Abstract
The crystal structure of the rare-cutting HNH restriction endonuclease PacI in complex with its eight base pair target recognition sequence 5'-TTAATTAA-3' has been determined to 1.9 Å resolution. The enzyme forms an extended homodimer, with each subunit containing two zinc-bound motifs surrounding a ββα-metal catalytic site. The latter is unusual in that a tyrosine residue likely initiates strand-cleavage. PacI dramatically distorts its target sequence from Watson-Crick duplex DNA basepairing, with every base separated from its original partner. Two bases on each strand are unpaired, four are engaged in non-canonical A:A and T:T base pairs, and the remaining two bases are matched with new Watson-Crick partners. This represents a highly unusual DNA binding mechanism for a restriction endonuclease, and implies that initial recognition of the target site might involve significantly different contacts from those visualized in the DNA-bound cocrystal structures.
Restriction endonucleases (REases) occur in all free-living bacteria and archaea and are believed to function to defend their hosts against invasion by foreign DNA, particularly from bacteriophage (Pingoud et al., 2005). REases vary in sequence, structure, oligomeric composition, substrate-specificity, and enzymatic behavior (Bujnicki, 2003). They range from compact monomers that act independently, to elaborate multifunctional protein assemblages, and typically recognize target sequences in duplex DNA ranging from four to eight specific base pairs in length. (Pingoud et al., 2005). These sequences can be symmetric or asymmetric, as well as continuous or discontinuous, depending upon the enzyme architecture.
Several distinct catalytic site motifs and mechanisms have been identified among restriction endonucleases, suggesting this enzymatic and biological function has evolved independently several times. The most common catalytic motif, that of the 'PD…(D/E)xK' nuclease superfamily, is the the most wide-spread and best understood (Kosinski et al., 2005). Alternative catalytic motifs, associated with quite different core protein folds, have been identified in many additional restriction endonucleases, including the ‘HNH’ (Cymerman et al., 2006; Jakubauskas et al., 2007; Saravanan et al., 2004) and the ‘GIY-YIG’ (Ibryashkina et al., 2007) motifs (both of which are more commonly associated with mobile homing endonucleases from bacteriophage) (Stoddard, 2005). All three of these catalytic lineages are also found in a much wider variety of enzymes involved in DNA metabolism and modification, including those responsible for DNA repair, recombination and fidelity (Cymerman et al., 2006; Dunin-Horkawicz et al., 2006; Kosinski et al., 2005). As well, isolated examples of two additional structural motifs (containing the phospholipase D and 'half-pipe' folds) have also observed for R.BfiI and R.PabI, respectively (Grazulis et al., 2005; Miyazono et al., 2007).
The conserved structural core surrounding the HNH motif is termed the 'ββα-metal' fold. This protein topology consists of two anti-parallel β-strands connected by a loop of variable length, flanked by an α-helix (Mehta et al., 2004),(Kuhlmann et al., 1999). A binding site for a single catalytic metal ion—typically magnesium—is embedded within this catalytic fold. In some instances, significant insertions of additional structural elements are observed within this motif (Eastberg et al., 2007; Stoddard, 2005). The ββα-metal fold can exist as an independently folded catalytic domain (as observed in colicins) or it can be fused to additional protein domains that dictate DNA binding specificity and cleavage activity.
The PD…(D/E)×K motif can be very well-suited for recognition of short DNA sequences with high fidelity, because the catalytic center is surrounded by a densely packed array of side chains that can contact neighboring base pairs in the major groove in a sequence-specific manner (Orlowski and Bujnicki, 2008). In contrast, the HNH motif and its associated ββα-metal fold appears less well-suited for this task. In order to target the scissile phosphate, the catalytic core motifs of these enzymes primarily interact with the DNA backbone where they contribute little to sequence-specificity and fidelity (Eastberg et al., 2007). Sequence-recognition by these enzymes is therefore usually carried out by additional protein domains that are tethered to the ββα-metal region, necessitating significant repackaging and augmentation of this catalytic motif.
Recently, the structure of the ββα-metal restriction endonuclease Hpy99I was determined in complex with its DNA substrate, 5' - CGWCG - 3', at 1.5 Å resolution (Sokolowska et al., 2009) (W=A or T). Hpy99I binds as a homodimer and forms a ring-like structure that encircles the DNA. The protein contacts all four C:G base pairs within both the minor and major groove, and contacts the central base pair (A:T or T:A) in only the minor groove. The DNA is slightly bent in the complex. All nucleotides in the target site are found in canonical Watson-Crick basepair interactions.
In contrast, PacI is a 'rare-cutting' homodimeric HNH restriction endonuclease found in the bacterium Pseudomonas alcaligenes. It recognizes the symmetric eight base pair duplex DNA sequence 5' – TTAAT/TAA - 3' and cleaves each strand between the internal thymine residues (as the position indicated by "/") to generate product fragments containing 2-base, 3’-overhangs (Roberts et al., 2010). PacI is one of the smallest REases known, comprising only 142 amino acids per subunit, eight of which are cysteines (Figure 1). Its gene resides within a super-integron, a chromosomal array that contains multiple gene cassettes each flanked by a large direct repeat sequence and mobilized by a common site-specific integrase (Vaisvila et al., 2001). Unlike the vast majority of REases, which are accompanied by DNA-methyltransferases that protect the cell’s own DNA from REase auto-digestion, PacI appears to be a solitary enzyme with no companion methyltransferase (see Supplementary Material). Host protection in this rare instance seems likely to depend not on the methylation of recognition sequences, but rather on the absence of such sequences in the P.alcaligenes genome.
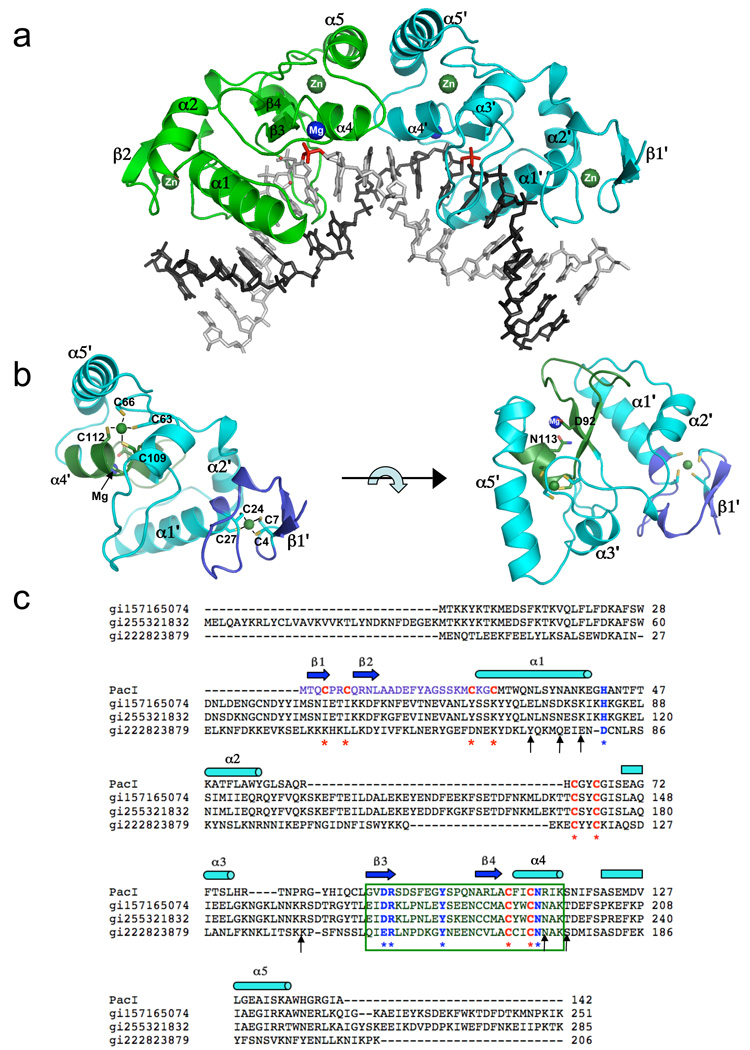
Figure 1. The structure of the PacI restriction endonuclease.
