Abstract
Calonectria pauciramosa is a pathogen of numerous plant hosts worldwide. Recent studies have indicated that it included cryptic species, some of which are identified in this study. Isolates from various geographical origins were collected and compared based on morphology, DNA sequence data of the β-tubulin, histone H3 and translation elongation factor-1α regions and mating compatibility. Comparisons of the DNA sequence data and mating compatibility revealed three new species. These included Ca. colombiana sp. nov. from Colombia, Ca. polizzii sp. nov. from Italy and Ca. zuluensis sp. nov. from South Africa, all of which had distinguishing morphological features. Based on DNA sequence data, Ca. brasiliensis is also elevated to species level.
Keywords: Calonectria, plant pathogens, sexual compatibility, systematics
INTRODUCTION
Several past studies have focused on the taxonomy of Calonectria spp. with small, 1-septate macroconidia and ellipsoidal to obpyriform vesicles (Crous et al. 1993, Overmeyer et al. 1996, Schoch et al. 1999, 2000). These Calonectria spp. were initially regarded as either Ca. morganii (= Cylindrocladium scoparium) or Ca. scoparia (= Cy. candelabrum) based on their morphological similarities. However, the anamorph state of Ca. morganii was circumscribed as having ellipsoidal to pyriform vesicles and Ca. scoparia having ellipsoidal to obpyriform vesicles by Crous et al. (1993). Later studies, incorporating DNA sequence data, have shown that Ca. morganii is restricted to the Northern Hemisphere and Brazil (Crous et al. 1993, Overmeyer et al. 1996, Schoch et al. 2000). In contrast, Ca. scoparia is found worldwide and forms part of a species complex consisting of four mating groups, each representing a different Calonectria species that includes Ca. pauciramosa (anamorph: Cy. pauciramosum), Ca. scoparia, Ca. mexicana (anamorph: Cy. mexicanum) and Ca. insularis (anamorph: Cy. insulare) (Schoch et al. 1999).
Calonectria pauciramosa has been reported worldwide on numerous plant hosts (Schoch et al. 1999, Koike et al. 1999, Koike & Crous 2001, Polizzi & Crous 1999, Polizzi 2000, Polizzi & Catara 2001, Polizzi & Vitale 2001, Crous 2002, Polizzi et al. 2006, 2007, 2009, Vitale et al. 2009), where it causes diseases such as cutting rot, damping-off, root rot and leaf blight. In South Africa and Australia, Ca. pauciramosa is regarded as the dominant pathogen in commercial forest nurseries (Crous 2002) and it is also found on various horticultural crops in commercial nurseries in Italy and the U.S.A. (Schoch et al. 2001, Crous 2002, Polizzi et al. 2006, 2007, 2009, Vitale et al. 2009).
Schoch et al. (2001) considered female fertility in populations of Ca. pauciramosa from various geographical regions to determine the ratio of mating types present, and based on these data suggested that Ca. pauciramosa was endemic to South America given that the ratio of both mating types approached 1:1. Furthermore, the study also indicated that Ca. pauciramosa isolates from California were represented by only one mating type, supporting the view that this represented an introduced pathogen. Isolates from Italy showed higher ratios of hermaphrodites and some variation was observed in the β-tubulin sequences. In contrast, South African isolates had close to a 1:1 mating type ratio and showed variation in β-tubulin sequence data (Schoch et al. 1999, 2001), indicating that this was either a native pathogen or that there had been multiple introductions into the country.
Initial investigations using DNA sequence comparisons and mating studies on Ca. pauciramosa isolates from South Africa and Colombia showed some variation amongst isolates. These findings and those of Schoch et al. (2001) suggested that Ca. pauciramosa might accommodate a number of cryptic species. The aim of this study was to consider the phylogenetic relationships, morphological characters and mating compatibility of available isolates of Ca. pauciramosa and to determine whether this species represented an assemblage of cryptic taxa.
MATERIALS AND METHODS
Isolates
Isolates of Ca. pauciramosa were obtained from culture collections (Table 1) or were isolated from infected plant material and soil samples following the methods of Crous (2002). For each isolate, single conidial cultures were prepared on 2 % (w/v) malt extract agar (MEA, Biolab, Midrand, South Africa). Representative strains are maintained in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa and the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands.
Table 1.
Isolates of Calonectria pauciramosa and other Calonectria species studied.
