Abstract
Since 2007, genome-wide association (GWA) studies have identified numerous well-supported, novel genetic risk loci for common cancers; however, there are concerns that this technology is reaching its limits. We provide an overview of GWA-identified genetic associations with solid tumors. We simulated the distribution of population risk alleles for colorectal, prostate, testicular, and thyroid cancers based on genetic variants identified in GWA studies. We also evaluated whether statistical power to detect typical genetic effects could be improved with studies performing GWA analyses of all available samples rather than multistage designs. Fifty-six eligible articles yielded 92 eligible associations between cancer phenotypes and genetic variants with a median per-allele odds ratio (OR) of 1.22 (interquartile range = 1.15–1.36). Half of the associations pertained to prostate, colorectal, or breast cancer. Individuals at the upper quartile of simulated risk had only 2.1- to 4.2-fold higher relative risk than those in the lower quartile. Comprehensive evaluation of currently available samples with GWA platforms would yield few additional variants with per-allele OR = 1.4, but many more variants with OR = 1.2 could be detected; statistical power to detect weak associations (OR = 1.07) would still be negligible. The GWA approach is effective in identifying common genetic variants with moderate effect; however, identifying loci with very small effects and rare variants will require major new efforts. At present, the utility of GWA-identified risk loci in risk stratification for cancer is limited.
Genome-wide association (GWA) studies have led to a paradigm shift in the discovery of gene–disease associations. Since 2007, several hundred GWA studies have been published, including several dozen on cancer phenotypes (1–56). As a result of this research effort, a large number of new gene–disease associations have been discovered pertaining to common genetic variants, with minor allele frequencies typically exceeding 5% in the general population (57–59). These discoveries include many associations with robust statistical support for influencing susceptibility to diverse cancers. However, despite the accumulating interesting findings, there is debate about the exact merits and future of such studies (60–64). Skeptics point out that discoveries of new associations with this expensive technology are not as numerous as originally expected, discovered genetic effects for common variants are small, the ability of the discovered variants to discriminate disease risk is minimal, and the true functional culprits linked to the discovered markers remain largely unknown. If so, GWA studies of common variants may be reaching their limits of discovery, and research efforts should shift to other approaches, such as genome sequencing and rare variant analysis (61). Alternatively, more optimistic investigators point out that the costs of GWA genotyping have decreased steeply over time, many robust associations continue to be discovered, disease risk may be adequately predicted if a large number of such markers are discovered for each disease, and the discovery of novel genetic risk loci creates opportunities for understanding the biological function of these loci and using this knowledge for translational purposes (62).
Whereas new sequencing technologies are already in use and will continue to grow in their applications, it is useful to review what we have found based on GWA studies targeting common genetic variants. An overview of the current evidence can provide a useful perspective on past achievements and future prospects of GWA studies and will help to assess the contribution these discoveries can make toward explaining the genetic risk of diverse malignancies.
In this review, we assembled a compendium of discovered associations with robust statistical support from cancer susceptibility GWA studies (excluding hematological malignancies). We addressed the following questions: Is the pace of new discoveries of associations between common variants and cancer accelerating or decelerating? How strong are the magnitudes of the discovered effects in terms of the genetic risk conferred and the frequency of the risk variants? How extensive might the discrimination of risk be if information from all identified risk variants is used? Finally, is the pattern of discovered effects reflective primarily of statistical power considerations and would it be possible to find many more similar associations if larger studies could be performed with the same platforms?
Compilation and Cleaning of Database of Associations
The review considered all single-nucleotide polymorphisms (SNPs) associated with any cancer, except for hematological malignancies, for which discovery of an association stemmed from a GWA study using an agnostic screening of more than 100 000 tagging SNPs across the human genome, and the P value for the test of association reached the genome-wide significance (GWS) threshold of less than 5 × 10−8 (65) in at least one publication. In an agnostic GWA study, there is no prior belief that one SNP should have more chances than another for being associated with the phenotype of interest. Therefore, for all SNPs, the same GWS threshold is applied that accounts for the extent of multiplicity of comparisons. Associations were considered eligible regardless of whether they reached GWS in the early stage at which the agnostic testing was performed or (as is more often the case) only after additional data were included from subsequent replication stages or from other GWA studies in a GWA meta-analysis (66). The review excluded pharmacogenetic studies and studies that evaluated only associations with various clinical or pathologic features of a specific cancer type (eg, grade, stage, or invasiveness).
The search for studies and eligible markers was based on the online catalog of GWA studies hosted by the National Human Genome Research Institute (NHGRI, last searched March 15, 2010; www.genome.gov/gwastudies). Details on the NHGRI catalog appear elsewhere (58,59). Briefly, the catalog is updated weekly to include all published studies that have performed genome-wide evaluations for human phenotypes and traits, and it lists associations with P values less than or equal to 10−5. Extracted data include study name, publication date (month, year), chromosomal region, potentially implicated gene(s), SNP with the strongest statistical support, risk allele, sample size (number of case patients and number of control subjects in the first and subsequent stages), frequency of the risk allele, respective effect size and 95% confidence interval, and P value.
For this review, a number of steps were taken to augment the information available from the NHGRI catalog and to confirm data quality. All potentially eligible articles in the catalog were retrieved, and information was collected on the ancestry of the studied populations. When possible, data regarding allele frequency and effect sizes were extracted separately based on population ancestry. For all associations with listed P values greater than or equal to 5 × 10−8 in the catalog, the data were scrutinized in the original publication to avoid missing statistically significant associations. We checked non-GWS P values in the original publications to identify whether they might have been corrected for multiple comparisons or some other identifiable error. In addition, other errors, inconsistencies, or missing values were corrected, and odds ratios (ORs) were consistently corrected, as needed, to reflect per-allele odds ratios in multiplicative models. The article reference lists were reviewed to identify articles with pertinent results of GWA studies that had not been indexed in the NHGRI catalog. Furthermore, to include recently published data, additional eligible articles were searched in the advance online publications of the journals that had published other eligible GWA studies until March 15, 2010.
For duplicate entries, for which the same SNP was found to be associated with the same cancer in two or more studies, only the earliest published study was retained. If both studies were published at exactly the same date, then the study with the largest total sample size was retained. The same rule was applied when two or more studies had found different SNPs in the same genetic locus, which were nevertheless perfect proxies (linkage disequilibrium measure D′ = 1.0, correlation coefficient r2 = 1.0). When two different SNPs were in linkage disequilibrium with r2 less than .8 and it had not been excluded that they might confer independent information, both were retained. When r2 was greater than or equal to .8, or there was evidence that the SNPs did not confer independent information, the same rules were applied as for duplicate entries or perfect proxies to select only one of the SNPs. When linked SNPs with r2 greater than or equal to .8 were listed as discovered in the same GWA study, only the one with higher population attributable fraction (that takes into account both the minor allele frequency and the OR of the association) (67) was retained. Pairwise values of r2 for linkage disequilibrium were obtained using the SNP Annotation and Proxy Search software (68) (http://www.broadinstitute.org/mpg/snap; Broad Institute, Boston, MA).
The review focused primarily on associations attaining a P value of less than 5 × 10−8, as analyzed by the primary authors. No effort was made to standardize analyses in terms of whether any adjustments were used or not (eg, age or sex). All associations are expressed as odds ratios per allele copy in log-additive (multiplicative) models. The chosen GWS threshold is not absolute, and it was used for operational purposes only. GWS may depend on the studied populations and their linkage disequilibrium structure, as well as the available sample size (65). The selected P value is a relatively lenient threshold, if one considers the multiplicity of analyses involving different phenotypes in such studies (69), but associations that reach such a P value have a very high chance of being genuine. Whereas some of the associations that had modestly higher P values than this GWS threshold may also be genuine, they may have to await further replication studies.