The protein is colored according to individual structural domains and motifs; the same color scheme is used in all panels. All figures were generated using the molecular graphics program PYMOL (DeLano, 2002). Panel a: The PacI homodimer is bound to ts palindromic DNA target sequence. The two protein subunits are colored green and cyan; the two bound zinc ions in each protein subunit are labeled and colored dark green, and the single bound divalent cation observed in each catalytic site is labeled 'Mg' and colored blue. The corresponding scissile phosphates, that yield 2 base, 3' cohesive overhangs when cleaved, are colored red. Panel b: A single subunit of PacI is shown, in two separate orientations. The left panel shows the same orientation as the cyan-colored subunit in panel a. The right panel shows the same subunit, rotated by 90° around the horizontal axis. In both panels, the ββα-metal motif is colored dark green, and the unique N-terminal region (which binds zinc number 1) is colored blue. On the left, the eight cysteine ligands to the two zinc ions are labeled. On the right, the single bound divalent cation (magnesium in the product complex) and the two residues involved in its coordination (D92 and N113) are labeled. Panel c: Sequence homology between PacI, and its three recognizable homologues (hypothetical protein sequences from Campylobacter concisus, cowae and lari, respectively). The ββα-metal motif is indicated by the box. Residues that directly contact DNA are indicated with asterisks arrows; catalytic residues of the HNH endonuclease motif are indicated with blue font and asterisks; zinc-binding cysteine residues are indicated with red font and asterisks. The N-terminal regions, that harbors the first bound zinc ion, is indicated with light blue font, corresponding to the coloring of the same region in panel b. See also supplemental Figure S1.
The length of the PacI recognition sequence (eight basepairs) places this enzyme in the company of NotI and SfiI, two other 'rare-cutting' endonucleases the co-crystal structures of which have been solved (Qiang and Schildkraut, 1987). NotI and SfiI belong to the PD…(D/E)×K catalytic site superfamily, and in contrast to PacI, recognize sequences composed entirely of G:C base pairs.
Bioinformatics analysis of PacI (Orlowski and Bujnicki, 2008), and independent analyses with online protein fold prediction servers such as PHYRE (Bennett-Lovsey et al., 2008) suggest the presence of an HNH-related catalytic site. The likely presence of this motif, combined with the opportunity to compare a ‘rare A:T-cutter’ to two ‘rare G:C-cutters’, led us to determine the structure of PacI bound to DNA. PacI displays little resemblance to either NotI or SfiI, and while it contains structural elements similar to those in Hpy99I, the arrangements of these elements and the overall folds of the two proteins are strikingly different. PacI binding induces an unusual distortion of its DNA target sequence that completely disrupts and reorganizes it normal Watson-Crick duplex structure.
Results
Overall protein structure and catalytic site
The structure of PacI was determined in complex with its eight base pair cognate target site, within the context of an 18 base pair synthetic DNA duplex. The structure was determined both in the presence of calcium (yielding a co-crystal structure containing uncleaved DNA that extended to 2.0 Å resolution) and in the presence of magnesium (resulting in a bound product complex that was visualized at 1.9 Å resolution). The two structures are virtually identical, with the exception of the presence of free 5' phosphate and 3' hydroxyl DNA product ends in the endonuclease catalytic sites in the presence of magnesium. Data collection and refinement statistics are provided in Table 1, and a detailed description of materials and methods is provided in Supplementary Information. Examples of the experimental electron density, calculated using phases derived by a combination of the multiple isomorphous replacement (MIR) and single anomalous dispersion (SAD) methods, are shown in Supplementary Figure S1.
Table 1.
Data collection and refinement statistics
| Data set Id | Native-1 | Native-2 | PtCl4-1 | PtCl4-2 | HgCN2 | PIP | WO4 |
|---|---|---|---|---|---|---|---|
| Wavelength (Å) | 1.5418 | 0.97741 | 1.0719 | 1.5418 | 1.5418 | 1.5418 | 1.5418 |
| Data collection | |||||||
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 |
| a (Å) | 36.86 | 36.86 | 37.09 | 37.32 | 36.93 | 36.89 | 37.83 |
| b (Å) | 115.75 | 115.75 | 115.16 | 114.08 | 116.08 | 117.79 | 116.05 |
| c (Å) | 114.37 | 114.37 | 114.83 | 114.32 | 114.19 | 113.89 | 114.14 |
| Resolution (Å) | 50-2.64 | 38-1.97 | 50-1.92 | 25-3.2 | 50-3.0 | 50-2.6 | 50-3.07 |
| Unique reflections | 7568 | 17385 | 19094 | 4343 | 5193 | 7947 | 5014 |
| Redundancy* | 6.6(4.7) | 10.7(4.2) | 13.5(11.1) | 6.9(7.1) | 13.4(8.9) | 7.1(7.0) | 11.0(7.8) |
| Completeness (%)* | 99.8(98.7) | 97.2(80.1) | 98.6(92.4) | 100(100) | 99.8(99.6) | 99.1(93.5) | 99.4(94.4) |
| I/σ* | 20.4(5.0) | 27.8(3.3) | 44.2(3.4) | 20.3(5.7) | 28.7(7.1) | 37.1(11.8) | 32.9(13.9) |
| Rmergea (%)* | 9.2(31.2) | 6.8(22.9) | 6.2(22.6) | 9.7(39.3) | 8.6(31.9) | 4.5(13.9) | 6.0(12.5) |
| B(iso)(Å2) | 47.4 | 26.25 | 29.04 | 60.4 | 61.1 | 58.7 | 53.5 |
| Refinement | |||||||
| Protein atoms# | 1108 | 1108 | 1108 | ||||
| DNA atoms# | 366 | 366 | 367 | ||||
| Heavy atoms | 2 Zn+2 | 2 Zn+2 | 2 Zn+2, Pt+2 | ||||
| Catalytic Metal ions | Ca+2 | Ca+2 | Mg+2 | ||||
| Cations | --- | --- | SO4−2 | ||||
| Solvent molecules | 76 | 94 | 114 | ||||
| R-factorb (%)* | 0.208(0.293) | 0.184(0.227) | 0.172(0.217) | ||||
| R-freeb (%)* | 0.278(0.319) | 0.217(0.361) | 0.201(0.285) | ||||
| Rmsd | |||||||
| Bond length (Å) | 0.012 | 0.013 | 0.011 | ||||
| Angles (°) | 1.667 | 1.541 | 1.305 | ||||
| Ramachandran (%) | |||||||
| Core region | 97.83 | 96.38 | 98.43 | ||||
| Allowed region | 1.45 | 2.90 | 1.57 | ||||
| Outliers | 0.72 | 0.72 | 0.00 |
Highest resolution shell values in parenthesis.