|
GenBank accession no. |
||||||||
|---|---|---|---|---|---|---|---|---|
| Species | Isolate | Mating type | β-tubulin | Histone H3 | Translation elongation factor-1α | Host | Country | Collector |
| Ca. brasiliensis | CBS 230.51T (= IMI 299576) | GQ267241 | GQ267259 | GQ267328 | Eucalyptus sp. | Brazil | T.R. Ciferri | |
| CBS 114257 | GQ267242 | GQ267260 | GQ267329 | Leaf litter | Brazil | A.C. Alfenas | ||
| CBS 116078 (= UFO 202) | GQ421772 | GQ421780 | GQ421788 | E. citriodora | Brazil | A.O. Carvalo | ||
| CMW 31505 (= CPC 2581) | GQ421775 | GQ421783 | GQ421791 | Prunus sp. | South Africa | C. Linde | ||
| CMW 31507 (= CPC 602) | GQ421773 | GQ421781 | GQ421789 | Eucalyptus sp. | Brazil | P.W. Crous | ||
| CMW 31508 (= CPC 1943) | GQ421774 | GQ421782 | GQ421790 | Leaf litter | Brazil | A.C. Alfenas | ||
| Ca. colombiana sp. nov. | CBS 111136 | Homothallic | FJ972424 | FJ972443 | FJ972493 | Soil | Colombia | M.J. Wingfield |
| CBS 115127T | Homothallic | FJ972423 | FJ972442 | FJ972492 | Soil | Colombia | M.J. Wingfield | |
| CBS 115638 | Homothallic | FJ972422 | FJ972441 | FJ972491 | Soil | Colombia | M.J. Wingfield | |
| CBS 115694 | Homothallic | FJ972425 | FJ972444 | FJ972494 | Soil | Colombia | M.J. Wingfield | |
| CMW 9058 | Homothallic | FJ972420 | FJ972439 | FJ972489 | Soil | Colombia | M.J. Wingfield | |
| Ca. colombiensis | CBS 112221 | AY725620 | AY725663 | AY725712 | Soil | Colombia | M.J. Wingfield | |
| Ca. insularis | CBS 114558 | AF210861 | FJ918526 | FJ918556 | Soil | Madagascar | P.W. Crous | |
| CBS 114559 | AF210862 | FJ918525 | FJ918555 | Soil | Madagascar | C.L. Schoch | ||
| Ca. mexicana | CBS 110918T | AF210863 | FJ972460 | FJ972526 | Soil | Mexico | M.J. Wingfield | |
| Ca. morganii | CBS 110666 | FJ918509 | FJ918527 | FJ918557 | Ilex vomitoria | U.S.A. | N.E. El-Gholl | |
| CBS 119669 | DQ521599 | DQ521601 | GQ421796 | Pistacia lentiscus | Italy | G. Polizzi | ||
| CBS 119670 | DQ521600 | DQ521602 | GQ421797 | Pistacia lentiscus | Italy | G. Polizzi | ||
| CMW 31506 (= CPC1722 = P94-4359) | AF210875 | GQ421787 | GQ421795 | Dodenaea vicosa | U.S.A. | N.E. El-Gholl | ||
| Ca. pauciramosa | CMW 1786 | Unknown | FJ972378 | FJ972445 | FJ972495 | Eucalyptus smithii | South Africa | M.J. Wingfield |
| CMW 2151 | Mat1-2 | FJ972400 | FJ972468 | FJ972517 | E. nitens | South Africa | M.J. Wingfield | |
| CMW 5683T | Mat1-2 | FJ918514 | FJ918531 | FJ918565 | E. grandis | South Africa | P.W. Crous | |
| CMW 7592 | Mat1-1 | FJ972380 | FJ972447 | FJ972497 | E. grandis | Uruguay | M.J. Wingfield | |
| CMW 7597 | Mat1-1 | FJ972406 | FJ972474 | FJ972523 | E. grandis | Uruguay | M.J. Wingfield | |
| CMW 7600 | Mat1-1 | FJ972405 | FJ972473 | FJ972522 | E. grandis | Uruguay | M.J. Wingfield | |
| CMW 7826 | Mat1-2 | FJ972392 | FJ972459 | FJ972509 | Soil | Australia | P.W. Crous | |
| CMW 7827 | Mat1-2 | FJ972385 | FJ972452 | FJ972502 | Soil | Australia | P.W. Crous | |
| CMW 7828 | Mat1-2 | FJ972391 | FJ972458 | FJ972508 | Soil | Australia | P.W. Crous | |
| CMW 7849 | Mat1-2 | FJ972383 | FJ972450 | FJ972500 | Erica sp. | U.S.A. | S.T. Koike | |
| CMW 7851 | Mat1-2 | FJ972382 | FJ972449 | FJ972499 | Mytrus communis | U.S.A. | S.T. Koike | |
| CMW 7852 | Mat1-2 | FJ972381 | FJ972448 | FJ972498 | M. communis | U.S.A. | S.T. Koike | |
| CMW 8061 | Mat1-2 | FJ972386 | FJ972453 | FJ972503 | Soil | Australia | P.W. Crous | |
| CMW 9151 | Mat1-2 | FJ972384 | FJ972451 | FJ972501 | Acacia mearnsii | South Africa | L. Lombard | |
| CMW 9172 | Mat1-2 | FJ972379 | FJ972446 | FJ972496 | A. mearnsii | South Africa | L. Lombard | |
| CMW 10148 | Mat1-2 | FJ972387 | FJ972454 | FJ972504 | Erica sp. | U.S.A. | S.T. Koike | |
| CBS 102296 | Mat1-2 | FJ972404 | FJ972472 | FJ972521 | Vriessea sp. | New Zealand | H.M. Dance | |