Analyses
Descriptives and Time Trends
Descriptives summarized the number of associations, the distribution of the odds ratios, risk allele frequencies, and minor allele frequencies. Moreover, the number of newly discovered associations each year (2007, 2008, 2009, and 2010 [until March 15th]) was evaluated based on the time of the first publication for each association. An increase in the number of discovered associations may simply be the result of an accumulation of new discoveries for different types of cancers that had not been evaluated before, rather than an addition of many more associations for cancers for which some associations were already reported by one or more GWA studies. Thus, these two categories were considered separately. When multiple GWA studies pertaining to the same cancer were published on exactly the same date, the one with the largest number of eligible entries in the catalog was considered the first for the purposes of this categorization. Odds ratios, risk allele frequencies, and minor allele frequencies were compared in the two categories of discoveries.
Distribution of Risk
For selected cancer types (colorectal, prostate, testicular, and thyroid), simulations were performed to generate the anticipated distribution of risk in the population, assuming that these SNPs would have independent effects. For each cancer, 25 000 individuals were simulated. For each SNP and each risk allele, its carrier status was randomly assigned across the simulated population with a frequency equal to the observed risk allele frequency in the GWA study that had discovered that association. Carrier status was considered to confer an increased risk of the disease equal to that observed in the article that had originally discovered the association. Allele frequency and odds ratios were simulated based on estimates from European ancestry populations. Data were too limited to perform simulations for other ancestry populations. These simulations were performed for two cancers with a high number of genetic loci discovered in GWA studies of populations of European ancestry (prostate and colorectal cancers) and the two cancers that had the two genetic loci with highest population attributable fraction (testicular and thyroid cancers). For other cancers, the discrimination of risk would likely be even more limited. For loci that had more than one linked SNP entered in the compendium, only the SNP with the lowest P value was considered. Discovered estimates may be slightly or modestly inflated because of winner’s curse (70,71). The winner's curse means that when associations are discovered based on crossing a significance threshold, their effect size (the OR) is expected to be on average inflated compared with the true value. However, this pertains primarily to the magnitude of the effect that emerges out of the original agnostic screening, and it should be less of an issue for the final effect size that emerges once the subsequent stage and replication samples have been included, as in the data used for the simulations. Conversely, the estimated effects may be slightly or modestly underestimated if the multiplicative allelic model is misspecified (72), if gene–gene interactions exist (73), or if each locus has additional variants that confer independent risk (64).
The distributions of risk across the simulated individuals were visualized, and the population relative risk (approximated by the multiplicative OR) was obtained for individuals in the upper vs lower decile and in the upper vs lower quartile of predicted risk (74). The mean of the simulated risk was set at 100, and the risk of subjects at the 10th, 25th, 50th, 75th, and 90th percentiles of risk was also estimated.
Statistical Power Considerations
The statistical power to discover associations with per-allele odds ratios of 1.40, 1.20, and 1.07 for risk allele frequencies f of 40% and 10% at α = 5 × 10−8 level of statistical significance was estimated for the sample sizes available in representative GWA investigations. In a GWA study with multiple stage design, the statistical power must be corrected by multiplying by the power of the first (and any intermediate) stage to select SNPs for the final-stage replication. To illustrate this principle, consider a GWA study with three stages. In the first stage, SNPs are selected only if they pass an α1 threshold (typically far less stringent than 5 × 10−8); in the intermediate stage, SNPs are selected only if they pass an α2 threshold; and in the subsequent final stage, SNP GWS is claimed for P less than 5 × 10−8 based on combined data from all stages with power Pgws when all data are combined. Then, the statistical power of that study is estimated as Pgws × P1 × P2, where P1 and P2 are the power estimates for the first and intermediate stages with α1 and α2, respectively. Statistical power calculations were performed for the largest study on breast cancer (42) and for a large meta-analysis of GWA and replication studies for colorectal cancer (15). For comparison, we also calculated the statistical power that would be achievable if the GWA approach were maximally used, and all available samples from all stages could be evaluated in a GWA platform with discovery claimed at α = 5 × 10−8. These analyses provided information on how many additional discoveries could be expected for odds ratios in the upper range of what has been observed so far for common cancers (OR = 1.40), for a typical value (OR = 1.20), and for the lowest value observed for common variants in common cancers (OR = 1.07).
Software
Statistical analyses were performed in Stata (College Station, TX), version 10.1 (75), and by using the PS program (William D. Dupont and Walton D. Plummer, http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize) (76) for statistical power calculations. P values for the comparison of groups by Mann–Whitney U test are two-tailed.
Eligible GWS Associations
The NHGRI catalog listed 54 eligible articles; two more cited articles were retrieved by perusing the full text of the 54 articles. The 56 eligible articles (1–56) were published between January 2007 and March 2010 and had a total of 173 listed associations. Of these, further screening excluded 24 duplicates, nine perfect proxies, seven associations with r2 greater than or equal to .8, 40 associations with P values between 1 × 10−5 and 5 × 10−8, and one association reaching the GWS threshold only in haplotypic analyses.