Rmerge = Σ|Ihi - <Ih> |/ΣIh, where Ihi is the ith measurement of reflection h, and <Ih> is the average measured intensity of reflection h.
R-factor/R-free = Σh|Fh(o) - Fh(c)|/Σh|Fh(o)|. Where R-free was calculated with 5% of the data excluded from refinement.
The overall structure of the endonuclease homodimer bound to its DNA target is shown in Figure 1a; two separate views of a single enzyme subunit are shown in Figure 1b. The overall core topology of the PacI subunit corresponds to "β1–β2–α2–α3–β4–α4–α5", with the β3–β4–α4 secondary structure elements comprising the ββα-metal catalytic site motif. This core topology is further extended by very short β-hairpin motifs on the protein surface that are involved in DNA contacts.
The PacI subunits display an extended structure containing a pair of bound zinc ions, each of which is coordinated by four cysteine residues. The first zinc ion is entirely sequestered within an N-terminal region (containing cysteines 4, 7, 24 and 27) that appears to be a unique feature of PacI: the only three homologues of PacI currently in Genbank (all from strains of the bacterium Campylobacter) display little sequence similarity to this region (Figure 1c). The second zinc ion is buried in the enzyme core, and is also coordinated by four cysteine residues (Cys 63, 66, 109 and 112). This zinc ion is located near the endonuclease catalytic site. Two of the cysteine residues involved in its coordination (Cys 109 and 112) extend from the α4 helix from the ββα-metal motif.
The overall structural organization of the PacI enzyme resembles, at a superficial level, the organization of the homodimeric HNH restriction endonuclease Hpy99I (Sokolowska et al., 2009), and more distantly resembles the homodimeric HNH homing endonuclease I-PpoI (Flick et al., 1998). All three proteins contain a catalytic ββα-metal motif and contain two structural zinc ions embedded within each protein subunit, and all three position their active sites across the minor groove to produce 3' overhangs. However, the extended architecture and DNA binding modes of these enzymes are very different from one another (Figure 2 and supplementary Figure S2), indicating that they appear to have independently acquired and then optimized similar structural strategies for stabilization and catalysis, presumably after their divergence from a common ancestral endonuclease.
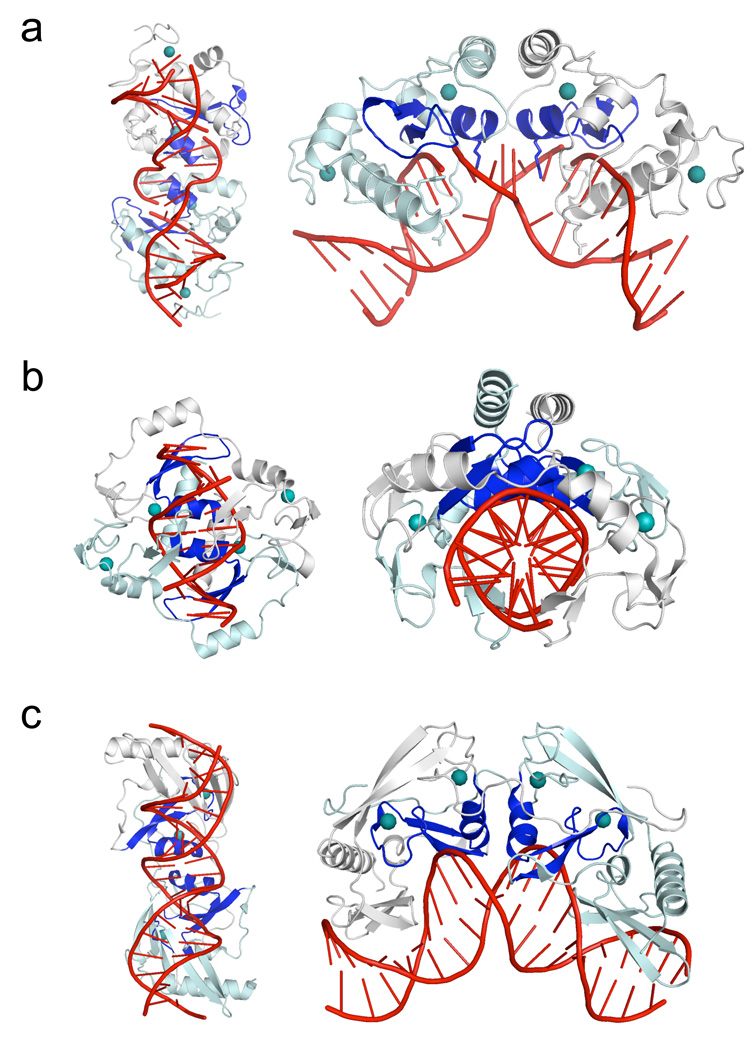
Figure 2. ββα-metal (HNH) endonucleases that contain two bound zinc ions per subunit.
For each endonuclease structure, the left panels show comparable orientations of the bound DNA targets, while the right panels show comparable orientations of the protein subunits (with the dyad symmetry axis running vertically and the protein subunits oriented to the left and right side of that axis). For all three structures, the ββα-metal catalytic motif is colored blue, and the bound zinc ions are shown as teal spheres. Panel a: The PacI restriction endonuclease. While the protein's overall organization of secondary structure elements, relative to the catalytic sites and bound zinc ions, is similar to Hpy99I, the overall architecture of the individual ββα-metal repeats, as well as the mode of DNA binding, is significantly different. Panel b: The core of the Hpy99I restriction endonuclease, which cleaves a five base pair CGWCG target, generating a 5-base, 3' overhang (Sokolowska et al., 2009). An N-terminal β-barrel domain (residues 1 to 53) that is not involved in DNA binding is removed for clarity. The remaining protein homodimer structure includes all catalytic and zinc-binding regions that correspond to those observed in PacI. Note that the orientation of the protein is roughly orthogonal to the major axis of the DNA duplex, which is relatively unbent. Panel c: The I-PpoI restriction endonuclease (a fourteen-base cutting homing endonuclease found in the eukaryotic amoeboid Physarum polycephalum) (Flick et al., 1998). The overall bend of the DNA target is similar to that displayed by PacI; however all base pairs are maintained in their original Watson-Crick base pairing arrangement. The protein fold, beyond the core ββα-metal motif, is completely different from that displayed by Hpy99I and PacI, indicating that divergence of these protein lineages may have occurred at an early stage, from a simple ββα-metal scaffold prior to subsequent elaboration and specialization. See also supplemental Figure S2.