| CBS 110945 | Mat1-1 | FJ972389 | FJ972456 | FJ972506 | Podocarpus sp. | South Africa | P.W. Crous | |
| CBS 111873 | Mat1-1 | FJ972399 | FJ972467 | FJ972516 | Prunus sp. | South Africa | C. Linde | |
| CBS 114861 | Mat1-1 | FJ972403 | FJ972471 | FJ972520 | Eucalyptus sp. | South Africa | P.W. Crous | |
| CBS 115670 | Mat1-1 | FJ972393 | FJ972461 | FJ972510 | Pinus sp. | South Africa | P.W. Crous | |
| CBS 115893 | Unknown | FJ972411 | FJ972430 | FJ972480 | ||||
| CMW 30819 | Mat1-2 | FJ972402 | FJ972470 | FJ972519 | E. grandis | South Africa | P.W. Crous | |
| CMW 30875 | Mat1-1 | FJ972390 | FJ972457 | FJ972507 | Eucalyptus sp. | South Africa | P.W. Crous | |
| CMW 30823 | Mat1-1 | FJ918515 | FJ918532 | FJ918566 | E. grandis | South Africa | P.W. Crous | |
| CMW 30814 | Unknown | FJ972408 | FJ972427 | FJ972477 | Eucalyptus sp. | Kenya | J. Roux | |
| CMW 30822 | Unknown | FJ972409 | FJ972428 | FJ972478 | Eucalyptus sp. | Kenya | J. Roux | |
| CMW30873 | Mat1-2 | FJ972388 | FJ972455 | FJ972505 | Eucalyptus sp. | South Africa | L. Lombard | |
| CMW 27203 | Mat1-2 | FJ972398 | FJ972466 | FJ972515 | Eucalyptus sp. | China | S. Chen | |
| CMW 27206 | Mat1-2 | FJ972396 | FJ972464 | FJ972513 | Eucalyptus sp. | China | S. Chen | |
| CMW 27283 | Mat1-2 | FJ972397 | FJ972465 | FJ972514 | Eucalyptus sp. | China | S. Chen | |
| CMW 30878 | Mat1-1 | FJ972401 | FJ972469 | FJ972518 | Prunus sp. | South Africa | C. Linde | |
| CMW 30818 | Mat1-2 | FJ972395 | FJ972463 | FJ972512 | Limonium sp. | New Zealand | I. Brice | |
| CMW 30817 | Unknown | FJ972394 | FJ972462 | FJ972511 | Rhododendron sp. | New Zealand | R.A.J. White | |
| CMW 30879 | Mat1-2 | FJ972407 | FJ972475 | FJ972524 | Azalea sp. | Germany | G. Hagedorn | |
| CMW 30815 | Unknown | FJ972410 | FJ972429 | FJ972479 | Eucalyptus sp. | South Africa | P.W. Crous | |
| Ca. polizzii sp. nov. | CBS 123402T | FJ972419 | FJ972438 | FJ972488 | Arbutus unedo | Italy | G. Polizzi | |
| CMW 7804 | FJ972417 | FJ972436 | FJ972486 | Callistemon citrinus | Italy | G. Polizzi | ||
| CMW 10151 | FJ972418 | FJ972437 | FJ972487 | A. unedo | Italy | G. Polizzi | ||
| Ca. scoparia | CMW 31000 | FJ972426 | FJ972476 | FJ97252 | Eucalyptus sp. | Brazil | A.C. Alfenas | |
| CMW 31001 | GQ421779 | GQ267246 | GQ267246 | Eucalyptus sp. | Brazil | A.C. Alfenas | ||
| CBS 116076 | GQ421776 | GQ421784 | GQ421792 | Eucalyptus sp. | Brazil | P.W. Crous | ||
| CBS 116081 | GQ421777 | GQ421785 | GQ421793 | Soil | Brazil | M.J. Wingfield | ||
| CMW 7578 | GQ421778 | GQ421786 | GQ421794 | E. grandis | Argentina | L. Lombard | ||
| Ca. spathulata | CBS 112689 | AF308463 | FJ918524 | FJ918554 | E. viminalis | Brazil | N.E. El-Gholl | |
| CBS555.92T | GQ267215 | GQ267261 | GQ267331 | Araucaria angustifolia | Brazil | C. Hodges | ||
| Ca. zuluensis sp. nov. | CMW 9115 | Homothallic | FJ972413 | FJ972432 | FJ972482 | Eucalyptus sp. | South Africa | L. Lombard |
| CMW 9188T | Homothallic | FJ972414 | FJ972433 | FJ972483 | Eucalyptus sp. | South Africa | L. Lombard | |
| CMW 9208 | Homothallic | FJ972412 | FJ972431 | FJ972481 | Eucalyptus sp. | South Africa | L. Lombard | |
| CMW 9215 | Homothallic | FJ972416 | FJ972435 | FJ972485 | Eucalyptus sp. | South Africa | L. Lombard | |
| CMW 9896 | Homothallic | FJ972415 | FJ972434 | FJ972484 | Eucalyptus sp. | South Africa | L. Lombard | |
| Cy chinense | CBS 112744 | AY725618 | AY725660 | AY725709 | Soil | China | M.J. Wingfield | |
| Cy. hawksworthii | CBS 111870T | AF333407 | DQ190649 | FJ918558 | Nelumbo nucifera | Mauritius | A. Peerally | |
| Cy. leucothoës | CBS 109166T | FJ918508 | FJ918523 | FJ918553 | Leucothoë axillaris | U.S.A. | N.E. El-Gholl | |
CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CMW: culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa; TEx-type cultures.
