Abstract
Williams–Beuren syndrome (WBS) is caused by a ~1.5 million base pair deletion at 7q11.23. A common inversion of the region, WBSinv-1, exists as a polymorphism but was also found in individuals with WBS-like features but no deletion, suggesting it could cause clinical symptoms. We performed a full clinical, developmental and genetic assessment of two previously reported individuals with clinical symptoms and WBSinv-1 but no 7q11.23 deletion. We also examined expression of genes at 7q11.23 in individuals in the general population who have WBSinv-1. We show that individuals with clinical symptoms and WBSinv-1 do not show significant clinical or psychological overlap with individuals with WBS. In addition, a 1.3 Mb duplication of part of the velocardiofacial syndrome region on chromosome 22q11.2 was found in one participant with WBSinv-1 and clinical symptoms. We also demonstrate that individuals with WBSinv-1 show normal expression of genes from the WBS region. These results suggest that WBSinv-1 does not cause clinical symptoms and we advise caution when diagnosing individuals with atypical presentation of rare syndromes. Whole genome analysis may reveal previously unidentified copy number variants that could contribute to syndromic features.
Keywords: Williams syndrome, chromosome inversion, genetic polymorphism, gene expression, copy number variant
INTRODUCTION
Williams–Beuren syndrome (WBS; OMIM #194050) is a multisystem developmental disorder caused by the hemizygous deletion of ~26 genes on chromosome 7q11.23 [Scherer and Osborne, 2006]. This region of chromosome 7 frequently undergoes genomic rearrangement due to the presence of low copy repeats (LCRs) that promote non-allelic homologous recombination during meiosis [Bayés et al., 2003]. Along with the 1.55 Mb deletion of 7q11.23 that results in WBS, duplication [Somerville et al., 2005; Osborne and Mervis, 2007] and inversion [Osborne et al., 2001; Bayés et al., 2003; Scherer et al., 2003] of the region have also been observed.
The classic clinical features of WBS are varied [Morris et al., 1988; Pober and Dykens, 1996; Morris, 2006a] (see Table I) but perhaps the most intriguing aspect of WBS is the unique cognitive and behavioral profile. Individuals with WBS usually have mild to moderate intellectual disability or learning disabilities with a mean composite IQ on the Kaufman Brief Intelligence Test of 69.32 and a standard deviation of 15.36 [Mervis and Becerra, 2006]. The WBS profile is characterized by relative strengths in verbal short term memory and language, alongside severe weakness in visuospatial construction [Mervis et al., 2000]. Approximately 65% of individuals with WBS have Attention Deficit-Hyperactivity Disorder (ADHD) and there is a high incidence of anxiety, especially specific phobia, combined with over-friendliness [Bellugi et al., 1990; Klein-Tasman and Mervis, 2003; Leyfer et al., 2006].
TABLE I.
Clinical and Neurobehavioral Features of Individuals With Williams–Beuren Syndrome and of Participants 1 and 2
| Williams–Beuren syndrome | Participant 1 | Participant 2 |
|---|---|---|
| Facies | ||
| Broad forehead | Normal forehead | Microcephaly |
| Bitemporal narrowing | Normal bitemporal area | Microcephaly |
| Low nasal root and bulbous nasal tip | Normal nose | Broad nose |
| Prominent earlobes | Low set and posteriorly rotated ears | Ears normally placed and formed |
| Periorbital fullness | Normal periorbital area | Normal periorbital area |
| Stellate iris | Normal iris | Normal iris |
| Malar flattening but full cheeks | Mild malar hypoplasia | Normal mala |
| Long philtrum | Normal philtrum | Normal philtrum |
| Full lips and wide mouth | Wide mouth (2 SD >mean) | Small mouth (25th centile) |
| Small jaw | Prominent jaw | Normal jaw |
| Small, widely spaced teeth | Normal sized teeth | Normal sized teeth |
| Normal palate | Normal palate | High, arched palate |
| Other physical | ||
| Low birth weight | Normal birth weight | Normal birth weight |
| Growth retardation | Normal growth | Growth retardation |
| Kidney and bladder abnormalities | Renal ultrasound normal | Incontinence |
| Kyphosis, lordosis, joint contractures, radio-ulnar synostosis | Lordosis, joint contractures | Joint contractures, bilateral hallux valgus. Thin tapered fingers and bilateral short fifth fingers |
| Radio-ulnar synostosis of the left elbow | ||
| Ectrodactyly of both feet | ||
| Hypercalcemia | Not tested | Not tested |
| Ocular problems: strabismus, hyperopia | Strabismus, myopia | Strabismus |
| Cardiovascular problems: SVAS | Normal echocardiogram | None reported |
| Cognitive abilities | ||
| Mild intellectual disability | Mild intellectual disability | Mild intellectual disability |
| Weakness in spatial skills and math | Relative strength in spatial skills and math | Relative strength in spatial skills |
| Relative strength in language | Relative weakness in verbal skills | Relative weakness in verbal skills |
| Behavior | ||
| Excessively social | Normal social interaction | Normal social interaction |
| Attention deficit hyperactivity disorder | No hyperactivity | No hyperactivity |
| Mild attention problems | Mild attention problems | |
| Hypersensitivity to sound and specific phobia of loud noises | Normal response to loud noises | Normal response to loud noises |
SD, standard deviation; SVAS, supravalvular aortic stenosis.