The remaining 92 associations were eligible for evaluation (Table 1). Of those, 81 were entirely independent and 11 referred to markers with modest linkage disequilibrium to others. These 81 associations pertained to 15 different types of cancer. More than half of the associations pertained to prostate cancer (n = 27, 26 independent loci), colorectal cancer (n = 11, 10 loci), or breast cancer (n = 11 loci); there were fewer associations for glioma (n = 8, 6 loci), basal cell skin cancer (n = 5 loci), melanoma (n = 5, 4 loci), lung cancer (n = 5, 4 loci), testicular cancer (n = 4, 3 loci), nasopharyngeal cancer (n = 4, 2 loci), and pancreatic cancer (n = 3 loci), whereas for the other five cancer types, there were only one or two discovered associations.
Table 1.
Eligible associations reaching genome-wide significance for cancer phenotypes
| Cancer | Locus | Reported genes* | Risk allele | First publication | Risk allele information on frequency and genetic effect |
|||
| fEur (fAs) (%)† | OR (95% CI) | P | PMID | |||||
| Basal cell carcinoma (skin) | 1q42.13 | RHOU | rs801114-G | October 12, 2008 (18) | 33 | 1.28 (1.19 to 1.37) | 6 × 10−12 | 18849993 |
| 1p36.13 | PADI4, PADI6, RCC2, ARHGEF10L | rs7538876-A | October 12, 2008 (18) | 35 | 1.28 (1.19 to 1.37) | 4 × 10−12 | 18849993 | |
| 12q12-13 | KRT5 | rs11170164-A | July 5, 2009 (4) | 8 | 1.35 (1.23 to 1.50) | 2 × 10−9 | 19578363 | |
| 9p21 | CDKN2A/B | rs2151280-C | July 5, 2009 (4) | 53 | 1.19 (1.12 to 1.26) | 7 × 10−9 | 19578363 | |
| 7q32 | Intergenic | rs157935-T | July 5, 2009 (4) | 68 | 1.23 (1.15 to 1.31)‡ | 6 × 10−10 | 19578363 | |
| Breast cancer | 10q26.13 | FGFR2 | rs2981582-A | May 27, 2007 (42) | 38 (30) | 1.26 (1.23 to 1.30) | 2 × 10−60 | 17529967 |
| 5q11.2 | MAP3K1 | rs889312-C | May 27, 2007 (42) | 28 (54) | 1.13 (1.10 to 1.16) | 7 × 10−20 | 17529967 | |
| 8q24.21 (region 3) | Intergenic | rs13281615-G§ | May 27, 2007 (42) | 40 (56) | 1.08 (1.05 to 1.11) | 5 × 10−12 | 17529967 | |
| 11p15.5 | LSP1 | rs3817198-C | May 27, 2007 (42) | 30 (14) | 1.07 (1.04 to 1.11) | 3 × 10−9 | 17529967 | |
| 16q12.1 | TNCR9, LOC643714 | rs3803662-A | May 27, 2007 (42) | 25 (60) | 1.20 (1.16 to 1.24) | 1 × 10−20 | 17529967 | |
| 2q35 | Intergenic | rs13387042-A† | May 27, 2007 (40) | 50 | 1.20 (1.14 to 1.26) | 1 × 10−13 | 17529974 | |
| 6q22.33 | ECHDC1, RNF146 | rs2180341-G | March 11, 2008 (29) | 21 | 1.41 (1.25 to 1.59) | 3 × 10−8 | 18326623 | |
| 6q25.1 | C6orf97 | rs2046210-A†,║ | February 15, 2009 (13) | (37) | 1.29 (1.21 to 1.37) | 2 × 10−15 | 19219042 | |
| 1p11.2 | Intergenic | rs11249433-C | March 29, 2009 (12) | 39 | 1.16 (1.10 to 1.23) | 7 × 10−10 | 19330030 | |
| 3p24 | SLC4A7, NEK10 | rs4973768-T | March 29, 2009 (52) | 46 (21) | 1.11 (1.08 to 1.13) | 4 × 10−23 | 19330027 | |
| 17q23 | COX11 | rs6504950-G | March 29, 2009 (52) | 73 (92) | 1.05 (1.01 to 1.09) | 1 × 10−8 | 19330027 | |
| Colorectal cancer | 8q24.21 (region 4) | Intergenic | rs6983267-G§ | July 8, 2007 (37) | 51 | 1.21 (1.15 to 1.27) | 1 × 10−14 | 17618284 |
| 18q21.1 | SMAD7 | rs4939827-T | October 14, 2007 (35) | 52 | 1.18 (1.12 to 1.23) | 1 × 10−12 | 17934461 | |
| 15q13.3 | Intergenic | rs4779584-T | December 16, 2007 (33) | 19 | 1.26 (1.19 to 1.34) | 4 × 10−14 | 18084292 | |
| 11q23.1 | Intergenic | rs3802842-C | March 30, 2008 (28) | 29 (30) | 1.11 (1.08 to 1.15) | 6 × 10−10 | 18372901 | |
| 8q23.3 | EIF3H | rs16892766-A | March 30, 2008 (27) | 7 | 1.25 (1.19 to 1.32) | 3 × 10−18 | 18372905 | |
| 10p14 | Intergenic | rs10795668-A | March 30, 2008 (27) | 67 | 1.12 (1.10 to 1.16) | 3 × 10−13 | 18372905 | |
| 8q24.21 (region 4) | POU5FIP1, HsG57825, DQ515897 | rs7014346-A§,¶ | March 30, 2008 (28) | 37 (22) | 1.19 (1.15 to 1.23) | 9 × 10−10 | 18372901 | |
| 19q13.11 | RHPN2 | rs10411210-C | November 16, 2008 (15) | 90 | 1.15 (1.10 to 1.20) | 5 × 10−9 | 19011631 | |
| 20p12.3 | Intergenic | rs961253-A | November 16, 2008 (15) | 36 | 1.12 (1.08 to 1.16) | 2 × 10−10 | 19011631 | |
| 14q22.2 | BMP4 | rs4444235-C | November 16, 2008 (15) | 46 | 1.11 (1.08 to 1.15) | 8 × 10−10 | 19011631 | |
| 16q22.1 | CDH1 | rs9929218-G | November 16, 2008 (15) | 71 | 1.10 (1.06 to 1.12) | 1 × 10−8 | 19011631 | |
| Esophageal cancer | 4q.23 | ADH6, ADH1B | rs1229984-G║ | August 18, 2009 (49) | (35) | 1.79 (1.69 to 1.88) | 8 × 10−24 | 19698717 |
| 12q24.12 | BRAP, ALDH2 | rs671-A‡,║ | August 18, 2009 (49) | (36) | 1.67 (1.58 to 1.76) | 3 × 10−24 | 19698717 | |
| Glioma | 9p21.3 | CDKN2A, CDKN2B | rs4977756-G§ | July 5, 2009 (5) | 60 | 1.24 (1.19 to 1.30) | 7 × 10−15 | 19578367 |
| 8q24.21 (region 7) | CCDC26 | rs4295627-G§,¶ | July 5, 2009 (5) | 83 | 1.36 (1.29 to 1.43) | 2 × 10−18 | 19578367 | |
| 5p15.33 | TERT | rs2736100-G | July 5, 2009 (5) | 49 | 1.27 (1.19 to 1.37) | 2 × 10−17 | 19578367 | |
| 11q23.3 | PHLDB1 | rs498872-T | July 5, 2009 (5) | 69 | 1.18 (1.13 to 1.24) | 1 × 10−8 | 19578367 | |
| 20q13.33 | RTEL1 (locus 1) | rs6010620-G§ | July 5, 2009 (5) | 77 | 1.28 (1.21 to 1.35) | 3 × 10−12 | 19578367 | |
| 8q24.21 (region 7) | CCDC26 | rs891835-G§ | July 5, 2009 (5) | 78 | 1.24 (1.17 to 1.30) | 8 × 10−11 | 19578367 | |
| 20q13.33 | RTEL1 (locus 2) | rs4809324-C§ | July 5, 2009 (6) | 10 | 1.60 (1.37 to 1.87) | 2 × 10−9 | 19578366 | |
| 9p21.3 | Intergenic | rs1412829-C¶ | July 5, 2009 (6) | 39 | 1.42 (1.27 to 1.58) | 2 × 10−10 | 19578366 | |
| Lung cancer | 15q25.1 | CHRNA3, CHRNA5, CHRNB4, PSMA4, LOC123688 | rs8034191-C | April 3, 2008 (25) | 34 | 1.30 (1.23 to 1.37) | 5 × 10−20 | 18385738 |
| 5p15.33 | CLPTM1L | rs401681-G¶ | November 2, 2008 (17) | 57 | 1.15 (1.09 to 1.19) | 8 × 10−9 | 18978787 | |
| 6p21.33 | BAT3, MSH5 | rs3117582-C | November 2, 2008 (17) | 13 | 1.24 (1.16 to 1.33) | 5 × 10−10 | 18978787 | |
| 5p15.33 | TERT | rs2736100-G¶ | October 15, 2009 (46) | 50 | 1.12 (1.08 to 1.16) | 2 × 10−10 | 19836008 | |
| 6p22.1 | TRNAA-UGC | rs4324798-A | October 15, 2009 (46) | 9 | 1.16 (1.09 to 1.24) | 2 × 10−8 | 19836008 | |
| Melanoma | 20q11.22 | CDC91L1 | rs910873-T | May 18, 2008 (22) | 9 | 1.75 (1.53 to 2.01) | 1 × 10−15 | 18488026 |