Sexual compatibility
A total of 57 single conidial Ca. pauciramosa-like isolates (Table 1), originating from various geographic regions and hosts were crossed in all possible combinations. Mating-tester strains CMW 30823 (= STE-U 416) and CMW 5683 (= STE-U 971) for Ca. pauciramosa defined by Schoch et al. (2001) were also crossed with these isolates. Matings were done as described in Schoch et al. (1999) on carnation leaf agar (CLA; Fisher et al. 1982, Crous et al. 1993) and on minimal salt agar (MSA; Guerber & Correll 2001, Halleen et al. 2006) with sterile toothpicks placed on the surface of the agar. Control tests, where isolates were crossed with themselves, were undertaken to determine whether strains had a heterothallic or homothallic mating system. The plates were stacked in plastic containers and incubated at 22 °C for 6 wk. Matings were regarded as successful when isolate combinations produced perithecia extruding viable ascospores.
DNA sequence comparisons
Calonectria pauciramosa-like isolates were grown on MEA for 7 d. Mycelium was then scraped from the surface of the cultures, freeze-dried, and ground to a powder in liquid nitrogen, using a mortar and pestle. DNA was extracted from the powdered mycelium as described by Lombard et al. (2008). Three loci including fragments of the β-tubulin (BT), histone H3 (HIS3) and translation elongation factor-1 alpha (TEF-1α) gene regions were sequenced. Primers used to sequence these regions were T1 (O'Donnell & Cigelnik 1997) and CYLTUB1R (Crous et al. 2004b) for the BT region, CYLH3F and CYLH3R (Crous et al. 2004b) for the HIS3 region and EF1-728F (Carbone & Kohn 1999) and EF2 (O'Donnell et al. 1998) for the TEF-1α region. The PCR reaction mixture used to amplify the different loci consisted of 2.5 units FastStart Taq polymerase (Roche Applied Science, U.S.A.), 1× PCR buffer, 1–1.5 mM MgCl2, 0.25 mM of each dNTP, 0.5 μm of each primer and approximately 30 ng of fungal genomic DNA, made up to a total reaction volume of 25 μL with sterile distilled water.
Amplified fragments were purified using High Pure PCR Product Purification Kit (Roche, U.S.A.) and sequenced in both directions. For this purpose, the BigDye terminator sequencing kit v. 3.1 (Applied Biosystems, U.S.A.) and an ABI PRISM™ 3100 DNA sequencer (Applied Biosystems) were used. All PCRs and sequencing reactions were performed on an Eppendorf Mastercycler Personal PCR (Eppendorf AG, Germany) with cycling conditions as described in Crous et al. (2006) for BT and HIS3. The same cycling conditions for HIS3 were used for TEF-1α amplifications.
The generated sequences were added to other sequences of closely related Calonectria spp. obtained from GenBank (http://www.ncbi.nlm.nih.gov) and these were assembled and aligned using Sequence Navigator v. 1.0.1 (Applied Biosystems) and MAFFT v. 5.11 (Katoh et al. 2005), respectively. The aligned sequences were then manually corrected where needed. Single nucleotide polymorphisms (SNP'S) were determined for each gene region analysed using DnaSP v. 5.00.07 (Librado & Rozas 2009).
To determine whether the DNA sequence datasets for the three gene regions were congruent, a 70 % reciprocal bootstrap method using Neighbour-Joining with Maximum Likelihood distance was employed (Mason-Gamer & Kellogg 1996, Gueidan et al. 2007). Models of evolution were estimated in Modeltest v. 3.7 (Posada & Crandall 1998) using the Akaike Information Criterion for each separate gene region. The bootstrap analyses were run in PAUP (Phylogenetic Analysis Using Parsimony v. 4.0b10, Swofford 2002) for 10 000 replicates. Resulting tree topologies were compared visually for conflicts between the separate gene regions. Phylogenetic relationships were estimated in PAUP, by heuristic searches based on 1 000 random addition sequences and tree bisection-reconnection was used, with the branch swapping option set on “best trees” only.
All characters were weighted equally and alignment gaps were treated as missing data. Measures calculated for parsimony included tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC). Bootstrap analysis (Hillis & Bull 1993) was based on 1 000 replications. All sequences for the isolates studied were analysed using the Basic Local Alignment Search Tool for Nucleotide sequences (BLASTN, Altschul et al. 1990). The phylogenetic analysis included 73 partial gene sequences per gene, representing 11 Calonectria and Cylindrocladium species (Table 1). Calonectria colombiensis (CBS 112221) and Cy. chinense (CBS 112744) were used as outgroup taxa (Lombard et al. 2009). Novel sequences were deposited in GenBank and all alignments in TreeBASE (http://www.treebase.org).
A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v. 3.1.1 (Ronquist & Huelsenbeck 2003). Models of nucleotide substitution for each gene were determined using Mrmodeltest (Nylander 2004) and included for each gene partition. Two analyses of four MCMC chains were run from random trees for 1 000 000 generations and sampled every 100 generations. Both runs converged on the same likelihood score and tree topology. Therefore, the first 1 000 trees were discarded as the burn-in phase of each analysis and posterior probabilities were determined from the remaining trees.