Due to the mechanism of unequal meiotic recombination, the vast majority of deletions of 7q11.23 span the same interval [Bayés et al., 2003]. There are, however, a few individuals with smaller deletions of the region, whose phenotypic features vary from isolated SVAS to classic WBS. The careful molecular and clinical examination of these individuals can help to correlate genotype and phenotype, with the aim of linking specific genes to clinical or cognitive/behavioral features of WBS [Morris, 2006b]. Other genomic rearrangements of the region may also aid in the identification of causative genes for WBS, particularly inversions since they could directly disrupt genes at the breakpoints. One such inversion, WBSinv-1, was initially identified in the parents of individuals with WBS [Osborne et al., 2001; Bayés et al., 2003; Hobart et al., 2004], and also in several individuals with WBS-like features but no deletion of 7q11.23 [Osborne et al., 2001]. WBSinv-1 has been shown to be present in between 25% and 33% of transmitting parents in WBS families, and in ~5% of the general population [Osborne et al., 2001; Bayés et al., 2003; Hobart et al., 2004; Scherer et al., 2005]. This suggests that WBSinv-1 is a predisposing chromosome rearrangement, increasing the chance of further unequal meiotic recombination in the germ cells of individuals in the general population who have this inversion [Hobart et al., 2004; Scherer et al., 2005].
The identification of WBSinv-1 in three individuals with WBS-like symptoms provided a means by which genes within the WBS interval could be disrupted without actually being deleted, either by direct interruption by the inversion breakpoints, or by alteration of gene expression due to re-location of regulatory elements such as enhancers or repressors. Indeed, one individual (Participant 1 in the current study) exhibits ectrodactyly due the presence of a 24 Mb inversion that disrupts the 7q21.3 region previously associated with split hand/foot malformation (SHFM) [Scherer et al., 1994]. However, the presence of the WBSinv-1 chromosome in unaffected parents, and in the general population, suggests either that the inversion is not fully penetrant or that WBSinv-1 is completely unrelated to the manifestation of clinical symptoms and the identification of WBSinv-1 in the individuals with WBS-like symptoms was coincidental.
To determine if there is any potentially pathogenic effect of the WBSinv-1 inversion, we have conducted a detailed clinical, developmental and genetic assessment of two individuals with WBSinv-1 and clinical symptoms reported in Osborne et al. [2001] and examined the expression of genes from the common WBS deletion region in individuals carrying the WBSinv-1 chromosome.
MATERIALS AND METHODS
Participants
Participants 1 and 2 were described previously as showing some features of WBS [Osborne et al., 2001]. The third individual originally reported was deceased. In the current study, a detailed clinical and developmental examination of each individual was performed by an experienced dysmorphologist (CAM) and developmental psychologist (CBM) who have extensive experience with WBS, and a detailed family and medical history was taken. Immediate family members were also examined for features of WBS.
The following developmental assessments were carried out: Wechsler Abbreviated Scale of Intelligence (WASI) [Wechsler, 1999], Differential Ability Scales (DAS) [Elliott, 1990], and Scales of Independent Behavior-Revised (SIB-R) [Bruininks et al., 1996]. The WASI is a standardized measure of intelligence that includes four subtests (verbal: vocabulary, similarities; performance: block design, matrices) and yields a verbal IQ, performance IQ, and full-scale IQ. This measure was used because it was appropriate for the full age range of individuals in the participants’ families (6–60 years). The Williams Syndrome Cognitive Profile (WSCP) [Mervis et al., 2000] that CBM developed and tested is based on an individual’s performance on the DAS, which measures intellectual ability. The DAS also includes screening tests for academic achievement. The SIB-R is a standardized measure of adaptive and maladaptive behavior. Four subscales of adaptive behavior (motor skills, social interaction and communication skills, personal living skills, community living skills) and three subscales of maladaptive behavior (internalized, asocial, externalized) are included.
All other study participants were from families that included a child with WBS. All participants were enrolled in studies approved by the Research Ethics Boards of the University of Toronto, the University of Louisville and the University of Nevada. Informed consent was obtained before any clinical, psychological or genetic studies were performed.
Inversion Testing
Three-color interphase fluorescence in situ hybridization (FISH) analysis was performed on both blood and transformed lymphoblastoid cell lines from each participant, according to previously described protocols [Osborne et al., 2006] using two probes located within the commonly deleted region, (RP5-1186P10 at the GTF2IRD1 locus and CTA-208H19 at the FZD9 locus) and one probe located telomeric to the WBS deleted region (CTB-139P11 at the HIP1 locus).
Expression Analysis
Expression analysis of genes from 7q11.23 was carried out as described previously, using total RNA extracted from transformed lymphoblast cell lines [Somerville et al., 2005]. Primer sequences are available on LROs laboratory web pages (http://www.utoronto.ca/osborne/). Real-time PCR experiments were normalized using hydroxymethylbilane synthase (HMBS), hypoxanthine-guanine phosphoribosyltransferase (HPRT), and TATA binding protein (TBP) as reference genes. Comparative expression ratios were calculated by dividing the averaged normalized values for each of the test genes by the normalized test gene values for the control group. All samples were run in triplicate and the experiment was repeated twice with consistent results. Comparative expression ratios for the WBSinv-1 and WBS deletion groups are expressed as a ratio of a normalized expression level of the test group (WBSinv-1 group, eight individuals from the general population who had one WBSinv-1 chromosome; WBS group, five individuals carrying the common 1.5 Mb deletion of 7q11.23) relative to the control group (eight individuals who tested negative for the WBSinv-1 chromosome). Pair-wise statistical comparison was performed using a two-tailed student t-test to look for differences in expression of each gene in the test groups relative to the control group. Probabilities of P <0.05 were considered significant.
