of histone H3 (H3K9Me3), a repressive histone modification, was decreased (Fig. 5G), consistent with the hypothesis that TIP60 is a repressor for the HPV promoter. These data show that TIP60 binds to HPV promoter and acetylates histone H4.
YY1 facilitate binding of TIP60 to the HPV promoter
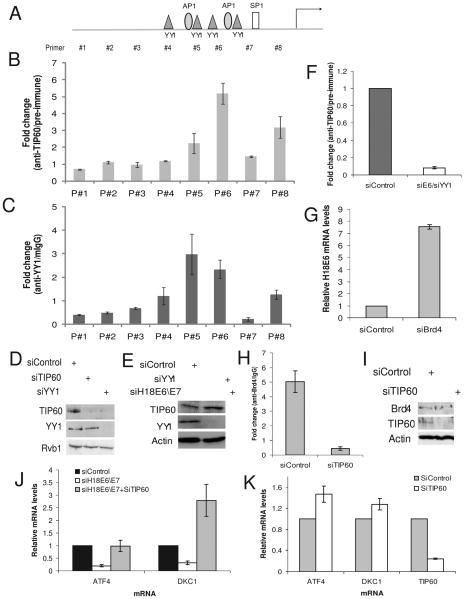
To understand how TIP60 is recruited to the HPV promoter we mapped TIP60's binding site by performing ChIP assay. For this, the HPV promoter was divided into eight regions (Fig. 6A and S7) and we found that TIP60 binds to the proximal region of the promoter (Fig. 6B). Interestingly, TIP60's binding region overlapped with the binding region of YY1 (Fig. 6C), a known repressor of HPV E6\E7 (Bauknecht et al., 1992; Lace et al., 2009; Ralph et al., 2006). To test if YY1 is required for binding of TIP60 to the HPV promoter we first depleted YY1 using siRNA and found that the level of TIP60 was also decreased (Fig. 6D). This is consistent with YY1's role in repression of HPV promoter so that depletion of YY1 will de-repress E6 expression, which in turn will degrade TIP60 (Fig. 6D). We, therefore, co-depleted E6 and YY1 so that the TIP60 protein would not disappear (Fig. 6E). Under these condition binding of TIP60 to the HPV promoter was significantly decreased (Fig. 6F). These data suggest that the YY1 is required to facilitate the binding of TIP60 to the HPV promoter.
Figure 6.
TIP60-YY1-Brd4 are involved in repressing HPV promoter. (A) Schematics of amplicon used to map the binding site of TIP60 and YY1 on HPV promoter. Details in Fig. S7. (B-C) TIP60 and YY1 bind to the proximal HPV promoter. Rest as in Fig. 5E. (D) Depletion of YY1 decreases TIP60. HeLa cells were transfected with indicated siRNA and lysates were probed for indicated antibodies. (E) Co-depletion of YY1 and E6 stabilizes TIP60. Rest as in (D). (F) YY1 is required for binding of TIP60 to the HPV promoter. ChIP assay was performed from chromatin prepared from control or YY1-E6 co-depleted cells (as in Fig. 6E). Binding of TIP60 was significantly decreased in absence of YY1. Rest as in Fig. 5E. (G) Increase in HPV18 E6 mRNA after depletion of Brd4 in HeLa cells. Rest as in (5A). (H) TIP60 augments binding of Brd4 on the HPV18 promoter. Brd4 occupancy on HPV18 promoter was determined using ChIP assay with anti-Brd4 antibody. Rest as in Fig. 5E. Depletion of TIP60 decreases binding of Brd4 to the HPV18 promoter. (I) TIP60 depletion does not affect Brd4 protein levels. Immunoblots with indicated antibodies shown in control and TIP60 depleted HeLa cells. (J) ATF4 and DKC1 are negatively regulated by TIP60. Knockdown of HPV18 E6 by siRNA represses ATF4 expression and this is dependent on TIP60 as depletion of TIP60 restores ATF4 level. ATF4 mRNA was measured by Q-RT-PCR and levels relative to actin mRNA are shown: mean ± S.D. of two experiments. (K) TIP60 mediated repression of ATF4 and DKC1 is HPV independent. HCT116 p53−/− cells were depleted of TIP60 using siRNA and levels relative to actin mRNA shown. Rest as in Fig. 6J. mean ± S.D. of two experiments.