Taxonomy
For morphological identification of the anamorphs, single conidial cultures were prepared on synthetic nutrient-poor agar (SNA; Nirenburg 1981, Lombard et al. 2009, 2010). Inoculated plates were incubated at room temperature and examined after 7d. Gross morphological characteristics were determined by mounting fungal structures in lactic acid and 30 measurements at ×1 000 magnification were made for each isolate. Teleomorph morphology was determined by mounting perithecia obtained from the sexual compatibility tests in Leica mountant (Setpoint Premier, Johannesburg, South Africa) and hand-sectioned with a Leica CM1100 cryostat (Setpoint Technologies) at -20 °C. The 10 μm sections were mounted in lactophenol or 3 % KOH. Gross morphological characteristics were observed as above. The 95 % confidence levels were calculated and extreme measurements of conidia are given in parentheses. For other structures, only the extremes are indicated. Optimal growth temperatures were determined for each isolate on MEA at 5–35 °C in 5 °C intervals in the dark. Colony colours were determined after 7 d on MEA at 25 °C in the dark, using the colour charts of Rayner (1970) for comparison. Descriptions, nomenclature, and illustrations were deposited in MycoBank (Crous et al. 2004a).
RESULTS
Sexual compatibility
Protoperithecia formed within 3 wk and successful matings produced perithecia with viable ascospores within 6 wk on both CLA and MSA. A total of 1 649 crosses were made using the 57 putative Ca. pauciramosa isolates and mating tester strains for Ca. pauciramosa. This resulted in 642 tests where perithecia produced viable ascospores. Self-self crosses indicated that 11 of the 57 isolates were self-fertile (homothallic). These included the Colombian isolates CBS 111041, CBS 111136, CBS 115127, CBS 115638, CBS 115694 and CMW 9058, and South African isolates CMW 9115, CMW 9188, CMW 9208, CMW 9215 and CMW 9896. Sixteen of the 57 putative Ca. pauciramosa did not cross with the mating tester strains for that species or with any other isolate included in this study. These included isolates CMW 7578 from Argentina; CBS 114257, CBS 116078, CBS 116076, CBS 116081, CMW 31505, CMW 31507 and CMW 31508, from Brazil; CMW 7804, CMW 10151 and CBS 123402 from Italy, CMW 30814 and CMW 30815 from Kenya; CMW 30817 from New Zealand; CMW 1786 and CMW 30815 from South Africa. The remaining 30 isolates produced perithecia containing viable ascospores when crossed with the Ca. pauciramosa mating tester strains and between them. This resulted in 203 successful heterothallic matings (Table 2).
Table 2.
Results of mating studies between isolates of Calonectria pauciramosa from various geographic regions.
| CBS 102296 | CBS 110945 | CBS 111873 | CBS 114861 | CBS 115670 | CMW 2151 | CMW 5683 | CMW 7592 | CMW 7597 | CMW 7600 | CMW 7826 | CMW 7827 | CMW 7828 | CMW 7849 | CMW 7851 | CMW 7852 | CMW 8061 | CMW 9151 | CMW 9172 | CMW 10148 | CMW 27203 | CMW 27206 | CMW 27283 | CMW 30817 | CMW 30818 | CMW 30819 | CMW 30823 | CMW 30873 | CMW 30875 | CMW 30878 | CMW 30879 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CBS 102296 | - | ||||||||||||||||||||||||||||||
| CBS 110945 | + | - | |||||||||||||||||||||||||||||
| CBS 111873 | + | - | - | ||||||||||||||||||||||||||||
| CBS 114861 | + | - | - | - | |||||||||||||||||||||||||||
| CBS 115670 | - | + | + | + | - | ||||||||||||||||||||||||||
| CMW 2151 | - | + | + | + | - | - | |||||||||||||||||||||||||
| CMW 5683 | - | + | + | + | - | + | - | ||||||||||||||||||||||||
| CMW 7592 | + | - | - | - | + | - | + | - | |||||||||||||||||||||||
| CMW 7597 | + | - | - | - | + | - | + | - | - | ||||||||||||||||||||||
| CMW 7600 | + | - | - | - | + | - | + | - | - | - | |||||||||||||||||||||
| CMW 7826 | - | + | + | + | - | + | - | + | + | + | - | ||||||||||||||||||||
| CMW 7827 | - | + | + | + | - | + | - | + | + | + | - | - | |||||||||||||||||||
| CMW 7828 | - | + | + | + | - | + | - | + | + | + | - | - | - | ||||||||||||||||||
| CMW 7849 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | |||||||||||||||||
| CMW 7851 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | ||||||||||||||||
| CMW 7852 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | |||||||||||||||
| CMW 8061 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | ||||||||||||||
| CMW 9151 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | |||||||||||||
| CMW 9172 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | ||||||||||||
| CMW 10148 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | |||||||||||
| CMW 27203 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | ||||||||||
| CMW 27206 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | |||||||||
| CMW 27283 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||||||
| CMW 30817 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||||||
| CMW 30818 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||||||
| CMW 30819 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||||
| CMW 30823 | + | - | - | - | + | - | + | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | ||||
| CMW 30873 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | |||
| CMW 30875 | + | - | - | - | + | - | + | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + | - | ||
| CMW 30878 | + | - | - | - | + | - | + | - | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + | - | - | |
| CMW 30879 | - | + | + | + | - | + | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - | + | + | - |
Isolates in bold indicate Ca. pauciramosa mating tester strains. + indicates formation of perithecia with viable ascospores; - indicates no perithecial formation
DNA sequence comparisons
Amplicons of approx. 500 bp were generated for the BT and TEF-1α gene regions and those for the HIS3 region were approx. 450 bp. Comparing the tree topologies of the 70 % reciprocal bootstrap trees indicated no conflicts. Subsequently, the datasets were combined and this resulted in a data set consisting of 1 529 characters including gaps. Of these characters, 1 151 were constant and parsimony-uninformative. The 378 parsimony-informative characters included in the parsimony analyses yielded eight most parsimonious trees (TL = 993, CI = 0.732, RI = 0.903, RC = 0.661), one of which is presented (Fig. 1). For Bayesian analyses, a HKY+I model was selected for BT, GTR+I+G model for HIS3 and a GTR+G model for TEF-1α and incorporated into the analyses. The consensus tree obtained for the Bayesian analyses confirmed the tree topology obtained with parsimony as well as bootstrap support (Fig. 1).