Copy Number Variation Analysis
Copy number variation (CNV) analysis was performed on Participant 1 and Participant 2 using SNP array analysis. Each DNA sample was genotyped with the Affymetrix GeneChip® Human Mapping NspI Array (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer’s instructions. The NspI Array scans were analyzed using dChip 2006 software (DNA Chip Analyzer)[Li and Wong, 2001] and copy number analysis performed essentially as described previously [Zhao et al., 2004, 2005]. The CNVs identified in each DNA sample were then compared with previously documented CNVs using the Database of Genomic Variants, a curated catalogue of structural variations in the human genome [Iafrate et al., 2004]. The CNV detected on chromosome 22q11.2 was confirmed using quantitative real-time PCR with primers located within the SERPIND1 and YPEL1 genes. Real-time PCR was carried out using a 7900HT genetic analyzer (Applied Biosystems, Foster City, CA) with 11 μl reactions, performed in triplicate, containing 5 ng of template for 40 cycles of amplification using Power® SYBR master mix (Applied Biosystems). The DNA copy number of each gene was obtained from a calibration curve that assumes the reference genome is diploid. Genomic ratios were determined by comparing absolute copy number of the two test genes to the reference gene, HMBS. Primer sequences were as follows: SERPD1e2-F 5′-CGGATCCAGCGTCTTAACAT-3′, SERPD1e2-R 5′-CCAACGGGTGCTATGAAGAT-3′, YPELe2-F 5′-GTC-CCAGCTGTGTGGACAGT-3′, YP-ELe2-R 5′-GCTGGC-CTCTCTGACAAAAG-3′.
RESULTS
Participant 1 Clinical Assessment
Medical and family history
Participant 1 was a female, delivered at term and her medical problems are summarized in Table I. Developmentally, she walked between 16 and 18 months, said her first words at age 1 year, and said sentences at age 3 years. A five-generation family history did not show any symptoms common to people with WBS, except for the occurrence of inguinal hernias in a maternal uncle. Her half sister, mother, and both maternal grandparents were examined and had no dysmorphic features.
Physical examination
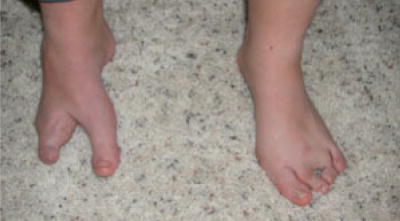
At examination, Participant 1 was 17 years of age. Her head circumference was at the 40th centile, and her cranial shape was dolichocephalic but her facial measurements were normal with the exception of the mouth width (Fig. 1). She had bilateral epicanthal folds and downslanting palpebral fissures. Her neck was mildly webbed and there was a low posterior hairline, although her hair pattern was normal. She had sloping shoulders. There was a tight heel cord on the right and her right leg was smaller than the left. She had bilateral ectrodactyly of the feet and her hands measured at the 70th centile (Fig. 2). A summary of her clinical presentation can be found in Table I.
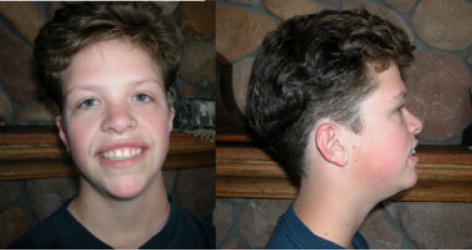
Fig. 1.
Participant 1 at age 17 years. Note prominent jaw, bilateral epicanthal folds, down slanting palpebral fissures and wide mouth with normal nose and philtrum. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Fig. 2.
Feet of participant 1. Note bilateral ectrodactyly. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Participant 1 did not meet the clinical criteria for WBS. She had some features that are not seen in WBS; specifically, down-slanted palpebral fissures, webbed neck, prominent jaw, down-turned corners of the mouth, and ectrodactyly. The features that she does have that can also be seen in WBS include 2 of 17 WBS facial features scored (WBS have >9): strabismus, and a wide mouth with bowed upper lip [Mervis and Morris, 2007]. She also had radioulnar synostosis, sloping shoulders, lordosis, and joint contractures (likely related to her ectrodactyly and leg length discrepancy).
Participant 2 Clinical Assessment
Medical and family history
Participant 2 was a female, delivered at term with initial respiratory distress. She had delayed motor development and was diagnosed with cerebral palsy (static encephalopathy). When she was evaluated for developmental delay at the age of 2 years, her head circumference was 43 cm, which was <2nd centile. At age 2.5 years, Participant 2 was noted to have increased tone in her lower extremities and a wide based gait. She had a past history of a seizure disorder, which resolved by the age of 12 years. She also had a history of chronic otitis media as a young child. She has had normal chromosome studies and a negative DNA test for Fragile X. At the age of 12 years, she was diagnosed with a growth hormone deficiency and had a positive result with growth hormone therapy. She has migraine headaches. A four-generation family history did not reveal any symptoms common to people with WBS. Participant 2’s older sister had Graves disease. Both her parents and her sister were examined and none had dysmorphic features.
Physical examination
When Participant 2 was examined at age 22 years, her height and weight were at the 5th centile, and her head circumference was 51.5 cm, which is <3rd centile. She had a low anterior hairline, mild upslanting of the palpebral fissures and hypotelorism with inner canthal distance, inter pupillary distance and outer canthal distance all <3rd centile. A summary of her clinical presentation can be found in Table I.
Participant 2 did not have any physical features that are typically associated with WBS. She had only 1 of 17 scored facial features for WBS (strabismus) [Mervis and Morris, 2007]. In her case, the joint contractures were related to her static encephalopathy and microcephaly.
Participants 1 and 2 Developmental Assessment
A summary of standard scores on intellectual and adaptive behavior assessments for Participants 1 and 2 is presented in Table II. Participant 1’s full-scale IQ was 0.43 SD below and Participant 2’s full-scale IQ was 1.07 SD below the mean for a group of 28 adolescents and young adults with WBS (CBM unpublished data). Both participants’ highest score on the four subtests was for Block Design, the subtest on which individuals with WBS typically have the most difficulty. Both maternal report, and the results of the DAS achievement screening tests indicate that Participant 1’s math skills are more advanced than her reading skills. In contrast, most people with WBS perform considerably better on reading than on math. Participant 2’s academic achievement was not tested; however, she showed an aptitude for remembering birth dates, including year of birth, and people’s ages. On one occasion, she corrected her mother regarding the age of an adult cousin; her mother checked and later confirmed the accuracy of Participant 1s correction. In contrast, individuals with WBS typically do not know the ages of their siblings (never mind their other relatives), and almost never know the year in which these people were born.