The backbone conformation and metal coordination exhibited by the 'ββα-metal catalytic core of PacI is similar to those observed in other HNH endonucleases (Kuhlmann et al., 1999) (Figure 3a and Supplementary Figures S2 and S3). Structure-based alignments of this region with five separate ββα-metal endonucleases (E9 colicin, endonuclease VII, Hpy99I, I-HmuI and I-PpoI) gives RMSD values for all backbone atoms of 1.5 to 1.8 Å, with corresponding sequence identities ranging from as low at 6.7% (I-PpoI) to as high as 24% (I-HmuI and EndoVII). A single divalent cation is bound within the PacI catalytic motif, where it is coordinated by aspartate 92 and by asparagine 113, and also interacts with the 3' oxygen and a nonbridging oxygen of the scissile phosphate. The distance from the metal to each DNA atom is approximately 2.5 Å.
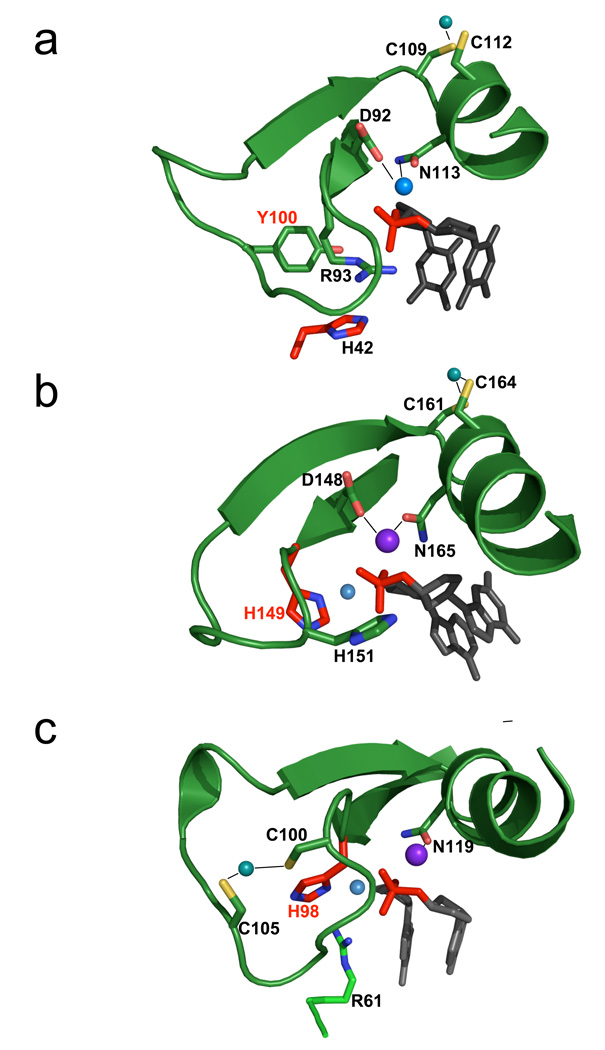
Figure 3. ββα-metal motifs and catalytic sites for HNH restriction and homing endonucleases.
The core catalytic motif, consisting of a two-stranded antiparallel beta-sheet and single alpha-helix, is shown in approximately the same orientation for (Panel a) PacI; (Panel b) Hpy99I and (Panel c) I-PpoI . The scissile phosphate is shown in red, flanked by its 5' and 3' nucleosides in gray. The histidine general base is colored and labeled with red, and the corresponding water nucleophile is a small light blue sphere. In all three catalytic sites, a single bound divalent metal ion (dark blue larger sphere) is coordinated by two asparagine/aspartate residues. An additional polar residue (participating in cleavage as a Lewis acid, whereby it stabilizes the phosphoanion transition state) is present in each catalytic site that is positioned to help satisfy the charge on the phosphate during cleavage (R93 for PacI; H151 for Hpy99I, and R61 for I-PpoI). Note that PacI displays a structural inversion in the positions of the histidine general base and the Lewis acid: His 42 is presented from a loop outside the ββα-metal motif, while R93 is found in the position that is usually occupied by an HNH histidine base. Finally, peripheral cysteine residues in all three ββα-metal motifs are involved in coordination of a structural zinc ion (C109 and C112 in PacI). However, the location of the bound zinc in the restriction endonucleases (panels a and b) differ from the homing endonuclease (panel c), indicating that these enzyme lineages may have diverged from a common ancestor prior to development of these metal binding sites. See also supplemental figure S3.
In spite of the structural similarity of the ββα-metal motif described above, the PacI catalytic site displays a significant departure from the typical HNH motif. The position within the ββα-metal motif that is normally the site of a histidine general base that activates the water nucleophile (corresponding to His 149 in Hpy99I and His 98 in I-PpoI; Figure 3b and 3c) is instead occupied by an arginine residue (Arg 93), that interacts with the 3' leaving group . In place of the usual histidine, a neighboring tyrosine residue (Tyr 100) is instead positioned to either assist in activation of a nucleophilic water (which is not observed), or perhaps to act directly as a nucleophile itself. The distance from the tyrosine hydroxyl group to the phosphorus atom is 4.5 angstroms in the uncleaved calcium-bound complex, and is 3.3 angstroms in the cleaved magnesium-bound complex (the distance is reduced in the cleaved complex due to rotation of the 5' phosphate group after cleavage). The nearest histidine residue (His 42) is located approximately over 8 Å from the scissile phosphate, and like Arg 93, is located closer to the leaving group than to the site of the nucleophilic attack. Thus, the PacI endonuclease displays a dramatic alteration and rearrangement of the usual side chains found in a ββα-metal catalytic motif, and perhaps a change in the actual cleavage mechanism.
To assess the relevance of Tyr 100, Arg 93 and His 42 in the PacI catalytic site, each was mutated by PCR and the mutant proteins were expressed in vitro and assayed for activity (Table 2). To the best of our ability to measure, the amounts of each protein construct generated in vitro, and then used in individual digest experiments, were comparable. Y100F was found to be inactive (less than 10−4 WT activity) indicating that the phenolic oxygen of this amino acid appears to be essential for catalysis . R93A and M were also inactive, but R93K displayed reduced activity, suggesting that a positively charged group in this position in the catalytic site is also essential. H42A displayed reduced activity (~10−2 WT activity). The putative metal-binding residues Asp 92 and Asn 113 were also mutated and assayed. D92L and N113L were inactive, whereas the D92A and N113A mutants displayed a low level of activity, indicating that the metal-binding residues are critical components of the catalytic site.
Table 2.