Binding of Brd4 to HPV promoter is TIP60-dependent
Acetylation of histone H4 is normally associated with activation of gene expression, but here we find that it correlates with the repression of E6 expression. To understand the mechanism of TIP60 mediated repression, we hypothesized that acetylated histones may act as a marker to recruit a repressor complex. E6 promoter is known to be repressed by E2 in both Brd4-dependent (Smith et al., ; Wu et al., 2006) or - independent manner (Schweiger et al., 2007). Brd4 depletion de-repressed E6 expression even though HeLa cells do not express E2 (Fig. 6G). As bromodomain containing proteins like Brd4 are known to bind acetylated proteins (Taverna et al., 2007), we hypothesized that TIP60 mediated acetylation of histone H4 is recognized by Brd4 and the recruitment of Brd4 represses E6 expression. Indeed, ChIP assay confirmed that Brd4 was bound to the HPV promoter in the absence of E2 and depletion of TIP60 decreased binding of Brd4 to the promoter (Fig. 6H). This decrease in Brd4 binding was not due to a change in cellular Brd4 levels after TIP60 depletion (Fig. 6I). These data suggest the existence of an E2-independent pathway to recruit Brd4 to the HPV promoter. YY1 recruits TIP60, which acetylates histone H4, which acts as a marker to recruit Brd4, resulting in the repression of E6 transcription.
TIP60 acts a repressor of cellular genes
Having uncovered this interplay between E6 and TIP60 at the HPV promoter we wondered whether cellular promoters are similarly impacted by HPV E6. TIP60 is reported to bind to cellular promoters such as ATF4 (activating transcription factor 4) and DKC1 (dyskeratosis congenita 1) (Martinato et al., 2008). Depletion of E6 indeed decreases ATF4 and DKC1 mRNA and this decrease was due to TIP60 stabilization since co-depletion of TIP60 and E6 de-repressed ATF4 and DKC1 (Fig. 6J, 1E and S8). Next, we investigated whether the TIP60 mediated repression was independent of HPV E6 by depleting TIP60 in p53−/− HCT116 colon cancer cell line. Depletion of TIP60 de-repressed both ATF4 and DKC1 (Fig. 6K) suggesting that TIP60 acts a repressor of these two cellular genes.
TIP60 is destabilized by both high-risk and low-risk HPV E6
The unexpected property of TIP60 to act as a repressor for E6 expression suggested to us that it might also inhibit expression of E6 from other HPV types and conversely, will be a target of E6 protein from other HPV types. Thus we investigated whether TIP60 is a target of high- and low-risk HPV E6. In contrast to p53, which is degraded only by the high-risk HPV E6 protein (Fig. S3, 4A and 7A), TIP60 is degraded by E6 from both high-risk (HPV18 and 16: Fig. 2A, 2B and 7B) and low-risk HPV (HPV11) and from the cutaneous HPV 8 (Fig. 7B).
Figure 7.