TABLE II.
Standard Scores on Intellectual and Adaptive Behavior Assessments for Participants 1 and 2 and for Adolescents and Young Adults With Williams–Beuren Syndrome
| Population mean ± SD | WBS mean ± SD | Participant 1 | Participant 2 | |
|---|---|---|---|---|
| WASIa | ||||
| Verbal IQ | 100 ± 15 | 71.9 ± 13.2 | 64 | 55 |
| Performance IQ | 100 ± 15 | 67.5 ± 12.7 | 61 | 58 |
| Full-scale IQ | 100 ± 15 | 67.6 ± 12.7 | 59 | 53 |
| DASb | ||||
| Pattern construction | 50 ± 10 | 23.2 ± 5.3 | 26 | 21 |
| Definitions | 50 ± 10 | 29.7 ± 8.8 | 29 | 20 |
| Similarities | 50 ± 10 | 30.1 ± 10.8 | 35 | 20 |
| Digit recall | 50 ± 10 | 34.6 ± 10.2 | 22 | 26 |
| Mean T (six core subtests) | 50 ± 10 | 28.3 ± 6.3 | 25.5 | 20.2 |
| SIB-Rc | ||||
| Adaptive behavior | ||||
| Motor skills | 100 ± 15 | 48.8 ± 13.1 | 48 | 27 |
| Social interaction and communication skills | 100 ± 15 | 70.6 ± 11.5 | 64 | 56 |
| Personal living skills | 100 ± 15 | 59.2 ± 11.7 | 69 | 38 |
| Community living skills | 100 ± 15 | 47.4 ± 14.2 | 57 | 16 |
| Broad independence | 100 ± 15 | 47.3 ± 11.5 | 52 | 23 |
| Maladaptive behavior | ||||
| Internalized | 0 ± 10 | −8.9 ± 8.6 | −17 | −3 |
| Asocial | 0 ± 10 | −9.2 ± 10.8 | 4 | 2 |
| Externalized | 0 ± 10 | 0.3 ± 6.4 | 1 | 3 |
| General | 0 ± 10 | −9.3 ± 6.9 | −6 | −1 |
Wechsler abbreviated scale of intelligence [Wechsler, 1999].
Differential ability scales [Elliott, 1990].
Scales of independent behavior-revised [Bruininks et al., 1996].
Neither participant exhibited any attention problems. Both were able to stay on task for the more than 2 hr it took to complete the testing, refusing offers of breaks, although both were reported to have difficulty staying on task in group situations. In contrast, most individuals with WBS have difficulty staying on task even in one-on-one situations. Neither participant showed any of the characteristic behavioral features seen in individuals with WBS. Participant 1 sat quietly while CBM and CAM spoke with her family, spoke only when asked a direct question and did not ask any personal questions. Participant 2 regarded CBM and CAM as strangers, spoke only when appropriate, did not ask any personal questions and stayed on topic during conversations.
Participant 1’s Broad Independence standard on the SIB-R adaptive behavior test [Bruininks et al., 1996] was in the range expected for WBS or any other syndrome associated with mild-to-moderate intellectual disability. Participant 2’s Broad Independence standard score was considerably lower than expected for individuals with WBS. Both participants’ overall maladaptive behavior scores were within the normal range.
To fit the WSCP, a person must meet all four of the following criteria on the Differential Ability Scales (DAS) [Elliott 1990] (met by 89% of individuals with WBS) [Mervis et al., 2000].
T for Digit Recall, Naming/Definitions, or Similarities >1st centile (T on at least one of these subtests ≥ 29)
Pattern Construction T <20th centile
Pattern Construction T <Mean T of the core subtests
Pattern Construction T <Digit Recall T
Participant 1 did not fit the WSCP because her DAS T scores did not fit criteria 3 and 4. Participant 2 did not fit the WSCP because her DAS T scores did not fit criteria 1 and 3. All of the members of both participants’ families who were available for testing had full-scale IQs in the average range, and none of them fit the WSCP.
Genetic Assessment
Inversion testing
Seven members of Participant 1’s family were available for testing using three-color interphase FISH. Her mother, half-sister, grandmother, and one great-aunt were positive for the WBSinv-1. The participant’s aunt, grandfather and one great-aunt were negative for the WBSinv-1. Three of Participant 2’s family members were available for testing using three-color interphase FISH. Her mother, father and sister were all negative for the WBSinv-1.
Expression analysis
Analysis of genes from within the common WBS deletion region showed no significant difference in expression between individuals without WBSinv-1 (n = 8), or the general population WBSinv-1 group (n = 8), with the exception of STX1A, which was significantly elevated in the WBSinv-1 group (P <0.04). In contrast, a group of individuals with the common WBS deletion (n = 5) showed levels of expression reduced to less than 50% for each gene tested as previously reported [Somerville et al., 2005; Merla et al., 2006]. The results of the expression analysis are summarized in Table III.
TABLE III.