Activity of PacI site-directed mutants
| PacI variant | Endonuclease activity |
Units per 25 µl in vitro reaction |
|---|---|---|
| Wild-type PacI | +++ | ~200 |
| Catalytic: | ||
| H42A | + | ~1 |
| R93K | +/− | ~0.2 |
| R93A,M | − | <0.01 |
| Y100F | − | <0.01 |
| Mg2+-binding: | ||
| D92A,L | +/− | <0.1 |
| N113A,L | +/− | <0.1 |
| Specificity: | ||
| N32A,T,L,D | − | <0.01 |
| N36A,T,L,D | − | <0.01 |
| K39A,M | ++ | >10 |
Endonuclease activities of wild-type PacI and of mutant derivatives. Mutants were constructed by two-step PCR, expressed in vitro using the PURExpress™ transcription/translation system, and assayed by DNA-digestion and gel electrophoresis, as described in the Supplementary Material. The standard 25 µl PURExpress™ reaction produced approximately 200 units of endonuclease activity from the wild-type PacI gene template (1 unit completely digests 1 µg of substrate DNA to completion in 1 h at 37°C). The limit of endonuclease activity detectable in this assay corresponded to 10−4-fold less than wild-type, or approximately 0.01 units. In most cases, several different mutants were constructed for each amino acid targeted for alteration. Mutants yielding the same result are grouped together on a single line in the table; thus, ‘N36A,T,L,D’, for example, signifies that Asn 36 was individually changed to Ala, Thr, Leu, and Asp, and all four mutant enzymes behaved similarly—in this case displaying no detectable endonuclease activity. Plus and minus symbols in column 2 indicate the relative levels of endonuclease activity observed across several independent experiments. These levels are quantified approximately with respect to wild-type in column 3.
The two bound zinc ions in PacI (each coordinated in a Cys4-Zn tetrahedral cluster) represent a widely distributed conserved structural motif, distinct from the conventional trinucleotide-specific Cys2-His2 ‘Zinc-finger’ domains of eukaryotic transcription factors (Supplementary Figure S3). The Cys4-Zn motif comprises a pair of CxxC sequences. The first two cysteines flank a loop, while the second two initiate an alpha helix (in some related sequences, the third Cys or the fourth Cys is replaced by His instead). The region between each CxxC pair varies in length and function, as does the helix. In many instances, this region includes catalytic residues that contribute to the HNH catalytic site. Approximately 200 HNH-like domains are aligned in pfam01844, and over one-third of these are embedded in Cys4-Zn motifs, indicating that this structural architecture is often associated with an HNH catalytic site. In contrast, this same region in the GATA family of transcription factors includes residues responsible for DNA sequence recognition (Bates et al., 2008) (Supplementary Figure S3, panel L).
DNA binding and recognition
The mode of DNA binding displayed by PacI is very unusual. The ββα-metal catalytic sites from each protein subunit straddle the minor groove at the center of the DNA target, resulting in an overall bend angle of approximately 90 degrees (Figures 1a and 4a). This results in a dramatic widening of the minor groove (to approximately 18 Å) and a corresponding reduction in the width of the opposing major groove. This bend is accompanied by a radical alteration of the DNA duplex: every base throughout the target site is unpaired from its original Watson-Crick partner (Figure 4b). Within the eight base pair target site, two bases on each strand are completely unpaired, four are engaged in non-canonical A:A and T:T base pairs, and the remaining two bases are matched with new Watson-Crick partners. This disruption of the DNA duplex is entirely localized to the PacI target site; the base pairs immediately outside the 5' - TTAATTAA - 3' sequence still display canonical B-form interactions. It does not appear that crystal contacts play a role in these features of the protein-DNA complex: the solvent content of the crystals is not unusual (about 55%) and the regions of the protein and DNA involved in contacts and recognition are not located near symmetry mates in the crystal lattice.
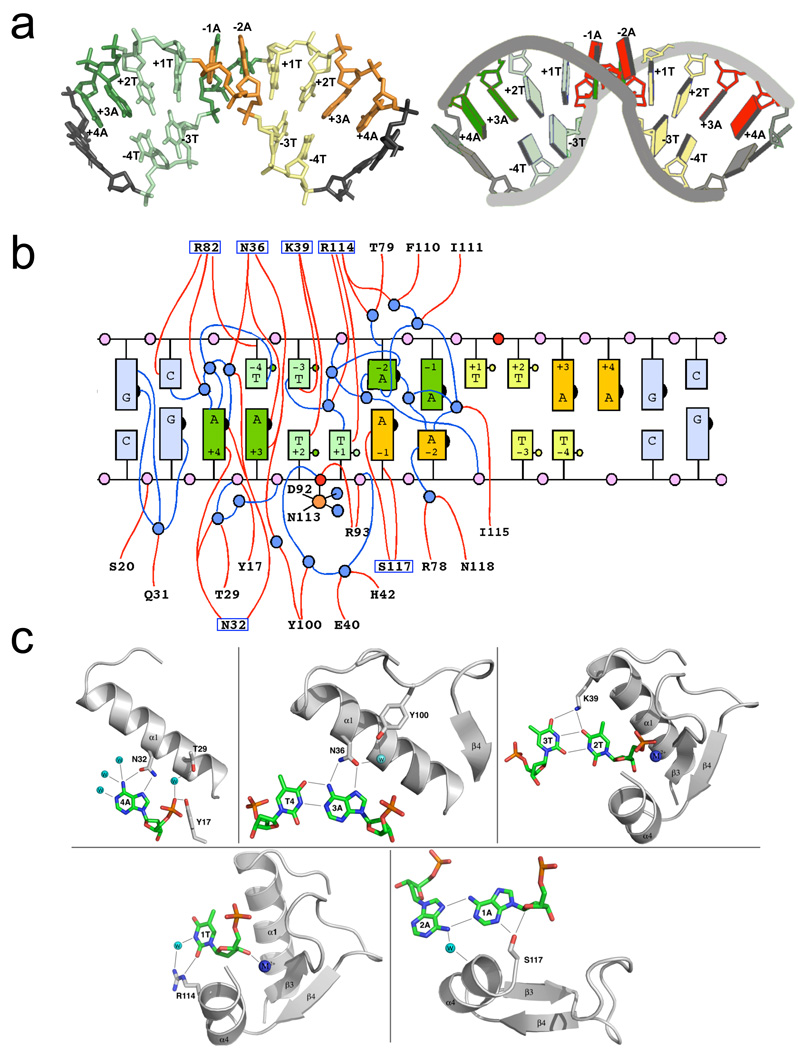
Figure 4. DNA recognition and binding by PacI.