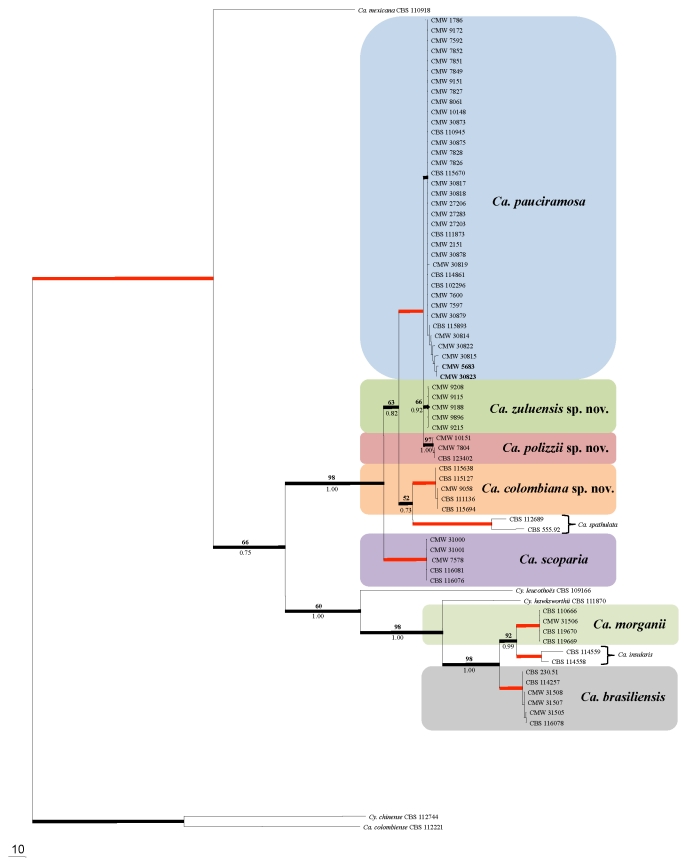
Fig. 1.
One of eight most parsimonious trees obtained from a heuristic search with 1 000 random addition of the combined BT, HIS3 and TEF-1α sequence alignments. Scale bar shows 10 changes and bootstrap support values from 1 000 replicates are shown above the nodes in bold. Bayesian posterior probability values are indicated below the nodes. Red lines indicate bootstrap support values of 100 and posterior probability values of 1.00. Thickened lines indicate branches in the strict consensus and Bayesian consensus tree. The tree was rooted to Ca. colombiensis (CBS 112221) and Ca. chinensis (CBS 112744). Mating tester strains of Ca. pauciramosa used in this study are indicated in bold.
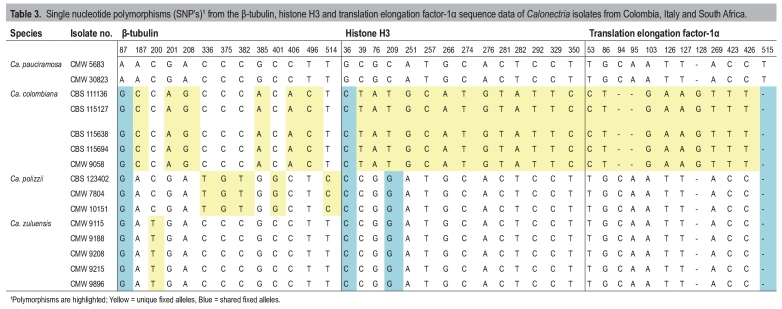
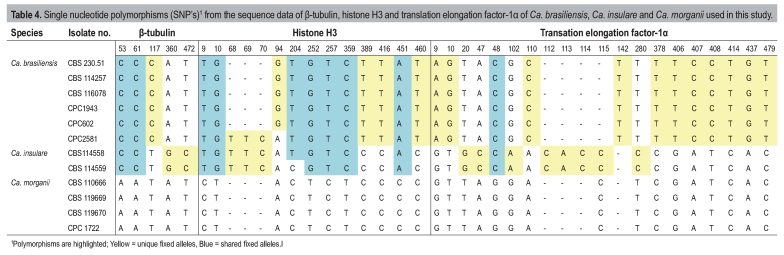
The majority of the Ca. pauciramosa isolates grouped together to form a monophyletic cluster with a bootstrap (BP) value of 100 and a Bayesian posterior probability (PP) value of 1.00. Within this cluster, two separate clades could be distinguished. The first (BP = 66, PP = 0.92) represented isolates obtained from South Africa (Table 1) and analyses of the SNP's (Table 3) showed one fixed allele for BT, two for HIS3 and one indel for TEF-1α. The second clade (BP = 97, PP = 1.00) represented isolates from Italy (Table 1) that were closely related to Ca. pauciramosa and have a number of shared fixed polymorphisms; five BT and two HIS3 (Table 3). Isolates from Colombia (Table 1) grouped together (BP = 100, PP = 1.00), separate from the Ca. pauciramosa cluster and SNP analyses show that six BT, 13 HIS3 and nine TEF-1α shared fixed alleles including three indels are characteristic for this group (Table 3). These isolates were closely related to Ca. spathulata. Isolates from Brazil grouped together with isolate CBS 230.51 (ex-type of Cy. brasiliensis; BP = 100, PP = 1.00), closely related to Ca. morganii and Ca. insularis, but separate from both of these species. Analyses of the SNP's for the isolates from Brazil compared to Ca. morganii and Ca. insularis also show several fixed alleles for these isolates, which include the ex-type culture of Cy. brasiliensis (CBS 230.51) (Table 4). The DNA sequence data for the three gene regions used in the present study showed 16 fixed alleles between Cy. brasiliensis, Ca. insularis and Ca. morganii (Table 4). An additional 10 fixed alleles were shared between Cy. brasiliensis and Ca. insularis and distinguished both species from Ca. morganii.