Expression Analysis of Genes From the WBS Region in Individuals Who Have WBS or Individuals in the General Population Who Have WBSinv-1
| Chromosome position | Gene | Comparative expression ratio (vs. control group n = 8) mean ± SEM |
||
|---|---|---|---|---|
| Individuals with WBS (n = 5) | Individuals with WBSinv-1 (n = 8) | |||
| 6.5 Mb cen | ASL | 0.606 ± 0.043** | 1.294 ± 0.089* | |
| 6 Mb cen | KCTD7 | 0.561 ± 0.073** | 1.067 ± 0.064 | |
| WBSinv-1 region | WBS common deletion region | BAZ1B | 0.101 ± 0.011** | 0.877 ± 0.049 |
| WBSCR18 | 0.291 ± 0.045** | 1.164 ± 0.093 | ||
| STX1A | 0.244 ± 0.031** | 1.188 ± 0.075* | ||
| LIMK1 | 0.320 ± 0.040** | 1.089 ± 0.041 | ||
| WBSCR1 | 0.450 ± 0.035** | 1.050 ± 0.050 | ||
| RFC2 | 0.324 ± 0.027** | 0.947 ± 0.046 | ||
| CYLN2 | 0.371 ± 0.028** | 0.993 ± 0.041 | ||
| GTF2I | 0.245 ± 0.030** | 1.176 ± 0.088 | ||
| WBSCR16 | 1.604 ± 0.200** | 1.217 ± 0.075* | ||
| Next gene tel | HIP1 | 0.714 ± 0.108 | 1.182 ± 0.209 | |
| 1 Mb tel | POR | 0.311 ± 0.022** | 1.194 ± 0.075 | |
| 1.2 Mb tel | MDH2 | 1.265 ± 0.139 | 1.081 ± 0.116 | |
t-test,
P <0.05,
P <0.001.
Although STX1A expression was statistically elevated in the WBSinv-1 group, the increase was modest (1.188 times the level in individuals without WBSinv-1) and in the opposite direction to the change in expression seen in the group with WBS (0.244 times the level in individuals without WBSinv-1). Several genes outside the WBS region exhibited altered expression in the WBS group, as previously reported [Merla et al., 2006], but did not show a similar decrease in the WBSinv-1 group.
Copy number variation analysis
The results of CNV analysis for Participant 1 showed the presence of three previously identified CNVs on chromosomes 9p24, 9p21, and 22q11.1, and a 1.3 Mb gain at 22q11.22 spanning the region between 19,428,100 and 20,742,400 Mb according to the March 2006 human reference sequence (NCBI Build 36) (Table IV). The 22q11.22 gain partially overlapped both known CNVs and microduplications of the region, but included a 248 kb gain spanning five known genes that did not overlap with CNVs found in control samples (Fig. 2). The chromosome 22 gain in Participant 1 was confirmed using real-time PCR, with ratios of 1.564 (±0.167) and 1.461 (±0.156) for SERPIND1 at the proximal end and YPEL1 at the distal end, respectively. Real-time PCR demonstrated that the CNV was not present in DNA from Participant 1’s mother. Her father’s DNA was not available for analysis. The results of CGH for Participant 2 showed the presence of two CNVs on chromosomes 7p14.3 and 17q21 previously identified in the general population. No other changes in copy number were identified (Table IV).
TABLE IV.
Copy Number Variant Analysis of Participants 1 and 2
| Participant | Cytogenetic band | Size of CNV (bp) | Type | Present in Database of Genomic Variants | Unique gene copy number alteration |
|---|---|---|---|---|---|
| 1 | 9p24.3 | 55,124 | Gain | Yes | — |
| 9p21.1 | 172,600 | Loss | Yes | — | |
| 22q11.1 | 140,300 | Gain | Yes | — | |
| 22q11.21 | 1,314,300 | Gain | Partial overlap | PIK4CA, SERPIND1, SNAP29, CRKL, AIFM3 | |
| 2 | 7p14.3 | 182,000 | Gain | Yes | — |
| 17q21.3 | 631,100 | Gain | Yes | — |
DISCUSSION
Molecular diagnosis of WBS includes testing for hemizygosity at 7q11.23 by FISH using a mixture of probes encompassing the elastin and LIM kinase 1 genes (Vysis, Inc., Des Plains, IL). In more than 95% of cases, there is a defined 1.55 Mb deletion but for the remaining individuals with a clinical diagnosis of WBS, there is no detectable chromosomal rearrangement [Lowery et al., 1995; Mari et al., 1995; Nickerson et al., 1995]. These individuals could constitute phenocopies of WBS with genetic mutations at other loci, or they could also have disruption of key genes at 7q11.23 without an easily detectable deletion. We previously reported on three individuals with WBS-like symptoms according to medical records, but no detectable deletion of 7q11.23 [Osborne et al., 2001]. All three individuals were found to carry an inversion of the WBS region (WBSinv-1), a rearrangement also identified in the parents of some children with WBS.
The breakpoints of the common WBSinv-1 are predicted to lie within the B-block segments of the centromeric and telomeric LCRs that are in an inverted orientation with respect to each other, since these sequences are more than 99.6% nucleotide identical over large stretches and more than 95% of the WBS deletions occur between B-blocks in a direct orientation [Bayés et al., 2003]. The centromeric and telomeric B-blocks do not contain any genes commonly deleted in WBS and because the LCRs have undergone extensive genomic rearrangement during primate evolution [Antonell et al., 2005], they are unlikely to contain key regulatory elements for such genes. Our analysis of the expression of genes from 7q11.23 confirms this prediction, since we found no evidence of significantly altered expression of any of the genes tested in a sample of individuals in the general population who have WBSinv-1 (Table III). Even genes many Mb from the critical region that exhibit altered expression in individuals with the WBS deletion showed normal expression in the WBSinv-1 group, suggesting that the inversion has a negligible effect on the surrounding chromosomal region.
We identified one individual who was homozygous for the WBSinv-1 chromosome. Interestingly, this individual was the parent of a child with WBS, but the child’s deletion originated in the other parent, who was heterozygous for WBSinv-1. Although we were not able to perform a clinical or developmental examination of this individual, he did not report any symptoms of WBS.