| 22q13.1 | Intergenic | rs2284063-G | July 5, 2009 (7) | 37 | 1.20 (1.14 to 1.28) | 2 × 10−9 | 19578364 | |
| 16q24.3 | MC1R | rs4785763-A¶ | July 5, 2009 (7) | 32 | 1.36 (1.28 to 1.45) | 6 × 10−22 | 19578364 | |
| 11q14.3 | TYR | rs1393350-A | July 5, 2009 (7) | 27 | 1.29 (1.21 to 1.38) | 2 × 10−14 | 19578364 | |
| 16q24.3 | MC1R | rs258322-A¶ | July 5, 2009 (7) | 9 | 1.67 (1.52 to 1.83) | 3 × 10−27 | 19578364 | |
| Nasopharyngeal | 6p22.1 | GABBR1 | rs29232-A║,¶ | August 5, 2009 (50) | (46) | 1.67 (1.48 to 1.88) | 9 × 10−17 | 19664746 |
| 6p21.33 | HLA-A | rs2517713-A║,¶ | August 5, 2009 (50) | (62) | 1.88 (1.65 to 2.15) | 4 × 10−20 | 19664746 | |
| 6p22.1 | HLA-F | rs3129055-G║,¶ | August 5, 2009 (50) | (31) | 1.51 (1.34 to 1.71) | 7 × 10−11 | 19664746 | |
| 6p21.33 | HCG9 | rs3869062-A║,¶ | August 5, 2009 (50) | (68) | 1.78 (1.55 to 2.05) | 9 × 10−16 | 19664746 | |
| Neuroblastoma | 6p22.3 | FLJ22536, FLJ44180 | rs6939340-G | May 9, 2008 (24) | 50 | 1.37 (1.27 to 1.49) | 9 × 10−15 | 18463370 |
| 2q35 | BARD1 | rs6435862-G | May 3, 2009 (11) | 29 | 1.68 (1.49 to 1.90) | 9 × 10−18 | 19412175 | |
| Ovarian cancer | 9p22.2 | Intergenic | rs3814113-T | August 2, 2009 (2) | 68 | 1.22 (1.16 to 1.27) | 5 × 10−19 | 19648919 |
| Pancreatic cancer | 9q34 | ABO | rs505922-C | August 2, 2009 (2) | 39 | 1.20 (1.12 to 1.28) | 3 × 10−8 | 19648918 |
| 1q32.1 | NR5A2 | rs3790844-T | January 24, 2010 (56) | 76 | 1.30 (1.19 to 1.41) | 2 × 10−10 | 20101243 | |
| 13q22.1 | Intergenic | rs9543325-C | January 24, 2010 (56) | 37 | 1.26 (1.18 to 1.35) | 3 × 10−11 | 20101243 | |
| Prostate cancer | 8q24.21 (region 1) | Intergenic | rs16901979-A§,† | April 1, 2007 (43) | 3 | 1.79 (1.53 to 2.11) | 1 × 10−12 | 17401366 |
| 8q24.21 (region 4) | Intergenic | rs6983267-G§,¶ | April 1, 2007 (43) | 50 | 1.26 (1.16 to 1.38) | 9 × 10−13 | 17401363 | |
| 8q24.21 (region 5) | Intergenic | rs1447295-A§ | April 1, 2007 (43) | 11 | 1.43 (1.30 to 1.58) | 2 × 10−14 | 17401363 | |
| 17q12 | TCF2 | rs7501939-C¶ | July 1, 2007 (39) | 58 | 1.19 (1.12 to 1.26) | 5 × 10−9 | 17603485 | |
| 17q12 | TCF2 | rs4430796-A¶ | July 1, 2007 (39) | 49 | 1.22 (1.15 to 1.30) | 1 × 10−11 | 17603485 | |
| 17q24.3 | Intergenic | rs1859962-G | July 1, 2007 (39) | 46 | 1.20 (1.14 to 1.27) | 3 × 10−10 | 17603485 | |
| 3p12.1 | Intergenic | rs2660753-T# | February 10, 2008 (31) | 11 | 1.18 (1.06 to 1.31)# | 3 × 10−8 | 18264097 | |
| 19q13.33 | KLK3 | rs2735839-G# | February 10, 2008 (31) | 85 | 1.20 (1.10 to 1.33)# | 2 × 10−18 | 18264097 | |
| 6q25.3 | SLC22A3 | rs9364554-T# | February 10, 2008 (31) | 29 | 1.17 (1.08 to 1.26)# | 6 × 10−10 | 18264097 | |
| 7q21.3 | LMTK2 | rs6465657-C# | February 10, 2008 (31) | 46 | 1.12 (1.05 to 1.20)# | 1 × 10−9 | 18264097 | |
| Xp11.22 | NUDT10, NUDT11, LOC340602, GSPT2, MAGED1 | rs5945572-A | February 10, 2008 (30) | 35 | 1.23 (1.16 to 1.30) | 4 × 10−13 | 18264098 | |
| 2p15 | EHBP1 | rs721048-A | February 10, 2008 (30) | 19 | 1.15 (1.10 to 1.21) | 8 × 10−9 | 18264098 | |
| 11q13.2 | Intergenic | rs10896449-G¶ | February 10, 2008 (32) | 52 | 1.28 (1.14 to 1.45) | 2 × 10−9 | 18264096 | |
| 10q11.23 | MSMB | rs10993994-T | February 10, 2008 (32) | 40 | 1.16 (1.04 to 1.29) | 7 × 10−13 | 18264096 | |
| 19q13.2 | Intergenic | rs8102476-C | September 20, 2009 (47) | 54 | 1.12 (1.08 to 1.15) | 2 × 10−11 | 19767754 | |
| 8q24.21 (region 3) | Intergenic | rs445114-T§ | September 20, 2009 (47) | 64 | 1.14 (1.10 to 1.19) | 5 × 10−10 | 19767754 | |
| 3q21.3 | Intergenic | rs10934853-A | September 20, 2009 (47) | 28 | 1.12 (1.08 to 1.16) | 3 × 10−10 | 19767754 | |
| 8q24.21 (region 2) | Intergenic | rs16902094-G§ | September 20, 2009 (47) | 15 | 1.21 (1.15 to 1.26) | 6 × 10−15 | 19767754 | |
| 11q13.2 | Intergenic | rs11228565-A¶ | September 20, 2009 (47) | 20 | 1.23 (1.16 to 1.31) | 7 × 10−12 | 19767754 | |
| 2p21 | THADA | rs1465618-A | September 20, 2009 (54) | 22 (67) | 1.11 (1.07 to 1.15) | 2 × 10−8 | 19767753 | |
| 2q31 | ITGA6 | rs12621278-A | September 20, 2009 (54) | 94 (76) | 1.36 (1.28 to 1.44) | 9 × 10−23 | 19767753 | |
| 4q22 | PDLIM5 | rs17021918-C¶ | September 20, 2009 (54) | 66 (63) | 1.13 (1.10 to 1.16) | 4 × 10−15 | 19767753 | |
| 4q22 | PDLIM5 | rs12500426-A†,¶ | September 20, 2009 (54) | 45 | 1.10 (1.07 to 1.14) | 1 × 10−11 | 19767753 | |
| 4q24 | TET2 | rs7679673-A | September 20, 2009 (54) | 43 (80) | 1.12 (1.09 to 1.15) | 3 × 10−14 | 19767753 | |
| 8p21 | NKX3.1 | rs1512268-A | September 20, 2009 (54) | 44 (37) | 1.18 (1.14 to 1.21) | 3 × 10−30 | 19767753 | |
| 11p15 | IGF2, IGF2A, INS, TH | rs7127900-A | September 20, 2009 (54) | 19 (9) | 1.27 (1.23 to 1.32) | 3 × 10−33 | 19767753 | |
| 22q13 | TTLL1, BIK, MCAT, PACSIN2 | rs5759167-T | September 20, 2009 (54) | 52 (34) | 1.17 (1.14 to 1.20) | 6 × 10−29 | 19767753 | |
| Testicular germ cell tumor | 12q21.32 | KITLG | rs995030-G§ | May 31, 2009 (9) | 80 | 2.55 (2.05 to 3.19) | 1 × 10−31 | 19483681 |
| 5q31.3 | SPRY4 | rs4624820-A | May 31, 2009 (9) | 54 | 1.37 (1.19 to 1.58) | 3 × 10−13 | 19483681 | |
| 6p21.31 | BAK1 | rs210138-G | May 31, 2009 (9) | 20 | 1.50 (1.28 to 1.75) | 1 × 10−13 | 19483681 | |
| 12q21.32 | KITLG | rs1508595-G¶ | May 31, 2009 (9) | 83 | 2.69 (2.10 to 3.44) | 3 × 10−30 | 19483681 | |
| Thyroid cancer | 9q22.33 | FOXE1 | rs965513-A | February 6, 2009 (14) | 34 | 1.75 (1.59 to 1.94) | 2 × 10−27 | 19198613 |
| 14q13.3 | NKX2-1 | rs944289-T | February 6, 2009 (14) | 57 | 1.37 (1.24 to 1.52) | 2 × 10−9 | 19198613 | |
| Urinary bladder cancer | 8q24.21 (region 6) | MYC, BC042052 | rs9642880-T§ | September 14, 2008 (19) | 45 | 1.22 (1.15 to 1.29) | 9 × 10−12 | 18794855 |
| 8q24.21 (region 8) | PSAC | rs2294008-T§ | August 2, 2009 (1) | 46 | 1.15 (1.10 to 1.20) | 2 × 10−10 | 19648920 | |
Reported gene(s) are those mentioned by the investigators of the primary studies, and they refer to genes for which the listed single-nucleotide polymorphisms are found or neighboring genes; it is not necessary that these genes contain the culprit variants. Information on the risk allele includes its frequency (f), per-allele odds ratio (OR) and 95% confidence interval (CI), the respective P value reported, and the Pub-Med identifier (PMID) of the first publication on this association (the one from which the f, OR, 95% CI, and P value are taken).