Table 3.
Single nucleotide polymorphisms (SNP's)1 from the β-tubulin, histone H3 and translation elongation factor-1α sequence data of Calonectria isolates from Colombia, Italy and South Africa.
Table 4.
Single nucleotide polymorphisms (SNP's)1 from the sequence data if β-tubulin, histone H3 and translation elongation factor-1α of Ca. brasiliensis, Ca. insulare and Ca. morganii used in this study.
Taxonomy
Isolates CMW 9115, CMW 9188, CMW 9208, CMW 9215 and CMW 9896 represent a distinct species closely related to Ca. pauciramosa, based on phylogenetic inference. Mating studies also showed that these isolates have a homothallic mating system, distinguishing them from Ca. pauciramosa. A similar situation was found for the isolates CBS 111136, CBS 115127, CBS 115638 and CBS 115694 from Colombia and they are also treated as a new species based on their homothallic mating system and phylogenetic inference. Furthermore, isolates CBS 123402, CMW 7804 and CMW 10151 from Italy are closely related to Ca. pauciramosa and failed to cross with the mating tester strains of that species. Morphological observations and DNA sequence data indicate that these isolates represent an undescribed taxon.
Species of Cylindrocladium (1892) represent anamorph states of Calonectria (1867) (Rossman et al. 1999). In this study, these fungi are described as new species of Calonectria, which represents the older generic name. This is irrespective whether the teleomorph states of these fungi have been found or not and follows the approach of Lombard et al. (2009, 2010).
Calonectria brasiliensis (Bat. & Cif.) L. Lombard, M.J. Wingf. & Crous, comb. nov. MycoBank MB 515110. Fig. 2. Basionym: Cylindrocladium brasiliensis (Bat. & Cif.) Peerally, (as braziliensis) CMI Descriptions of Pathogenic Fungi and Bacteria 427. 1974.
Fig. 2.
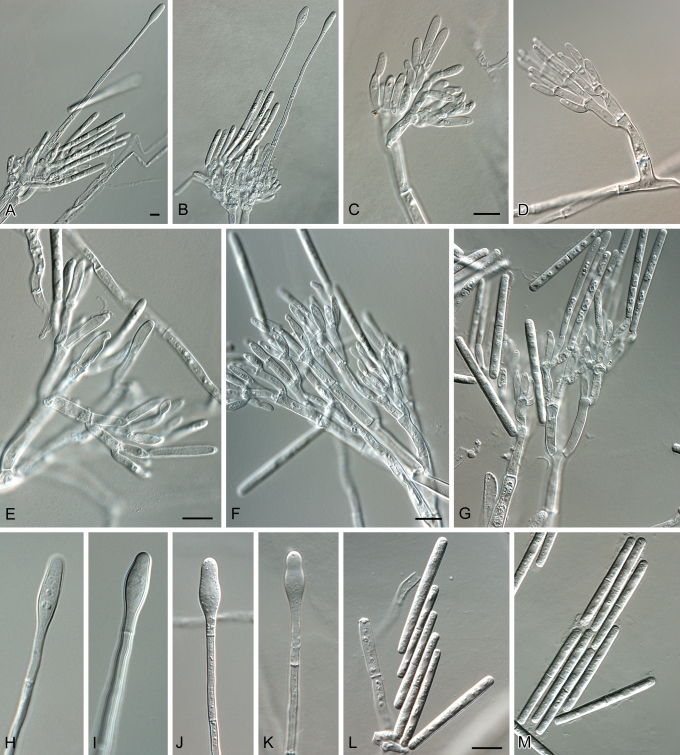
Calonectria brasiliensis. A–B. Macroconidiophores. C–G. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. H–K. Ellipsoidal to obpyriform vesicles. L–M. One-septate macroconidia. Scale bars = 10 μm.