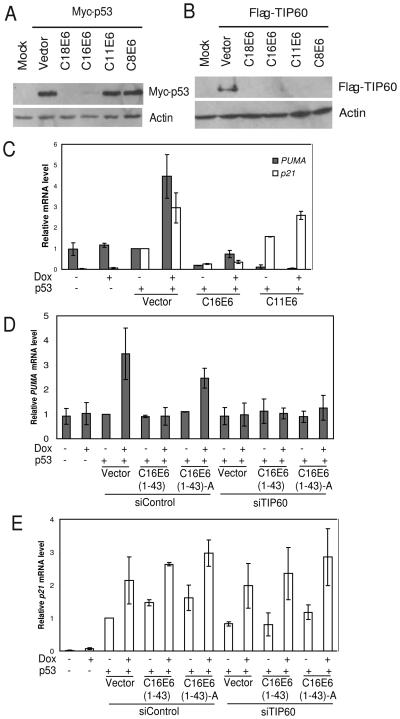
TIP60 is destabilized by both high- and low-risk HPV E6. (A) p53 is degraded only by high risk (HPV18 and 16) HPV E6. 293T were transfected with indicated plasmids and cells were harvested 48 hrs after transfection. Lysates were resolved on SDS-PAGE and probed for indicated antibodies. (B) TIP60 is degraded by both high- (HPV16 and HPV-18) and low-risk (HPV 11) HPV E6 and by the cutaneous HPV (HPV8) E6. Rest as in (A). (C) p53 dependent activation of the apoptotic gene PUMA is attenuated by low-risk HPV. H1299 cells were transfected with indicated plasmids and doxorubicin (5 μM) added 24 hrs after transfection. Cells were harvested 18 hrs post doxorubicin treatment. Relative mRNA levels of PUMA or p21 after normalizing to actin are shown: mean ± S.D. of two experiments. (D-E) TIP60 is required for p53-dependent expression of PUMA, but not p21, after DNA damage. Depletion of TIP60 using siRNA in H1299 cells abrogates induction of pro-apoptotic gene, PUMA. Rest as in (C).
Destabilization of TIP60 by HPVs abrogates p53-dependent activation of proapoptotic genes
p53 regulates transcription of a number of genes, among which PUMA and BAX are pro-apoptotic genes. The ability of p53 to bind to promoters of PUMA and BAX depends on its acetylation at K120 by TIP60 (Sykes et al., 2006; Tang et al., 2006) or phosphorylation at S46, indirectly promoted by TIP60 (Li et al., 2009). As TIP60 is destabilized by E6 even from low-risk HPVs, we wondered whether E6 protein from low-risk HPV will attenuate the p53-dependent apoptotic pathways. To test this hypothesis, we transfected H1299 (p53 null) cells with p53 along with plasmids expressing either the vector, HPV16 E6 (high-risk), or HPV11 E6 (low-risk) and induced apoptotic pathway by adding doxorubicin. Doxorubicin activated p53-dependent expression of PUMA and BAX in vector transformed cells (Fig. 7C and S9). However, expression of PUMA was not observed when high- or low-risk HPV E6 was co-expressed. Since high-risk E6 degrades p53, the failure to induce PUMA was expected. Failure to induce PUMA when low-risk HPV E6 is present is more interesting and suggests that destabilization of TIP60 by low-risk E6 affects the ability of p53 to up-regulate PUMA. Consistent with published results on the function of TIP60, the attenuation of p53-dependent activation was specific to pro-apoptotic genes. For example, p21, a p53-target that prevents G1-S transition after DNA damage, was still induced (Fig. 7C).
To test whether TIP60 destabilization is responsible for the attenuation of PUMA activation, cells were co-transfected with a plasmid expressing the N-terminal region of E6 (1-43), which degrades TIP60, but not p53 (Fig. 4). Although p53 was present and activated p21 (Fig. 7E) it was unable to activate PUMA (Fig. 7D, bars 5 and 6). Cotransfection of E6 (1-43)-A, the charge-less mutant that does not degrade TIP60, still allowed induction of PUMA (Fig. 7D, bars 7 and 8). Finally, siRNA of TIP60 blocks PUMA activation seen with the charge-less mutant (Fig. 7D, bars 13 and 14). Collectively, these results are consistent with the hypothesis that HPV E6 suppresses p53-dependent pro-apoptotic pathways by degrading TIP60. This is particularly important in low-risk HPV, which do not degrade p53.
Discussion
TIP60's role as a co-activator of gene expression is well documented and this activity is primarily through its recruitment to the promoter and acetylation of histones. TIP60 is also a haplo-insufficient tumor suppressor (Gorrini et al., 2007). Here we have identified (i) that the TIP60 tumor suppressor is destabilized by E6, a viral oncogene, (ii) TIP60 destabilization is targeted by E6 of both high- and low-risk HPV, (iii) YY1 facilitates binding of TIP60 to the promoter, (iv) TIP60 acts as a repressor of the major viral promoter, (v) this repression is through TIP60's ability to regulate acetylation of histone H4 to facilitate Brd4 recruitment to the HPV promoter, (vi) TIP60 represses cellular genes and, (vii) even low-risk HPV E6 attenuates p53-dependent activation of apoptotic pathways by destabilizing TIP60. Collectively, these findings provide new insight into the roles of the E6 proteins during the viral life cycle common to both high- and low-risk HPV types.