The risk allele frequency is shown in parentheses for populations of Asian ancestry (fAs) when evaluated separately from those of European ancestry (fEur), unless the investigators considered that the association was likely to be present or much stronger in only one type of population that alone crossed the genome-wide significance threshold (European ancestry for rs13387042, where the association was not replicated in populations of Japanese, Hawaiian, and African American ancestry; Asian ancestry for rs2046210, where the association was much weaker and of borderline nominal significance in populations of European ancestry with OR = 1.15 and 95% CI = 1.03 to 1.28; European ancestry for rs16901979, where the association was much weaker in a population of African American ancestry with OR = 1.34 and 95% CI = 1.09 to 1.64, although the risk allele frequency was much higher than that in the European populations [42% vs 3%]; and European ancestry for rs12500426, where the association was not statistically significant in populations of Asians and African Americans). For the same reason, whenever data were available from populations of more than one ancestry, the odds ratio, 95% confidence interval, and P value shown refer to all populations combined except for rs13387042 and rs16901979 (European ancestry only) and rs2046210 (Asian ancestry only). For rs505922, the data include also a small cohort of Asian subjects and few African Americans, but the total population is more than 94% of European ancestry. For rs1465618, rs12621278, rs170211918, rs12500426, rs7679673, rs1522268, rs7127900, and rs5779167, the data included small populations of Asian, Hawaiian, Latino, and African American ancestry. Whenever odds ratio estimates pertain to both European and other ancestry populations combined, the odds ratio estimate limited to European ancestry populations is very similar (difference of ≤1%, data not shown).
Differential effect according to paternal and maternal alleles (OR = 1.40 and 1.09, respectively).
The eight regions of 8q24.21 are independent loci (r2 < .03), and the same applies to the two loci of the RTEL1 gene (r2 = .03).
For the seven associations on breast, esophageal, and nasopharyngeal cancers, only data on Asian descent populations were available.
For pairs of polymorphisms in the same locus, linkage disequilibrium r2 values are .43 for rs7014346 and rs6983267; .32 for rs891835 and rs4295627; .78 for rs1412829 and rs4977756; .18 for rs258322 and rs4785763; .78 for rs7501939 and rs4430796; .73 for rs1508595 and rs995030; .23 for rs29232 and rs3129055; .09 for rs25117713 and rs3869062; .02 for rs401681 and rs2736100; .23 for rs10896449 and rs11228565; and .31 for rs12500426 and rs17021918.
Data from stage II only because investigators considered that stage I estimates may be not be representative of the general population (enriched samples used).
The discovered genetic loci are scattered across the genome, but some clustering is also obvious (Table 1). The most profound example is the 8q24.21 area, to which 12 of the 92 associations map. This area includes eight independent loci (regions) associated with the following cancers: breast, colorectal, and prostate cancers (five independent loci), bladder cancer (two independent loci), and glioma. Each of these eight loci is robustly associated with one type of cancer, but there is also some clustering even within the same region. For example, region 4 contains genetic markers associated with both prostate and colorectal cancers, with additional weaker evidence for association with ovarian cancer (77). Two other chromosomal bands each have genetic markers for two different cancers (2q35 for breast cancer and neuroblastoma and 5p15.33 for lung cancer and glioma), but these pertain to distant independent loci that localize to different genes or intergenic areas.
Pace of Discovery
Of the 92 associations, 36 came from articles that were the first to discover GWS associations for the respective cancer; the other 56 had appeared in subsequent publications. Across the 92 associations, the pace of discovery was accelerated between 2007 and 2009, with 15 associations in 2007, 25 in 2008, and 50 in 2009, but only two in the first 10 weeks of 2010. Of those, associations for cancers for which no previous discoveries had been made accounted for 10, six, 20, and zero discoveries in the four years, respectively. Among the 56 discoveries that appeared in subsequent publications after a GWA study had already found one or more variants for a specific cancer type, five were published in 2007, 19 in 2008, 30 in 2009, and two in the first 10 weeks of 2010.
Population Ancestry
Eighty-three associations had been studied in European ancestry populations, whereas data were limited for Asian and African ancestry populations (details in the footnote of Table 1). For 69 associations, GWS was attained based on European ancestry data. For 16 associations (rs2981582, rs889312, rs13281615, rs3817198, rs3803662, rs3802842, rs505922, rs1465618, rs12621278, rs17021918, rs7679673, rs1512268, rs7127900, rs5557167, rs3790844, and rs9543325), GWS was attained by combining data from both European and other populations, and for seven associations (rs2046210, rs1299984, rs671, rs3869062, rs3129055, rs2517713, and rs29232), GWS was attained from data on populations of Asian ancestry. Data for formal cross-ancestry comparisons are limited, although several examples were noted of divergent ancestry-specific risk allele frequencies and odds ratio estimates (Table 1).
Allele Frequencies and Odds Ratios
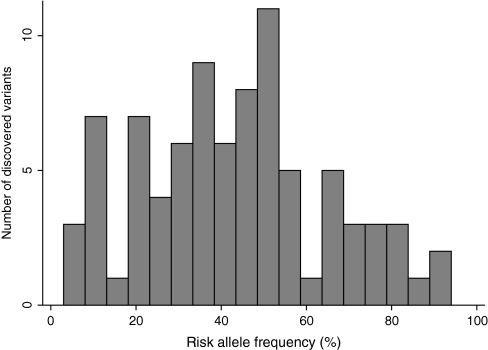
The risk alleles (based on European ancestry control populations) tended to have relatively high frequencies (median = 43%, interquartile range [IQR] = 28%–54%), and the distribution had an inverse U-shape, with many associations having risk allele frequencies in the range of 25%–55% and fewer having small or high-risk allele frequencies (Figure 1). The median minor allele frequency was 33% (IQR = 20%–43%). Risk alleles were more likely to be minor rather than major alleles (57 vs 28). Seven associations had risk allele frequency of 10% or less, whereas two had risk allele frequency greater than 90%.
Figure 1.
Distribution of allele frequencies of the discovered risk alleles based on European control populations.
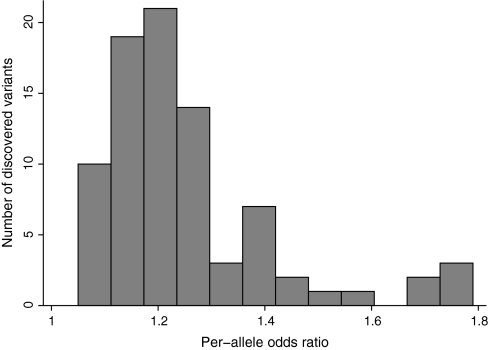
Per-allele odds ratios for the discovered variants were modest, with a median of 1.22 and IQR = 1.15–1.36 (Figure 2). With one exception, the eight odds ratios exceeding 1.50 had been documented for relatively less common cancers (testicular, thyroid, neuroblastoma, melanoma, and glioma), whereas only one association of prostate cancer had an odds ratio exceeding 1.79, and even that one pertained to a variant with low risk allele frequency in European ancestry populations (3%). The strongest odds ratio for any of the other three common cancers (lung, breast, and colorectal) was 1.41. The median odds ratio for the seven associations that were identified in Asian populations was 1.67, and the cancers studied were breast, esophageal, and nasopharyngeal.
Figure 2.
Distribution of per-allele odds ratios for the discovered variants. Two outliers with odds ratios greater than 1.8 are not shown.