Panel a: Conformation of the bound DNA target site. The individual bases of the 'TTAATTAA' target are colored both by their position in the original DNA half-site (left half-site = green; right = yellow) and by their identity (dark bases are adenine; light bases are thymine). The bound DNA is shown in a stick representation on the left, and in a cartoon representation, generated by the program 3DNA (Zheng et al., 2009), on the right. Those bases that lie outside the eight base pair target site are colored grey. Panel b: Cartoon representation of the base pairing interactions between the two target site strands and the contacts between the protein, DNA and solvent molecules. The individual bases are colored as shown in panel (a) above. The numbering of the bases corresponds to the position in the original unbound target sequence, with bases −4T to −1A corresponding to the 5' half (left half) of the target site along one DNA strand, and bases +1T through +4A corresponding to the 3' (right) half of the target site along the same strand. In the bound complex, all bases in the target are removed from their original Watson-Crick partners, so that some bases are unpaired (+1T and +4A on each strand), some are found in noncanonical A:A and T:T base pairs (+2T, −1A, −2A and −3T on each strand) and some are found in new Watson-Crick base pairs (−4T with +3A from each strand). The scissile phosphates on each strand are red; well-ordered water molecules are blue. Protein residues (from only one of the two protein subunits, for clarity) that are involved in direct or water-mediated contacts to the DNA are indicated; those that form direct contacts to DNA bases are boxed. Panel c: Structural interactions between each unique DNA base or base pair in a single half-site to solvent and protein residues. The numbering of bases is consistent with panels a and b above. See also supplemental figure S4.
The bound conformation of the DNA target was analyzed using the online program 3DNA (Zheng et al., 2009) (Supplementary Figure S4). The perturbation of the DNA structure results from significant distortion of the individual ribose moieties and the corresponding glycosidic bonds between sugar C1' carbons and the corresponding nucleotide bases. Only three ribose sugars on each strand (corresponding to −4T, −2A and +3A) are found in their original C2'-endo pucker, while the remaining sugars are predominantly flipped into a C1'-exo conformation. The chi angles linking the ribose C1' carbons to the N1 nitrogen of the thymines, or to the N9 nitrogen of the adenines, deviate from their nominal B-form values by as much as +/− 40°, leading to a rotation of individual bases that allows non-canonical A:A or T:T base pairing. These base pairs still exhibit two intra-strand hydrogen bonds (Figure 4c), linking the thymine-thymine pairs via the O2-N3 and N3-O4 atoms of their pyrimidine rings, and the adenine-adenine pairs via the N6-N1 and N7-N6 atoms of their purine rings. These base pair interactions, while rarely observed in DNA duplexes, are often found in folded RNA structures (Olson et al., 2009).
The deformation of the DNA in PacI is accompanied by a significant unwinding of the duplex at base step −2A in each DNA half-site (Supplementary Figure S4). That base, which is engaged in an A:A base pair with its −1A partner, exhibits a −40° tilt and unstacking from its neighboring (symmetry-related) A:A base pair. The local unwinding of each DNA half-site at these A:A base pairs is complemented by local over winding of the adjacent base steps and base pairs, allowing the rearranged DNA to maintain an overall duplex architecture. Although the DNA backbone and its base pairing interactions exhibits a dramatic rearrangement in the bound protein complex, all the individual base pairs (both Watson-Crick and non-Watson-Crick) exhibit near normal values of propeller twist and buckle angles.
The PacI-DNA complex is further notable for the paucity of direct contacts between the protein and the nucleotides. The two unpaired bases in each half site (+1T and +4A) are in direct contact with amino acid side chains: +4A interacts in the major groove with Asn 32, and +1T interacts in the minor groove with Arg 114 (Figures 4b and 4c). The adenines in the reorganized A:T base pairs ( involving +3A in each DNA half-site) interact in the major groove with Asn 36. One adenine in each A:A base pair makes a nonspecific contact to Ser 117 in the minor groove, and the O4 groups of both thymines in each T:T base pair contact Lys 39 in the major groove. To assess the importance of the major groove contacts, Asn 32, Asn 36, and K 39 were changed by PCR to various other amino acids and the mutant proteins expressed in vitro and assayed (Supplementary Table S1). Mutation of Asn 32 or Asn36 abolished activity (<10−4 WT activity) indicating that these two amino acids are essential. Mutation of Lys 39 had little effect indicating that this amino acid is unimportant in spite of the hydrogen bonds it forms with the T:T base pairs.
Thus, across the eight nucleotides in each DNA half-site, PacI makes only eight direct hydrogen bond contacts: six in the major groove (N32 and N36 to adenine bases and K39 to thymine bases) and two more in the minor groove. This represents a radical departure from the usual strategy of restriction endonucleases which, under strong selective pressure for absolute cleavage fidelity, usually make more direct contacts than are strictly necessary for high fidelity sequence recognition.
Discussion
Diversity of site-specific HNH endonuclease scaffolds
The overall organization of PacI is superficially similar to the HNH restriction endonuclease Hpy99I (Sokolowska et al., 2009), and more distantly related to the HNH (His-Cys box) homing endonuclease I-PpoI (Flick et al., 1998). All are homodimers containing one ββα-metal motif and two bound zinc ions per subunit. However, close examination of these three enzymes, which recognize target sites ranging from 5 base pairs to 14 base pairs in length, indicates that their folded structures, as well as their DNA binding modes and recognition mechanisms, differ significantly (Figure 2). Whereas the core of the Hpy99I protein forms a structure that encircles and binds almost orthogonally across and around its target site (with the helices from the catalytic site ββα-metal motif aligned almost perpendicular with the DNA duplex axis), PacI displays an elongated fold that associates with one face of the DNA target, with the two subunits and the ββα-metal motif aligned nearly parallel to the DNA duplex. The structure of the I-PpoI homing endonuclease is even more divergent: that protein relies upon extended β-sheet structures for the completion of the core protein fold and for formation of its DNA binding surface. Based on these observations, it seems likely that these site-specific HNH endonucleases are distantly related, but probably all descended from a common ββα-metal ancestor. That predecessor protein may have consisted of a nonspecific endonuclease folded around the common catalytic motif (perhaps resembling modern colicin nucleases).
The details of the active site organization of PacI also indicate a significant divergence from the usual architecture and mechanism that is observed for an HNH active site (Figure 3). The presence of a tyrosine side chain at the position usually occupied by an imidazole base and nucleophilic water, combined with the requirement of the tyrosine phenolic oxygen for catalysis, indicates that this side chain might act as a direct nucleophile in DNA strand cleavage (although a covalently trapped phosphotyrosyl intermediate has not been observed in either of the structures determined in this study, and a role as a general base in more traditional mechanism involving water-mediated hydrolysis cannot be ruled out). While such a mechanism has not been observed previously for a restriction endonuclease, the BfiI enzyme is a member of the phospholipase D family of nucleases, that includes many enzymes that proceed via a phosphotyrosyl covalent intermediate, and is known to form a phospho-histidyl covalent intermediate during strand cleavage (Sasnauskas et al., 2010).
DNA binding and perturbation
The appropriate balance of specificity, fidelity and affinity for protein-DNA interactions is one of the most fundamental of biological requirements. Restriction endonucleases reside at one end of the spectrum of possible DNA recognition behaviors: they cleave relatively short DNA sequences that usually occur frequently within both the host genome and invasive DNA sequences , and display extremely high fidelity that spares the host from off-target cleavage (Pingoud et al., 2005).