Participants 1 and 2, who were originally reported as exhibiting symptoms of WBS based on a review of medical records, did not fit any of the diagnostic criteria for WBS, suggesting that the presence of the WBSinv-1 chromosome and clinical symptoms in these individuals is coincidental. In an attempt to identify other chromosome anomalies that might account for their clinical symptoms, we undertook CNV analysis. We failed to identify any alterations in copy number in Participant 2 that had not been previously reported in the general population, leaving the etiology of her symptoms unknown. CNV analysis of DNA from Participant 1, however, revealed a previously undescribed gain spanning a 1.3 Mb segment within the region that is commonly duplicated in dup(22)(q11.2q11.2) syndrome [Ensenauer et al., 2003; Yobb et al., 2005]. Most of the chromosome 22q11.22 gain seen in this participant is overlapping with CNVs seen in numerous control samples [Locke et al., 2006; Simon-Sanchez et al., 2007; Wong et al., 2007], but, because the gain includes genes not contained within common CNVs, and because it spans at least three distinct CNVs, this genomic variant may contribute to the phenotypic features seen in Participant 1 (Fig. 3).
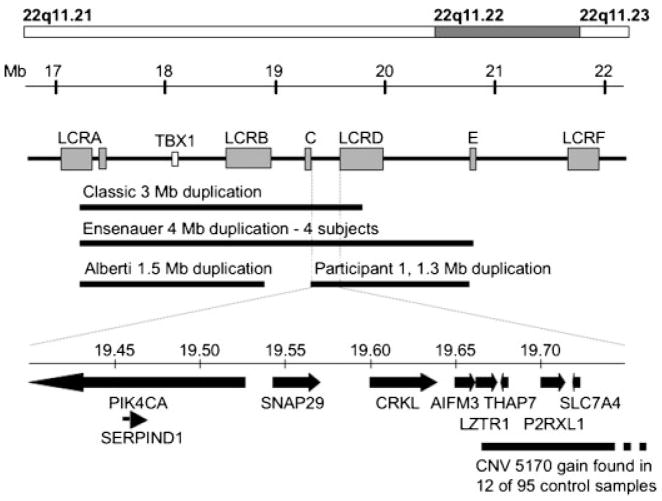
Fig. 3.
Summary of microduplications and copy number variants overlapping with the chromosome 22q11.21 gain identified in Participant 1. The 22q11.21 gain and other published microduplications of the region [Ensenauer et al., 2003; Yobb et al., 2005; Alberti et al., 2007] are shown beneath a schematic of the velocardiofacial syndrome region. Low copy repeats are labeled according to Jalali et al. [2008] and the position of the TBX1 gene is shown. The regions of overlap between the duplication identified in Participant 1, other published microduplications [Ensenauer et al., 2003; Alberti et al., 2007] and previously identified copy number variants (CNVs) [Iafrate et al., 2004] are shown below, with a common duplicated interval expanded below to show the gene content.
The phenotypic presentation of dup(22)(q11.2q11.2) syndrome is variable but there are features that frequently occur in conjunction with the common 3 Mb duplication (velopharyngeal insufficiency, cleft palate, hearing loss, cognitive deficits, motor delay, poor growth, characteristic dysmorphism) [Ensenauer et al., 2003; Portnoi et al., 2005; Yobb et al., 2005]. The pharyngeal malformations are thought to be linked to duplication of TBX1, since hemizygous deletion of Tbx1 in mice causes similar anomalies [Arnold et al., 2006]. Participant 1 does not exhibit the typical phenotype of individuals with dup(22)(q11.2q11.2), but is not duplicated for the region spanning TBX1. She does, however, have some overlapping features with dup(22)(q11.2q11.2), such as bilateral mixed hearing loss, cognitive deficits, mild motor delay, down-slanted palpebral fissures, strabismus and radioulnar synostosis, although this last malformation has also been reported in SHFM, which is a concurrent disorder in this participant [Debeer et al., 2004]. It is possible, therefore, that the dup(22)(q11.2q11.2) syndrome is a contiguous gene duplication disorder and that gene(s) contributing to the features seen in Participant 1 are contained within the 1.3 Mb duplicated segment, most likely in the segment that overlaps with the more common 3 Mb duplication (Fig. 3).
The emergence of comparative genomic hybridization and SNP array analysis as tools for the global analysis of copy number across the genome, has revealed a startling number of variants present in the general population, many of which alter gene copy number and expression [Rodriguez-Revenga et al., 2007]. SNP arrays have recently been used to identify novel CNVs associated with syndromic disorders [Rodriguez-Revenga et al., 2007] and it will be important in the future to examine individuals’ genomes for CNVs that may be contributing to their phenotypic presentation, rather than attributing symptoms to already identified variants.
The two participants discussed in this article were initially reported, based on their medical records, to have features of WBS [Osborne et al., 2001]. In their medical records, both participants were described as having WBS-like facial features, a WBS-like behavior profile and developmental delay. In contrast, our assessment by professionals who have had many years of experience with both individuals who have WBS and children with other developmental disabilities, did not identify any significant overlap between the presentation of these two participants and that of individuals with WBS. These findings underscore the importance of experienced clinical and psychological assessments in cases where a specific diagnosis is suspected, but the presentation is atypical. Obviously this is not always possible, but the development of syndrome-specific matrices for facial features [Hammond et al., 2005], growth parameters [Martin et al., 2007] cognitive or behavioral profiles [Mervis and Klein-Tasman, 2000] or for overall clinical presentation [Sugayama et al., 2007] should help with accurate diagnosis.
Acknowledgments
We thank the participants and their families for their involvement. This work was supported by grants from the Canadian Institutes of Health Research (LRO), Sick Kids Foundation (LRO), Genome Canada/Ontario Genomics Institute (LRO and SWS) and the National Institute of Neurological Disorders and Stroke (R01 NS35102 to CBM and CAM). EJY is supported by the CIHR-University of Toronto Collaborative Graduate Training Program in Molecular Medicine. SWS is a CIHR Investigator and an International Scholar of the Howard Hughes Medical Institute. We thank The Centre for Applied Genomics for technical assistance.