There was evidence that the 30 associations that came from articles that were the first to publish GWS associations for the respective cancer had stronger effect sizes than the 55 associations discovered in subsequent publications (median OR = 1.28, IQR = 1.21–1.37 vs OR = 1.18, IQR = 1.12–1.25, P < .001 by two-sided Mann–Whitney U test). There was no statistically significant difference in the risk allele frequencies or minor allele frequencies (P = .32 and .68, respectively).
Anticipated Distribution of Risk
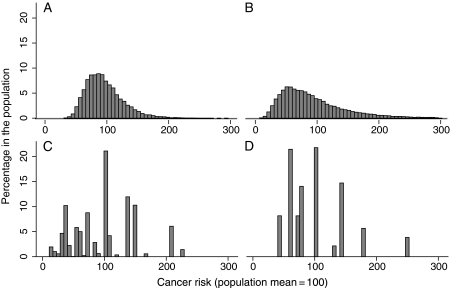
For the simulation studies of cancer risk in colorectal, prostate, testicular, and thyroid cancers, 10, 26, three, and two risk variants, respectively, were considered (Figure 3). The two cancers (colorectal and prostate) with a substantial number of variants have smooth distributions that are left-skewed and have long tails corresponding to individuals who have various possible combinations of a substantial number of susceptibility alleles (Figure 3, A and B). The other two cancers (testicular germ cell and thyroid) have more clearly discrete categories of risk corresponding to the many fewer possible combinations of risk alleles (Figure 3, C and D). The differences in risk across individuals are generally not large for any of the four cancers (Table 2). Individuals at the upper vs lower decile have 2.8-fold higher risk in colorectal cancer, 6.9-fold higher risk in prostate cancer, 8.0-fold higher risk in testicular cancer, and 4.5-fold higher risk in thyroid cancer (Table 2). The risk in the upper vs lower quartile of simulated risk is 2.1-fold higher in colorectal cancer, 4.1-fold higher in prostate cancer, 4.2-fold higher in testicular cancer, and 4.1-fold higher in thyroid cancer (Table 2).
Figure 3.
Distribution of the risk in a simulated sample of European descent individuals for which the mean risk in the population is set at a value of 100. A) Colorectal, B) prostate, C) testicular germ cell, and D) thyroid cancers.
Table 2.
Distribution of risk in simulated populations based on the combined multiplicative effect of all variants discovered in genome-wide association studies
| Cancer type | Variants considered | Risk in percentile of simulated risk* |
RR in upper vs lower decile (95% CI) | RR in upper vs lower quartile (95% CI) | ||||
| 10th | 25th | 50th | 75th | 90th | ||||
| Colorectal | 10 | 66.4 | 78.3 | 95.6 | 113.3 | 139.6 | 2.75 (2.41 to 3.14) | 2.10 (1.91 to 2.31) |
| Prostate | 26 | 43.2 | 58.7 | 84.8 | 123.1 | 174.4 | 6.85 (6.02 to 7.79) | 4.08 (3.67 to 4.53) |
| Testicular | 3 | 39.8 | 54.5 | 101.4 | 138.9 | 152 | 7.99 (7.06 to 9.03) | 4.19 (3.89 to 4.52) |
| Thyroid | 2 | 58.9 | 58.9 | 80.8 | 131.8 | 141.4 | 4.45 (3.96 to 5.01) | 4.09 (3.75 to 4.46) |
Risks are shown standardized against the mean simulated risk in the population (mean = 100). CI = confidence interval; RR = relative risk.
Statistical Power Considerations
We used two published GWA studies as a working example to calculate statistical power in a multistage design for breast cancer and colorectal cancer. The multistage design used for the breast cancer GWA study had good statistical power (0.65) to detect an OR = 1.40 for f = 40%, but statistical power was very limited to detect an OR = 1.40 for f = 10% or an OR = 1.20 (0.23 and 0, respectively) even for a very common variant (Table 3). For the colorectal cancer GWA meta-analysis in which a much larger sample was genotyped in the first stage, the statistical power was excellent (0.8 for all) for detecting common variants with f = 40% and OR ≥ 1.20 and those with f = 10% and OR = 1.40 but not those with f = 10% and OR = 1.20 (0.08).
Table 3.
Statistical power analyses for actual multistage genome-wide association (GWA) studies and for hypothetical studies in which all available samples would be subjected upfront to genotyping in a GWA platform*
| Type of cancer | Sample, No. of case patients/No. of control subjects | Alpha level | Statistical power |
|||||
|
f = 10% |
f = 40% |
|||||||
| OR = 1.4 | OR = 1.2 | OR = 1.07 | OR = 1.4 | OR = 1.2 | OR = 1.07 | |||
| Breast cancer (42) | ||||||||
| Stage I | 390/364 | .052 | 0.33 | 0.13 | 0.06 | 0.65 | 0.25 | 0.08 |
| Stage II | 3990/3916 | 2 × 10−5 | 0.71 | 0.04 | 0 | 1 | 0.40 | 0 |
| Stage III† | 26 240/26 858 | 5 × 10−8 | 1 | 0.85 | 0 | 1 | 1 | 0.02 |
| Power of multistage design | 0.23 | 0 | 0 | 0.65 | 0.10 | 0 | ||
| Hypothetical achievable power‡ | 26 240/26 858 | 5 × 10−8 | 1 | 0.85 | 0.001 | 1 | 1 | 0.02 |
| Colorectal cancer (15) | ||||||||
| Stage I | 6780/6843 | 10−5 | 0.97 | 0.13 | 0 | 1 | 0.80 | 0.01 |
| Stage II§ | 20 186/20 855 | 5 × 10−8 | 1 | 0.60 | 0 | 1 | 1 | 0.02 |
| Power of multistage design | 0.97 | 0.08 | 0 | 1 | 0.80 | 0 | ||
| Hypothetical achievable power‡ | 20 186/20 855 | 5 × 10−8 | 1 | 0.60 | 0 | 1 | 1 | 0.02 |
The power of a statistical test represents the probability of rejecting a false null hypothesis (ie, finding an association when one truly exists) and depends on sample and effect size. The breast cancer GWA study (42) discovered six variants, of which one had an odds ratio (OR) = 1.20–1.40, five had OR = 1.07–1.20, and none had odds ratio less than 1.07 or greater than 1.40. The colorectal cancer meta-analysis of GWA studies (74) increased the total number of associations to 11, of which three had OR = 1.20–1.40, eight had OR = 1.07–1.20, and none had odds ratio less than 1.07 or greater than 1.40. f = minor allele frequency.
In this stage, all data from stage I and stage II and new replication data were combined; therefore, power calculations include all data.
Power achieved if all available samples from all stages could be evaluated in a GWA platform with discovery claimed at α = 5 × 10−8 level of significance.
In this stage, replication data were combined with data from stage I. Power calculations include all data. Calculations do not consider the possibility of better power if an enriched sampling design is used, for example, as in the colorectal cancer study (15]).
In the breast cancer study, excellent statistical power (0.85) would be achieved (hypothetical achievable power, Table 3) for OR ≥ 1.20, even for f = 10%, if all available samples from all stages (n = 26 240 case patients and 26 858 control subjects) could be evaluated in a GWA platform, with discovery claimed at α = 5 × 10−8 level of statistical significance. Hypothetical achievable statistical power for colorectal cancer (n = 20 186 case patients and 20 855 control subjects) would be 1.0 for f = 40% and OR = 1.2 and 1.4, although it drops to 0.6 for f = 10% and OR = 1.2. The available total sample sizes do not provide any statistical power to identify risk variants with weak associations (OR = 1.07), even if all samples were genotyped in a GWA platform (Table 3).
A comparison of the statistical power in the performed multistage designs vs the hypothetical achievable power in a study genotyping all available samples in a GWA platform (Table 3) shows that substantial increments in the number of discovered variants could occur for breast cancer for variants with f = 40% and modest odds ratio (1.20) and for f = 10% and either modest or larger odds ratio (1.2–1.4). For colorectal cancer, there would be few additional discovered variants, if any, for very common alleles (f = 40%) and also no increased yield for less common alleles (f = 10%) and OR = 1.4, but there would be potential to discover additional variants with f = 10% and OR = 1.2.