Protein-DNA recognition specificity is thought to depend upon a combination of direct readout of the nucleotide bases through contacts between the protein and the DNA that can be direct and/or water-mediated, and the additional 'indirect' exploitation of DNA conformational preferences by inducing a DNA structural perturbation or bend that is favored by a limited number of possible DNA sequences (Jones et al., 1999; Luscombe et al., 2001; vonHippel, 2007). Direct readout of DNA sequences is most effective within the major groove which is physically accessible and also provides chemically distinct combinations of hydrogen-bond partners from the four possible base pairs.
However, many specific DNA binding proteins augment these contacts with additional interactions made within the minor groove (completely encircling the DNA), and in some cases can achieve specificity entirely via interactions within and across the minor groove (Bewley et al., 1998; Rohs et al., 2009). DNA-binding proteins that contact minor groove structural elements may rely on a combination of DNA bending and surface complementarity (for example, as displayed by the TATA binding protein) (Kim et al., 1993) and may also read out local sequence-dependent shape and charge characteristics of the minor groove (as observed for a variety of DNA-binding proteins, including the nucleosome core particle and the Drosophila Hox protein SCR) (Rohs et al., 2009). In these examples, the DNA is dramatically deformed, but the canonical Watson-Crick base pairing of the complementary strands is still preserved.
Because of extreme pressure to maintain high fidelity of recognition, restriction endonucleases are notable for their propensity to fully exploit multiple avenues of DNA readout and specificity (a behavior that can be termed 'recognition overkill'). For example, the MunI restriction endonuclease (a PD‥(D/E)×K enzyme which recognizes the six base pair sequence 5' - GTTAAC - 3') establishes 16 direct hydrogen bonds to these bases in the major groove, 10 direct contacts to phosphates, and it induces a significant distortion between the central base pairs in the sequence (Deibert et al., 1999). That protein displays approximately four direct contacts per base pair--an accomplishment that is facilitated by its core fold, in which the catalytic sites are surrounded by a densely packed array of polar side chains that can fully read out the DNA target's sequence and its shape.
In contrast, PacI is one of the smallest known restriction endonucleases, yet it recognizes a longer (eight basepair) target site while being folded around nonspecific catalytic motif that primarily interacts with the phosphate backbone. Evidently, PacI achieves a similarly high level of recognition-fidelity while forming far fewer hydrogen bonds to the nucleotides. Such a minimal protein-DNA interface—in which less than 50% of potential hydrogen bond partners within the target's major groove are engaged in direct contacts with the protein— would typically be expected to correspond to greatly reduced fidelity of recognition (Chevalier et al., 2003).
The rearrangement of the DNA conformation and its interstrand base pair contacts may represent a mechanism that significantly increases specificity of recognition by PacI, without requiring an investment by the enzyme in a large number of base-specific contacts. It is known that the act of unstacking and/or unpairing consecutive base pairs can result in unfavorable increases in free energy of binding. Computational and direct biophysical analyses indicate that this energetic cost can differ by several kcal/mol per base step, depending on the sequence context of the bases involved (Delcourt and Blake, 1991; Hobza and Sponer, 2002). The sequestration of individual bases into unpaired conformations in the PacI complex and the partial unstacking of flanking base pairs, may therefore greatly favor the correct target sequence for binding and cleavage over closely related DNA sequences
While no sequence-specific DNA binding proteins or endonucleases have displayed the extreme basepair deformation and reorganization displayed by PacI, some restriction endonudleases have been observed to unpair and 'flip out' individual bases, while also greatly distorting the DNA backbone conformation. For example, the Ecl18kI enzyme flips the adenine and thymine bases from each strand of its cognate 5'-CCNGG-3' target site and sequesters both into protein binding pockets, as part of a mechanism that dramatically kinks the DNA and greatly reduces the value for the rise between the flanking inner C:G basepairs, thus decreasing the distance between scissile phosphates by several angstroms (Bochtler et al., 2006). A similar deformation is seen for the PspGI restriction endonuclease (Szczepanowski et al., 2008), and is presumably a common feature of many such REases that utilize base-flipping as part of their recognition mechanism.
While the mechanism of nucleic acid recognition displayed by the PacI endonuclease appears very extreme as compared to most sequence-specific DNA-binding proteins , the distortion of the substrate and the contacts formed by the protein are in fact quite similar to the pattern of RNA recognition exhibited by archaeosine tRNA-guanine transglycosylase, which modifies a guanine base in the 'D arm' of its tRNA substrate. That enzyme disrupts all of the normal basepair and tertiary interactions in the tRNA D arm, leading to reorganization of the tRNA helical strucure and association of the G15 base with the enzyme active site (Ishitani et al., 2003).
Initial cognate site recognition
Finally, the observation of such a dramatic reorganization of the PacI target site, involving removal of each base from its complementary partner, begs the question of how the initial moment of cognate site recognition is related to the subsequent formation of the catalytic enzyme-substrate (ES) complex that is visualized in typical enzyme-DNA co crystal structures. A long history of biophysical studies of protein-DNA recognition (recently revisited and reviewed in (Halford, 2009)) indicates that DNA-binding proteins sample potential DNA binding sites by rapidly associating and dissociating from non-cognate DNA sequences (a process greatly accelerated by non-specific orientation and interaction between the oppositely charged molecules), while also sliding back and forth across regions covering approximately 50 base pairs around each initial 'landing site' in a limited 1-dimensional search of nearby DNA sequences.
It is generally assumed that the contacts made within the initial encounter complex between a specific DNA-binding protein and its correct cognate target site are similar to those found in the enzyme-substrate complex, with additional conformational changes driven by the binding energy derived in the initial encounter with the cognate site. In this model, additional specificity of recognition, beyond that which is engendered by the contacts made between protein and DNA bases, can be derived by sequence-specific conformational preferences of the DNA. This model also allows for the possibility that the unbound sequence of a cognate DNA target site might be predisposed to physically sample a conformation that is similar to its final bound state, which would also enhance recognition and high affinity binding.
However, the structure of the PacI endonuclease in complex with its cognate target site indicates that for this enzyme, and perhaps for other highly specific DNA binding proteins, the structure of the initial specific encounter complex might differ significantly from the subsequent biologically or catalytically active complex. Its seems unlikely that the 5' - TTAATTAA- 3' sequence recognized by PacI is predisposed to sample a conformation, in the absence of bound protein, in which several bases are completely unpaired from their Watson-Crick partners and the flanking base pairs are significantly unstacked. When examining the current collection of crystallographic structures of protein-DNA complexes, many examples can be found where the number of observable contacts between protein and DNA bases do not obviously correspond to the actual specificity of the binding interaction. While such observations may be explained at least in part by the contribution of indirect readout to affinity and specificity, it may be that some specific DNA-protein binding events may be driven by the initial, transient formation of atomic contacts in the cognate complex that are difficult to visualize using traditional crystallographic methods, and are then significantly rearranged to produce the final catalytically or biologically active state.