The mechanism of HPV E6-mediated destabilization of TIP60 is independent of E6AP, a cellular ubiquitin ligase known to be involved in E6-mediated degradation of the tumor suppressor p53. Mdm2 and Cul3 ubiquitin ligases are known to be involved in degradation of TIP60 (Bhoumik et al., 2008; Legube et al., 2002). However, depletion of E6AP, Mdm2 and Cul3 did not stabilize TIP60 in HeLa cells suggesting that these ligases do not co-operate with HPV E6 for destabilizing TIP60 (data not shown). HPV E6 destabilizes its targets by acting as an adaptor and facilitates interaction of the target protein with cellular ubiquitin ligases. Previous studies suggest that HIV-tat could act as an adaptor protein and facilitate destabilization of TIP60 through p300 and Mdm2 (Col et al., 2005). However, we did not see any stabilization of TIP60 in HPV positive cells after depleting Mdm2 or p300 (data not shown). This could be because we failed to deplete these proteins sufficiently, or because a novel E3 ligase is utilized by E6 to target TIP60 to proteasomes. Another possibility is that TIP60 is destabilized by proteasomes in an ubiquitin independent pathway as reported for several other proteins (Mao et al., 2008).
The inability of p53 to induce expression of PUMA in cells expressing the low-risk HPV E6, or N-terminal (1-43) region of E6 highlights the significance of TIP60 in the activation of apoptotic pathway (Fig. 7E). TIP60 acetylates p53 on lysine 120 and targets p53 to promoters of pro-apoptotic genes (Sykes et al., 2006; Tang et al., 2006). An alternate hypothesis suggests that binding of TIP60 to p53 is sufficient to recruit HIPK2, a homeodomain-interacting protein kinase-2 which targets p53 to pro-apoptotic genes by phosphorylating Ser46 (Li et al., 2009). We expect both these pathways to be abrogated by TIP60 degradation. In addition, TIP60 is also involved in activation of the checkpoint pathway through acetylation of ATM (Sun et al., 2005), so that destabilizing TIP60 would also attenuate the checkpoint pathway response. Our results suggest that simply stabilizing p53 in HPV-infected cancers by inhibiting E6AP may not be sufficient to cure cancers without concordantly restoring TIP60.
Although TIP60 and hisone H4 acetylation is normally thought of as an activator of gene expression, we provide evidence that TIP60 can also repress viral and cellular promoters. For the viral promoter, this represson is a direct effect of TIP60 binding and activity with the acetylated H4 recruiting a bromodomain containing cellular repressor, Brd4. While this paper was under review, a genome wide siRNA screen reported that four subunits of the TIP60\NuA4 complex (EP400, EPC1, Brd8 and YEATS\GAS41), along with Brd4, are required for repressing the HPV promoter by HPV E2 (Smith et al.). Thus mechanisms similar to that shown here for the E2-independent repression of the HPV promoter, may also be at play in the E2-dependent repression pathways.
The mechanisms underlying the few occasions when TIP60 represses a cellular promoter have been studied (reviewed in (Sapountzi et al., 2006)). We speculate that the Bromodomain-dependent mechanism reported here may also explain the repression of some cellular promoters by TIP60 and the de-repression by E6. Involvement of TIP60 in regulating transcription of cellular genes suggests that by degrading TIP60, E6 could alter the differentiation program of keratinocytes. Inactivation of the p130, a pRB-related pocket protein, by the high- and low-risk HPV E7 proteins promote S phase re-entry by the differentiated keratinocytes to facilitate viral DNA amplification (Genovese et al., 2008; Zhang et al., 2006) while the viral E6 proteins of high- and low-risk HPV inactivate the transcriptional activity of the major tumor suppressor p53 by interfering with its acetylation by p300 (Thomas and Chiang, 2005). This is the first instance of a cellular protein being targeted for degradation by both low- and high-risk HPV E6, raising the possibility that degradation of the tumor suppressor TIP60 is equally important for the life cycle and pathogenesis of the virus and in the cellular deregulation seen in HPV-associated cancers and benign papillomas.