Use of familial case patients can reduce the sample size required to detect a common variant by more than twofold (78) depending on whether effect sizes are stronger in familial cancer. For colorectal cancer, 922 familial case patients were included in stage I of the design. This may increase the statistical power to identify a variant with f = 40% and OR = 1.2 from 0.8 to 0.88 using the same inflation as suggested by the original meta-analysis (15). However, the statistical power for a variant with f = 10% and the same effect size would still be very small (0.19). If we assume that all case patients considered in the first design stage of the colorectal cancer study were familial case patients, then the statistical power to detect an association with f = 10% and OR = 1.20 would be inflated to 0.60. However, even with all samples available for genotyping, the statistical power to detect weak associations (OR = 1.07) would still be negligible. Similar changes would be seen in statistical power calculations for breast cancer.
Conclusions
GWA studies in cancer have effectively identified several genetic loci with strong statistical evidence for association with particular cancer types. These loci have not been previously identified through linkage or candidate gene studies. However, the explanatory power of these loci to predict individual cancer risk is limited by their modest effect sizes. Thus, despite the early success of the GWA approach, GWA-identified loci explain only a small proportion of the overall variability in cancer susceptibility.
The effect sizes of the loci identified in cancer GWA studies (average OR of 1.20) are smaller than the effect sizes identified in a recent NHGRI analysis of GWA-identified loci for diverse phenotypes (48). This difference may reflect the different spectrum of considered phenotypes. The SNP trait associations with the largest odds ratios in the NHGRI catalog (58) generally pertain to physical traits (eye color, hair color), biochemical traits (lipid levels, immunoglobulin E levels), or pharmacogenomic effects (eg, warfarin dosage and hemorrhagic risk). Compared with these phenotypes, it is likely that cancer is a more heterogeneous phenotype with a more complex genetic architecture and more modest genetic effects conferred by single variants. Another potential explanation is the expected difference in effect sizes between the first wave of GWA studies and subsequent GWA studies. For a given disease, the earliest GWA studies often identify the largest effects, and subsequent studies identify additional loci that often have a smaller effect size.
Given the initial success of cancer GWA studies, is it reasonable to expect a similar yield from future GWA studies, or has the GWA approach reached the stage of diminishing returns? We observed a steady, or even accelerating, rate of new discoveries between January 2007 and late 2009 but very few new associations in early 2010. Statistical power analyses suggest that if all existing GWA samples were analyzed for well-studied cancers such as breast and colorectal cancers, there would be reasonable statistical power to detect odds ratios in the range of 1.2, which are currently missed by multistage designs. However, extending GWA efforts to discover risk variants with weak associations (ie, ORs ∼ 1.07) will require sample sizes orders of magnitude larger than even the most comprehensive efforts to date. Thus, whereas comprehensive analysis of existing GWA samples for well-studied cancers will likely identify the bulk of common variants with odds ratios of 1.2 or higher, extending the GWA approach to identify variants of very small effect will require substantial new efforts.
Such efforts would likely depend on the coordinated work of international research consortia and efforts to establish additional very large prospective cohorts (79). Whereas the development of research consortia has been critical to the success of many current GWA efforts, much larger sample sizes would be needed to identify variants with very small effects. However, because the contribution of such variants to the genetic architecture is not well known, it is not clear that even very large sample sizes would be enough to make personalized cancer risk prediction viable based on current GWA platforms alone (80).
Simulations considering all GWA-discovered common genetic variants for four cancers showed that the predicted cancer risk of individuals differed only 2.1- to 4.2-fold between the upper and lower quartiles of risk. The number of available variants did not matter as much as the effect sizes of these variants. Thus, risk discrimination was best for testicular cancer, for which only three independent variants were considered in the calculations, whereas discrimination was slightly worse for prostate cancer with 26 variants, and 10 variants for colorectal cancer achieved very limited discrimination because they all had small effect sizes. These findings are in agreement with other studies that have modeled the risk of populations for specific types of cancers (81–86). In theoretical studies of the effect of multiple breast cancer loci, the risk discrimination was similar in magnitude to our simulated results. An initial report of the cumulative effect of five loci for prostate cancer reported somewhat higher odds ratios (83), but subsequent replication efforts have demonstrated more modest results (84). In a more recent multivariable analysis of prostate cancer including 22 variants in the Icelandic population (47), the top 1.3% of highest risk had only 2.5-fold higher risk of prostate cancer than the general population. Even this modest estimate may be overfit to the Icelandic data and needs independent validation.
The question of the predictive value of genetic information in disease prediction at the individual and population levels remains an area of considerable debate (87,88). Our results, along with the others cited above, suggest that for individual disease prediction, the prognostic importance of GWA-identified cancer susceptibility loci is limited. For population screening efforts, such information may eventually become useful in specific contexts, but more data are needed on the predictive performance of evolving prognostic models in independent data samples, the incremental benefit of adding genetic information to preexisting risk models, risk reclassification analyses, and the potential benefits and harms of genetic-based population screening strategies (89,90).
As the coverage of genotyping chips continues to improve, the ability to identify risk variants in previously under-interrogated regions may lead to new discoveries without an increase in sample sizes. Moreover, genome coverage for European and Asian ancestry populations is quite good with current chip technology, but coverage for non-European and non-Asian populations is still less than optimal (91,92). The vast majority of studies have addressed European descent populations, with limited data on other ancestral groups. Therefore, it is possible that additional loci will be discovered by performing GWA studies in non-European ancestry groups. Despite these caveats, it appears unlikely that the current “prediction gap” between the predictive power of GWA-discovered common variants and the anticipated genetic proportion of disease variance will be closed by additional GWA studies focusing on common variants alone.
There are several potential explanations for this prediction gap. One obvious possibility is that a substantial proportion of genetic risk variability is due to uncommon and rare variants (93,94). The advent of next-generation sequencing for discovery of functional variants both within and outside of GWA-identified loci is under way. If much of the genetic architecture is because of rare variants with small or modest effects, deciphering this architecture may be very difficult, if not impossible. Second, more in-depth evaluation of interesting loci may reveal a number of additional independent signals. The more obvious example in this regard is the 8q24 region where the evaluation of more tag SNPs identified up to eight independent SNPs in five different linkage disequilibrium blocks to be associated with prostate cancer susceptibility (55). Third, we still do not know the functional implications of most of the discovered markers. Understanding function has not been easy, but it may point to regulation of genes at a distance from the identified variants, which may lead to further focused searching for additional variants. For example, there is preliminary evidence that the associations of rs6983267 in the 8q24 region with colorectal cancer may reflect an impact on Wnt signaling and differential binding of transcription factor 7–like 2, with an impact on either the MYC proto-oncogene or on other more remote gene targets (95–97). Fourth, the assumption that risk alleles simply contribute in an additive fashion to individual disease risk may be incorrect. If this is the case, accurate estimates of risk will depend on the identification of more accurate models of genetic risk and even potentially complex gene–gene and gene–environment interactions. Genetic effects may be different in populations with different environmental exposures. A number of systems biology approaches have been used to model complexity, including computational approaches using high-throughput genetic variation data and gene expression data to generate network models of interacting genes (98–100). Such approaches represent sophisticated attempts to capture the connectivity pattern of the underlying disease biology, but these efforts are in early stages, and their ultimate impact on individual disease prediction remains unclear. Finally, the proposed estimates of the heritability and genetic contribution of some cancers may need to be reviewed again. Some of the higher estimates may be inflated, and thus, all genetic factors may only explain a small or modest fraction of cancer risk.