Experimental Procedures
A detailed description of materials and methods is provided in Supplementary Information. Briefly, the gene encoding PacI was isolated from Pseudomonas alcaligenes chromosomal DNA and re-introduced into P.alcaligenes on a plasmid vector, resulting in a 48-fold increase in endonuclease expression. A 100-L culture was grown of this over expressing strain, from which 57 mg of homogeneous PacI was purified by FPLC column chromatography. The specific activity, monitored by conventional DNA-digestion and agarose gel electrophoresis, was approximately 6×105 units per mg. Crystals of the protein-DNA complex, using a synthetic 18 base pair DNA duplex corresponding to sequence 5' - GAGGCTTAATTAAGCCGC - 3' and a complementary bottom strand were grown by hanging drop geometry against a crystallization buffer containing 18 to 22% polyethylene glycol 3000 (PEG3K) 100 mM sodium citrate, pH 5.5 and either10 mM MgCl2 or 2 mM CaCl2. The structure of the complex in the presence of magnesium was determined using a combination of the multiple isomorphous replacement (MIR) and single anomalous dispersion (SAD) methods, using five independently generated heavy atom derivatives (two separate PtCl4 soaks, and one each of HgCN2, PIP and WO4). In-house wild-type and heavy atom MIR datasets, using a rotating anode generator, extended to approximately 2.6 Å resolution. In addition, a single SAD dataset (extending to 1.9 Å resolution) from a platinum-soaked was collected at the Advanced Light Source using beamline 5.0.2. The combination of in-house and synchrotron data was used to determine and refine the structure of the magnesium-bound, cleaved product complex. Subsequently, a second 2.0 Å resolution dataset of an unsoaked, wild-type crystal in the presence of calcium was also collected and refined, yielding a corresponding model of the uncleaved protein-DNA complex. Data and refinement statistics are provided in Table 1.
Coordinates and Data Deposition
The X-ray structure factor amplitudes and corresponding refined coordinates for the PacI/DNA complex, in the form of calcium-bound uncleaved DNA and magnesium-bound cleaved DNA structures, have been deposited in the RCSB database for immediate release (PDB ID code 3LDY and 3M7K). Requests for the PacI-overexpression clone should be direct to New England Biolabs (xus@neb.com).
Supplementary Material
Acknowledgements
X-ray data was collected at the Advanced Light Source (ALS) synchrotron facility at the Lawrence Berkeley National Laboratory (University of California) on beamline 5.0.2 with the assistance of ALS staff. We thank members of the laboratories of Roland Strong and Adrian Ferre-D'Amare for advice and assistance during structure determination. This work was supported by funding from the NIH to BLS (R01 GM49857) and by funding from the Fred Hutchinson Cancer Center to the Program in Structural Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Gronenborn AM, Clore gM. Minor groove-binding architectural proteins: Structure, function and DNA recognition. Ann Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. Embo J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the "midnight zone" of homology. Curr Protein Pept Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J Mol Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- Cymerman IA, Obarska A, Skowronek KJ, Lubys A, Bujnicki JM. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006;65:867–876. doi: 10.1002/prot.21156. [DOI] [PubMed] [Google Scholar]
- Deibert M, Grazulis S, Janulaitis A, Siksnys V, Huber R. Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 Å resolution. EMBO Journal. 1999;18:5805–5816. doi: 10.1093/emboj/18.21.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. The PYMOL molecular graphics system. San Carlos CA: DeLano Scientific; 2002. [Google Scholar]
- Delcourt SG, Blake RD. Stacking energies in DNA. J Biol Chem. 1991;266:15160–15169. [PubMed] [Google Scholar]
- Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastberg JH, Eklund J, Monnat R, Jr, Stoddard BL. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007;46:7215–7225. doi: 10.1021/bi700418d. [DOI] [PubMed] [Google Scholar]
- Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc Natl Acad Sci U S A. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- Hobza P, Sponer J. Toward true DNA base-stacking energies: MP2, CCSD(T), and complete basis set calculations. J Am Chem Soc. 2002;124:11802–11808. doi: 10.1021/ja026759n. [DOI] [PubMed] [Google Scholar]
- Ibryashkina EM, Zakharova MV, Baskunov vB, Bogdanova ES, Nagornykh MO, Denmukhamedov MM, Melnik BS, Kolinski A, Gront D, Feder M, et al. Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct Biol. 2007;7:48–56. doi: 10.1186/1472-6807-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- Jakubauskas A, Giedriene J, Bujnicki JM, Janulaitis A. Identification of a single HNH active site in type IIS restriction endonuclease Eco31I. J Mol Biol. 2007;370:157–169. doi: 10.1016/j.jmb.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Heyningen Pv, Berman HM, Thornton JM. Protein-DNA interactions: a structural analysis. J Mol Biol. 1999;287 doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- Kim JL, Nikolov DB, Burley SK. Co-crystal struture of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Letters. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Katta K, Krishnaswamy S. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Science. 2004;13:295–300. doi: 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Watanabe M, Kosinski J, Ishikawa K, Kamo M, Sawasaki T, Nagata K, Bujnicki JM, Endo Y, Tanokura M, Kobayashi I. Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 2007;35:1908–1918. doi: 10.1093/nar/gkm091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WK, Esguerra M, Xin Y, Lu XJ. New information content in RNA base pairing deduced from quantitative analysis of high-resolution structures. Methods. 2009;47:177–186. doi: 10.1016/j.ymeth.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:1–13. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang BQ, Schildkraut I. NotI and SfiI: restriction endonucleases with octanucleotide recognition sequences. Methods Enzymol. 1987;155:15–21. doi: 10.1016/0076-6879(87)55005-4. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasnauskas G, Zakrys L, Zaremba M, Cosstick R, Gaynor JW, Halford SE, Siksnys V. A novel mechanism for the scission of double-stranded DNA: BfiI cuts both 3'–5' and 5' –3' strands by rotating a single active site. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp1194. Advance Access published January 4, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowska M, Czapinska H, Bochtler M. Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009;37:3799–3810. doi: 10.1093/nar/gkp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonuclease structure and function. Quarterly Reviews of Biophysics. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Szczepanowski RH, Carpenter MA, Czapinska H, Zaremba M, Tamulaitis G, Siksnys V, Bhagwat AS, Bochtler M. Central base pair flipping and discrimination by PspGI. Nucleic Acids Res. 2008;36:6109–6117. doi: 10.1093/nar/gkn622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisvila R, Morgan RD, Posfai J, Raleigh EA. Discovery and distribution of super-integrons among pseudomonads. Mol Microbiol. 2001;42:587–601. doi: 10.1046/j.1365-2958.2001.02604.x. [DOI] [PubMed] [Google Scholar]
- vonHippel PH. From "Simple" DNA-Protein interactions to the macromolecular machines of gene expression. Ann Rev Biophys Biomol Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Lu XJ, Olson WK. Web 3DNA--a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.