Experimental Procedures
Cell culture and siRNA transfection
HeLa (cervical cancer cells expressing HPV-18 E6 and E7) and 293T (human embryonic kidney cells expressing adenovirus E1A, E1B and SV40 T-antigen) were cultured in DMEM containing 10% donor calf serum whereas H1299 and C33A cells were cultured in 10% fetal bovine serum. HCT116 p53−/− cells was cultured in McCoy's 5A media containing 10% fetal bovine serum. Primary human keratinocytes were recovered from neonatal foreskin cultured in keratinocyte serum–free medium. The generation of HPV18 genomic plasmid or a mutant genomic plasmid lacking the E6*I intron coding sequence by Cre-loxP mediated in vivo recombination in primary human keratinocytes have been described (Wang et al., 2009). Transfected cells were selected with G418 for 4 days and lysates were prepared after 1 or 2 passages.
For siRNA transfection, transfection was carried out using 30 nM of annealed siRNA duplex (Invitrogen) with Lipofectamine-2000 RNAi max (Invitrogen) reagent following manufacturer's instruction. Cells were split 1:2 after 48 hrs of transfection to maintain the cells in log phase. Target sequences of oligonucleotides used are provided in the supplemental material.
Plasmid constructs, transfections and luciferase assay
Construction of human TIP60 expressing plasmid is described elsewhere (Jha et al., 2008) and the plasmids used to express various HPV E6 genes were under control of CMV promoter. Details about plasmids expressing p53 and various E6 mutants used are in the supplemental material. 293T or H1299 cells were transfected with indicated plasmid using Lipofectamine-2000 (Invitrogen) following manufacturer's instruction. Cells were harvested after 48 hrs of transfection.
Luciferase assay was performed by transfecting either control or HPV major early promoter upstream to firefly luciferase gene, along with renilla luciferase expressing plasmid. Cells were harvested after 24 hrs of transfection and processed for luciferase assay following manufacturer's instructions (Promega).
Real time PCR and chromatin immunoprecipitation (ChIP) assay
Total RNA was isolated by TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions and used for cDNA synthesis by Superscript III (Invitrogen). The cDNAs were used as templates for real time PCR using SYBR Green PCR master mix (Applied Biosystems). Primers used for RT-PCR are in supplemental material.
ChIP assay was performed as described previously (Karnani et al., 2007). Primers used for ChIP-PCR are as follows; H18E6 promoter as in supplemental material (Fig. S7), NUC (Frank et al., 2003) and ACHR (Frank et al., 2003).
Antibodies and western blotting
For Western blotting, cells were lysed in IPH buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.5% v/v NP40, 1 mM dithiothreitol, 20 mM NaF and protease inhibitor mix (Sigma)] and sonicated three times. Lysates were immunoblotted with indicated antibodies. Details about the antibodies used in this study are in supplemental material.
In vitro degradation assay
In vitro degradation assay was performed as described previously (Scheffner et al., 1990). E6 was produced using rabbit reticulocyte lysate. 1, 2 or 3 μl of the rabbit reticulocyte lysate with or without E6 in a 10 μl reaction volume were incubated with TIP60 complex purified from cells, as described previously (Jha et al., 2008). MG132 was included in the indicated reactions.