Grant sponsor: Canadian Institutes of Health Research; Grant sponsor: Sick Kids Foundation; Grant sponsor: Genome Canada/Ontario Genomics Institute; Grant sponsor: National Institute of Neurological Disorders and Stroke; Grant number: R01 NS35102.
References
- Alberti A, Romano C, Falco M, Calì F, Schinocca P, Galesi O, Spalletta A, Di Benedetto D, Fichera M. 1.5 Mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet. 2007;71:177–182. doi: 10.1111/j.1399-0004.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- Antonell A, de Luis O, Domingo-Roura X, Perez-Jurado LA. Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23. Genome Res. 2005;15:1179–1188. doi: 10.1101/gr.3944605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JS, Werling U, Braunstein EM, Liao J, Nowotschin S, Edelmann W, Hebert JM, Morrow BE. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development. 2006;133:977–987. doi: 10.1242/dev.02264. [DOI] [PubMed] [Google Scholar]
- Bayés M, Magano LF, Rivera N, Flores R, Pérez-Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Med Genet. 1990;6:115–125. doi: 10.1002/ajmg.1320370621. [DOI] [PubMed] [Google Scholar]
- Bruininks R, Woodcock R, Weatherman R, Hill BK. Scales of independent behavior—Revised. Itasca, IL: Riverside Publishing; 1996. [Google Scholar]
- Debeer P, Vandenbossche L, de Ravel TJ, Desloovere C, De Smet L, Huysmans C, Thoelen R, Vermeesch J, Van de Ven WJ, Fryns JP. Bilateral complete radioulnar synostosis associated with ectrodactyly and sensorineural hearing loss: A variant of SHFM1. Clin Genet. 2004;65:153–155. doi: 10.1111/j.0009-9163.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- Elliott C. Differential ability scales. San Antonio, TX: Psychological Corporation9; 1990. [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, Smith WE, Simon-Fayard E, Alexander AA, Kulharya AS, Ketterling RP, Clark RD, Jalal SM. Micro-duplication 22q11.2, an emerging syndrome: Clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Hutton TJ, Allanson JE, Buxton B, Campbell LE, Clayton-Smith J, Donnai D, Karmiloff-Smith A, Metcalfe K, Murphy KC, Patton M, Pober B, Prescott K, Scambler P, Shaw A, Smith AC, Stevens AF, Temple IK, Hennekam R, Tassabehji M. Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet. 2005;77:999–1010. doi: 10.1086/498396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobart HH, Gregg RG, Mervis CB, Robinson BF, Kimberley KW, Rios CM, Morris CA. Heterozygotes for the micro-inversion of the Williams-Beuren syndrome region have an increased risk for affected offspring. [Abstract 891] Presented at The American Society of Human Genetics; 2004; Toronto, Canada. 2004. http://www.ashg.org/genetics/ashg/annmeet/2004/menu-annmeet-2004.shtml. [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jalali GR, Vorstman JA, Errami A, Vijzelaar R, Biegel J, Shaikh T, Emanuel BS. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2008;29:433–440. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Tasman BP, Mervis CB. Distinctive personality characteristics of 8-, 9-, and 10-year-olds with Williams syndrome. Dev Neuropsychol. 2003;23:269–290. doi: 10.1080/87565641.2003.9651895. [DOI] [PubMed] [Google Scholar]
- Leyfer OT, Woodruff-Borden J, Klein-Tasman BP, Fricke JS, Mervis CB. Prevalence of psychiatric disorders in 4 to 16-year-olds with Williams syndrome. Am J Med Genet Part B Neuropsychiatr Genet. 2006;141B:615–622. doi: 10.1002/ajmg.b.30344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery MC, Morris CA, Ewart A, Brothman LJ, Zhu XL, Leonard CO, Carey JC, Keating M, Brothman AR. Strong correlation of elastin deletions, detected by FISH, with Williams syndrome: Evaluation of 235 patients. Am J Hum Genet. 1995;57:49–53. [PMC free article] [PubMed] [Google Scholar]
- Mari A, Amati F, Mingarelli R, Giannotti A, Sebastio G, Colloridi V, Novelli G, Dallapiccola B. Analysis of the elastin gene in 60 patients with clinical diagnosis of Williams syndrome. Hum Genet. 1995:444–448. doi: 10.1007/BF00191804. [DOI] [PubMed] [Google Scholar]
- Martin ND, Smith WR, Cole TJ, Preece MA. New height, weight and head circumference charts for British children with Williams Syndrome. Arch Dis Child. 2007;92:598–601. doi: 10.1136/adc.2006.107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Howald C, Henrichsen CN, Lyle R, Wyss C, Zabot MT, Antonarakis SE, Reymond A. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervis CB, Becerra AM. Language and communicative development in Williams syndrome. Ment Retard Dev Disabil Res Rev. 2006;13:3–15. doi: 10.1002/mrdd.20140. [DOI] [PubMed] [Google Scholar]
- Mervis CB, Klein-Tasman BP. Williams syndrome: Cognition, personality, and adaptive behavior. Ment Retard Dev Disabil Res Rev. 2000;6:148–158. doi: 10.1002/1098-2779(2000)6:2<148::AID-MRDD10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Mervis CB, Morris CA. Williams syndrome. In: Mazzocco MMM, Ross JL, editors. Neurogenetic developmental disorders: Variation of manifestation in childhood. Cambridge, MA: MIT Press; 2007. pp. 199–262. [Google Scholar]
- Mervis CB, Robinson BF, Bertrand J, Morris CA, Klein-Tasman BP, Armstrong SC. The Williams syndrome cognitive profile. Brain Cogn. 2000;44:604–628. doi: 10.1006/brcg.2000.1232. [DOI] [PubMed] [Google Scholar]