It is difficult to tell upfront whether a new GWA study for a cancer that has not been previously examined in this framework will yield no major discoveries or several gene variants with substantive effects. For example, pancreatic cancer susceptibility was initially assessed in a large GWA study with almost 2000 case patients, as many control subjects, and with replication effort in another 12 datasets with even larger sample size, but the yield was only one variant with very modest odds ratio of 1.20 (3). Conversely, four variants were discovered for nasopharyngeal carcinoma starting with a small GWA investigation of less than 300 case patients and control subjects (50). Nevertheless, once a first effort has been made, the initial yield is suggestive of what might be expected to be found with further studies and larger sample sizes with the same platforms. It is also theoretically possible that more consistent phenotypic ascertainment and definition of case patients and more exhaustive screening of latent disease in control subjects may also increase the power of discovering new variants. However, this should be done without eroding sample size from excessive exclusions, and it is often difficult, if not impossible, to go back and reascertain case patients and control subjects in these populations. In all, when current GWA platforms do not yield substantial further discoveries with large sample sizes, other types of genetic variation (eg, rare variants) and technologies (eg, sequencing) would have priority in further research efforts.
In summary, the GWA approach in cancer has been successful for its intended purpose of identifying common genetic variants associated with cancer risk. For common solid tumors such as breast, colon, and prostate cancers, future GWA efforts with ever larger samples are likely to identify some additional risk variants. However, the effect size of most of these variants will likely be smaller than what has already been discovered, and the predictive value of such variants is likely to be very limited. Of course, the benefits of the GWA approach are not limited to personalized genomic risk prediction, and the wealth of newly identified genetic risk loci has opened new avenues for basic science investigation. The success of GWA studies in breast, colon, and prostate cancers suggests that the extension of the GWA approach to other cancers will likely be fruitful if similar sample sizes can be collected. Finally, complementary approaches to GWA, including high-throughput sequencing, may afford valuable new insights into the genetic architecture and underlying biology of cancer susceptibility.
Funding
National Institutes of Health (UL1 RR025752) (Principal investigator: Harry Selker).
Footnotes
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41(9):991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song H, Ramus SJ, Tyrer J, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41(9):996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, Sulem P, Masson G, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41(8):909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanetsky PA, Mitra N, Vardhanabhuti S, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41(7):811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapley EA, Turnbull C, Al Olama AA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41(7):807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng CC, Yew PY, Puah SM, et al. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. J Hum Genet. 2009;54(7):392–397. doi: 10.1038/jhg.2009.49. [DOI] [PubMed] [Google Scholar]
- 11.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41(4):460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40(12):1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey SN, Gudbjartsson DF, Sulem P, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40(11):1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 19.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40(11):1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100(18):1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galvan A, Falvella FS, Spinola M, et al. A polygenic model with common variants may predict lung adenocarcinoma risk in humans. Int J Cancer. 2008;123(10):2327–2330. doi: 10.1002/ijc.23789. [DOI] [PubMed] [Google Scholar]
- 22.Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40(7):838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kibriya MG, Jasmine F, Argos M, et al. A pilot genome-wide association study of early-onset breast cancer. Breast Cancer Res Treat. 2009;114(3):463–477. doi: 10.1007/s10549-008-0039-9. [DOI] [PubMed] [Google Scholar]
- 24.Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 26.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40(5):623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 28.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40(5):631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40(3):281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40(3):316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 32.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 33.Jaeger E, Webb E, Howarth K, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40(1):26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 34.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99(24):1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 35.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39(11):1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 36.Murabito JM, Rosenberg CL, Finger D, et al. A genome-wide association study of breast and prostate cancer in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8(suppl 1) doi: 10.1186/1471-2350-8-S1-S6. S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 38.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 39.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 40.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 41.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 44.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 45.Spinola M, Leoni VP, Galvan A, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251(2):311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 46.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85(5):679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spain SL, Cazier JB, Consortium CORGI, Houlston R, Carvajal-Carmona L, Tomlinson I. Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res. 2009;69(18):7422–7429. doi: 10.1158/0008-5472.CAN-09-0659. [DOI] [PubMed] [Google Scholar]
- 49.Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137(5):1768–1775. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 50.Tse KP, Su WH, Chang KP, et al. Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. Am J Hum Genet. 2009;85(2):194–202. doi: 10.1016/j.ajhg.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69(16):6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41(10):1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41(10):1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 56.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 58.Hindorff LA, Junkins HA, Manolio TA. A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies. Accessed March 15, 2010. [Google Scholar]
- 59.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kraft P, Hunter DJ. Genetic risk prediction—are we there yet? N Engl J Med. 2009;360(17):1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 61.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 62.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 63.Chanock S. High marks for GWAS. Nat Genet. 2009;41(7):765–766. doi: 10.1038/ng0709-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32(2):179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- 66.Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10(2):191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rockhill B, Newman B, Weinberg C. Use and misuse of population attributable fractions. Am J Public Health. 1998;88(1):15–19. doi: 10.2105/ajph.88.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 70.Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80(4):605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19(5):640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 72.Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JPA. Discovery properties of genome-wide associations discovered from cumulatively combined data sets. Am J Epidemiol. 2009;170(10):1197–1206. doi: 10.1093/aje/kwp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cordell HG. Genome-wide association studies: detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ioannidis JP, Lau J. Heterogeneity of the baseline risk within patient populations of clinical trials: a proposed algorithm. Am J Epidemiol. 1998;148(11):1117–1126. doi: 10.1093/oxfordjournals.aje.a009590. [DOI] [PubMed] [Google Scholar]
- 75.Stata Corp. Stata Statistical Software: Release 10. College Station, TX: StataCorp LP; 2007. [Google Scholar]
- 76.Dupont WD, Plummer WD. Power and sample size calculations: a review and computer program. Controlled Clinical Trials. 1990;11:116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
- 77.Ghoussaini M, Song H, Koessler T, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100(13):962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: implication for design of association studies. Genet Epidemiol. 2003;25(3):190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 79.Collins FS, Manolio TA. Merging and emerging cohorts: necessary but not sufficient. Nature. 2007;445(7125):259. doi: 10.1038/445259a. [DOI] [PubMed] [Google Scholar]
- 80.Ioannidis JPA, Loy EY, Poulton R, Chia KS. Researching genetic versus non-genetic determinants of disease: a comparison and proposed unification. Sci Transl Med. 2009;1(7) doi: 10.1126/scitranslmed.3000247. 7ps8. [DOI] [PubMed] [Google Scholar]
- 81.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 82.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 84.Sun J, Chang BL, Isaacs SD, et al. Cumulative effect of five genetic variants on prostate cancer risk in multiple study populations. Prostate. 2008;68(12):1257–1262. doi: 10.1002/pros.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamada H, Penney KL, Takahashi H, et al. Replication of prostate cancer risk loci in a Japanese case-control association study. J Natl Cancer Inst. 2009;101(19):1330–1336. doi: 10.1093/jnci/djp287. [DOI] [PubMed] [Google Scholar]
- 86.Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ransohoff DF, Khoury MJ. Personal genomics: information can be harmful. Eur J Clin Invest. 2010;40(1):64–68. doi: 10.1111/j.1365-2362.2009.02232.x. [DOI] [PubMed] [Google Scholar]
- 88.Gulcher J, Stefansson K. Genetic risk information for common diseases may indeed be already useful for prevention and early detection. Eur J Clin Invest. 2010;40(1):56–63. doi: 10.1111/j.1365-2362.2009.02233.x. [DOI] [PubMed] [Google Scholar]
- 89.Khoury MJ, McBride CM, Schully SD, et al. The scientific foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009;11(8):559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grosse SD, Rogowski WH, Ross LF, et al. Population screening for genetic disorders in the 21st century: evidence, economics, and ethics. Public Health Genomics. 2010;13(2):106–115. doi: 10.1159/000226594. [DOI] [PubMed] [Google Scholar]
- 91.Pe’er I, de Bakker PI, Maller J, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38(6):663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 92.Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16(5):635–643. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- 93.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8(1) doi: 10.1371/journal.pbio.1000294. e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 2010;26(3):132–141. doi: 10.1016/j.tig.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeager M, Xiao N, Hayes RB, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124(2):161–170. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41(8):885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 97.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2009;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2009;18(6–7):389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]