Supplementary Material
Acknowledgements
We thank the members of the Dutta laboratory for helpful comments. This work was supported by RO1 GM084465 to A.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauknecht T, Angel P, Royer HD, zur Hausen H. Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: interaction with the transcriptional repressor YY1. Embo J. 1992;11:4607–4617. doi: 10.1002/j.1460-2075.1992.tb05563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoumik A, Singha N, O'Connell MJ, Ronai ZA. Regulation of TIP60 by ATF2 modulates ATM activation. J Biol Chem. 2008;283:17605–17614. doi: 10.1074/jbc.M802030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. Embo J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Schneider S, Bohl J, Jiang Y, Beaudet A, Vande Pol S. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306:87–99. doi: 10.1016/s0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- Genovese NJ, Banerjee NS, Broker TR, Chow LT. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J Virol. 2008;82:4862–4873. doi: 10.1128/JVI.01202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Grm HS, Banks L. Degradation of hDlg and MAGIs by human papillomavirus E6 is E6-AP-independent. J Gen Virol. 2004;85:2815–2819. doi: 10.1099/vir.0.80035-0. [DOI] [PubMed] [Google Scholar]
- Handa K, Yugawa T, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J Virol. 2007;81:1379–1389. doi: 10.1128/JVI.01712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM, Livingston DM. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. Embo J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Bohl J, Brimer N, Kinter M, Vande Pol SB. Degradation of tyrosine phosphatase PTPN3 (PTPH1) by association with oncogenic human papillomavirus E6 proteins. J Virol. 2007;81:2231–2239. doi: 10.1128/JVI.01979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Kuballa P, Matentzoglu K, Scheffner M. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J Biol Chem. 2007;282:65–71. doi: 10.1074/jbc.M605117200. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Lace MJ, Yamakawa Y, Ushikai M, Anson JR, Haugen TH, Turek LP. Cellular factor YY1 downregulates the human papillomavirus 16 E6/E7 promoter, P97, in vivo and in vitro from a negative element overlapping the transcription-initiation site. J Gen Virol. 2009;90:2402–2412. doi: 10.1099/vir.0.012708-0. [DOI] [PubMed] [Google Scholar]
- Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. Embo J. 2002;21:1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lin S, Wang X, Lian G, Lu Z, Guo H, Ruan K, Wang Y, Ye Z, Han J, Lin SC. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol. 2009;11:1128–1134. doi: 10.1038/ncb1927. [DOI] [PubMed] [Google Scholar]
- Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Nakagawa S, Yano T, Takizawa S, Nagasaka K, Nakagawa K, Minaguchi T, Wada O, Ooishi H, Matsumoto K, et al. Involvement of a cellular ubiquitin-protein ligase E6AP in the ubiquitin-mediated degradation of extensive substrates of high-risk human papillomavirus E6. J Med Virol. 2006;78:501–507. doi: 10.1002/jmv.20568. [DOI] [PubMed] [Google Scholar]
- Munger K, Howley PM. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Huibregtse JM. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pim D, Thomas M, Javier R, Gardiol D, Banks L. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene. 2000;19:719–725. doi: 10.1038/sj.onc.1203374. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- Ralph WM, Jr., Liu K, Auborn KJ. CCAAT/enhancer-binding protein beta represses human papillomavirus 11 upstream regulatory region expression through a promoter-proximal YY1-binding site. J Gen Virol. 2006;87:51–59. doi: 10.1099/vir.0.81207-0. [DOI] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schneider-Gadicke A, Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. Embo J. 1986;5:2285–2292. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger MR, Ottinger M, You J, Howley PM. Brd4-independent transcriptional repression function of the papillomavirus e2 proteins. J Virol. 2007;81:9612–9622. doi: 10.1128/JVI.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai A, Nguyen ML, Wagstaff J, Jiang YH, Lambert PF. HPV16 E6 confers p53-dependent and p53-independent phenotypes in the epidermis of mice deficient for E6AP. Oncogene. 2007;26:3321–3328. doi: 10.1038/sj.onc.1210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa H, Jin MH, Shimizu K, Akutsu N, Shino Y, Simizu B. Transcription-modulatory activity of full-length E6 and E6*I proteins of human papillomavirus type 16. Virology. 1994;203:36–42. doi: 10.1006/viro.1994.1452. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, White EA, Sowa ME, Powell ML, Ottinger M, Harper JW, Howley PM. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0914818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23:181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc Natl Acad Sci U S A. 2006;103:437–442. doi: 10.1073/pnas.0510012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.