- Morris CA. The dysmorphology, genetics, and natural history of Williams-Beuren syndrome. In: Morris CA, Lenhoff H, Wang P, editors. Williams-Beuren syndrome: Research, evaluation, and treatment. Baltimore, MD: Johns Hopkins University Press; 2006a. pp. 3–17. [Google Scholar]
- Morris CA. Genotype-phenotype correlations in Williams-Beuren syndrome. In: Morris CA, Lenhoff HM, Wang PP, editors. Williams-Beuren syndrome. Baltimore, MD: Johns Hopkins University Press; 2006b. pp. 59–82. [Google Scholar]
- Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: Physical characteristics. J Pediatr. 1988;113:318–326. doi: 10.1016/s0022-3476(88)80272-5. [DOI] [PubMed] [Google Scholar]
- Nickerson E, Greenberg F, Keating MT, McCaskill C, Shaffer LG. Deletions of the elastin gene at 7q11.23 occur in approximately 90% of patients with Williams syndrome. Am J Hum Genet. 1995;56:1156–1161. [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Mervis CB. Rearrangements of the Williams-Beuren syndrome locus: Molecular basis and implications for speech and language development. Expert Rev Mol Med. 2007;9:1–16. doi: 10.1017/S146239940700035X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Joseph-George AM, Scherer SW. Williams-Beuren syndrome diagnosis using fluorescence in situ hybridization. Methods Mol Med. 2006;126:113–128. doi: 10.1385/1-59745-088-X:113. [DOI] [PubMed] [Google Scholar]
- Pober BR, Dykens EM. Williams syndrome: An overview of medical, cognitive, and behavioral features. Child Adolesc Psych Clinics N Am. 1996;5:929–943. [Google Scholar]
- Portnoi MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, Finkel L, Roger G, Ducrocq S, Gold F, Taillemite JL, Marlin S. 22q11.2 duplication syndrome: Two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet Part A. 2005;137A:47–51. doi: 10.1002/ajmg.a.30847. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Revenga L, Mila M, Rosenberg C, Lamb A, Lee C. Structural variation in the human genome: The impact of copy number variants on clinical diagnosis. Genet Med. 2007;9:600–606. doi: 10.1097/gim.0b013e318149e1e3. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Osborne LR. Williams-Beuren syndrome. In: Lupski J, Stankiewicz P, editors. Genomic disorders: The genomic basis of disease. Totowa, NY: Humana Press Inc; 2006. pp. 221–236. [Google Scholar]
- Scherer SW, Poorkaj P, Massa H, Soder S, Allen T, Nunes M, Geshuri D, Wong E, Belloni E, Little S, Zhou L, Becker D, Kere J, Ignatius J, Niikawa N, Fukushima Y, Hasegawa T, Weissenbach J, Boncinelli E, Trask B, Tsui1 L-C, Evans JP. Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Genet. 1994;3:1345–1354. doi: 10.1093/hmg/3.8.1345. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Cheung J, MacDonald JR, Osborne LR, Nakabayashi K, Herbrick JA, Carson AR, Parker-Katiraee L, Skaug J, Khaja R, Zhang J, Hudek AK, Li M, Haddad M, Duggan GE, Fernandez BA, Kanematsu E, Gentles S, Christopoulos CC, Choufani S, Kwasnicka D, Zheng XH, Lai Z, Nusskern D, Zhang Q, Gu Z, Lu F, Zeesman S, Nowaczyk MJ, Teshima I, Chitayat D, Shuman C, Weksberg R, Zackai EH, Grebe TA, Cox SR, Kirkpatrick SJ, Rahman N, Friedman JM, Heng HH, Pelicci PG, Lo-Coco F, Belloni E, Shaffer LG, Pober B, Morton CC, Gusella JF, Bruns GA, Korf BR, Quade BJ, Ligon AH, Ferguson H, Higgins AW, Leach NT, Herrick SR, Lemyre E, Farra CG, Kim HG, Summers AM, Gripp KW, Roberts W, Szatmari P, Winsor EJ, Grzeschik KH, Teebi A, Minassian BA, Kere J, Armengol L, Pujana MA, Estivill X, Wilson MD, Koop BF, Tosi S, Moore GE, Boright AP, Zlotorynski E, Kerem B, Kroisel PM, Petek E, Oscier DG, Mould SJ, Döhner H, Döhner K, Rommens JM, Vincent JB, Venter JC, Li PW, Mural RJ, Adams MD, Tsui LC. Human chromosome 7: DNA sequence and biology. Science. 2003;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Gripp KW, Lucena J, Nicholson L, Bonnefont JP, Pérez-Jurado LA, Osborne LR. Observation of a parental inversion variant in a rare Williams-Beuren syndrome family with two affected children. Hum Genet. 2005;117:383–388. doi: 10.1007/s00439-005-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Pérez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugayama SM, Leone C, Chauffaille MD, Okay TS, Kim CA. Williams Syndrome: Development of a new scoring system for clinical diagnosis. Clinics. 2007;62:159–166. doi: 10.1590/s1807-59322007000200011. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler abbreviated scale of intelligence. San Antonio, TX: Psychological Corporation; 1999. [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, Gallo N, Morrow BE, Shaffer LG, Babcock M, Chernos J, Bernier F, Sprysak K, Christiansen J, Haase S, Elyas B, Lilley M, Bamforth S, McDermid HE. Microduplication and triplication of 22q11.2: A highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li C, Paez JG, Chin K, Jänne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, Gray JW, Sellers WR, Meyerson M. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, Sugarbaker D, Chen F, Rubin MA, Jänne PA, Girard L, Minna J, Christiani D, Li C, Sellers WR, Meyerson M. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]