≡ Cylindrocladium scoparium var. brasiliensis Bat. & Cif., (as brasiliense) Boletim de SA.I.C. Pernambuco 18: 188–191. 1951.
Teleomorph unknown. Conidiophores with a stipe bearing a penicillate suite of fertile branches, stipe extensions, and terminal vesicles. Stipe septate, hyaline, smooth, 63–103 × 7–14 μm; stipe extensions septate, straight to flexuous, 204–266 μm long, 6–7 μm wide at the apical septum, terminating in an ellipsoidal to obpyriform vesicle, 7–11 μm diam. Conidiogenous apparatus 58–90 μm long, and 81–103 μm wide; primary branches aseptate or 1-septate, 25–34 × 5–8 μm; secondary branches aseptate, 14–25 × 4–7 μm; tertiary branches aseptate, 8–20 × 3–5 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 8–12 × 2–4 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (35–)36–40(–41) × 3–5 μm (av. = 38 × 3.5 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Specimens examined: Brazil, Ceara State, Eucalyptus sp., Sept. 1948, T.R. Ciferri, ex-type culture CBS 230.51 = IMI 299576 = CMW 23671; Aracruz, Eucalyptus sp., June 1998, A.C. Alfenas, CBS 114257 = CMW 32949; Rio de Janeiro, Corymbia citriodora sub. sp. citriodora, A.O. Carvalho, CBS 116078 = CMW 32950; Champion nursery, Eucalyptus sp., P.W. Crous, CPC 602 = CMW 31507; Aracruz, Eucalyptus sp., P.W. Crous, CPC 1943 = CMW 31508.
Culture characteristics: Colonies fast growing (30–45 mm diam after 7 d) with optimal growth temperature at 25 °C (growth at 10–30 °C) on MEA, reverse amber to sepia-brown after 7 d; sparse white aerial mycelium with sparse sporulation; chlamydospores moderate throughout the medium, forming microsclerotia.
Substrate: Eucalyptus spp.
Distribution: Brazil.
Notes: Based on morphological observations, Crous & Wingfield (1994) reduced Ca. brasiliensis to synonymy with Ca. morganii. However, phylogenetic inference in this study has shown that the ex-type culture of Ca. brasiliensis (CBS 230.51) is distinct from Ca. morganii (CBS 110666, CBS 119669, CBS 119670 and CMW 31506). Morphological observations in this study also indicated that conidia of Ca. brasiliensis (av. 38 × 3.5 μm) are smaller than those of Ca. morganii (av. 45 × 4 μm). Calonectria brasiliensis only produces up to three branches per conidiophore, whereas Ca. morganii can have up to six branches per conidiophore.
Calonectria colombiana L. Lombard, Crous & M.J. Wingf., sp. nov. MycoBank MB515065, Fig. 3.
Fig. 3.
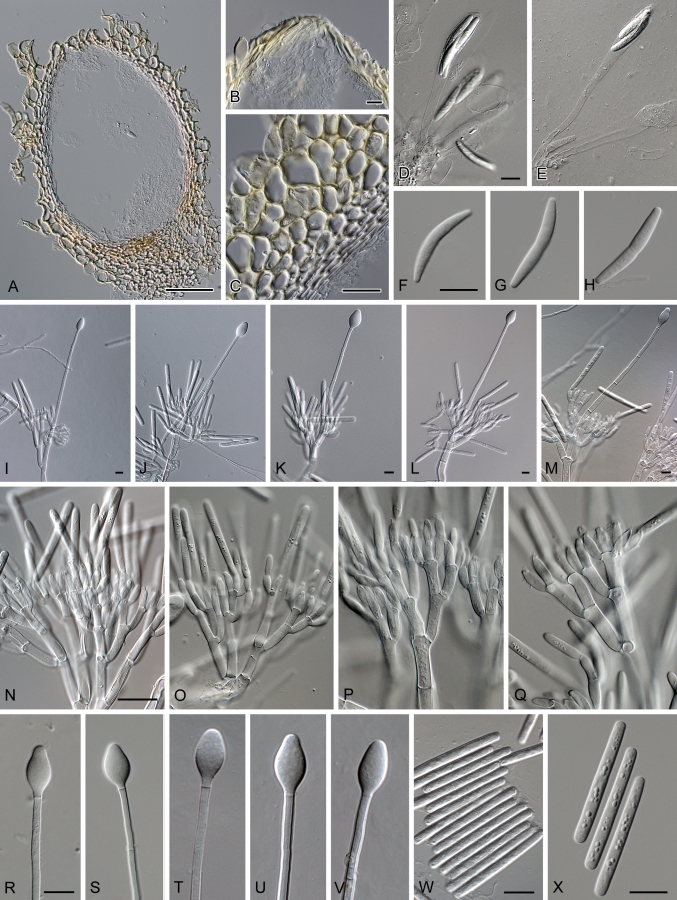
Calonectria colombiana. A. Perithecium. B. Ostiolar region of perithecium. C. Vertical section through perithecium, showing wall structure. D–E. Asci. F–H. Ascospores. I–M. Macroconidiophores. N–Q. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. R–V. Obpyriform to ellipsoid vesicles. W–X. One-septate macroconidia. Scale bars: A = 70 μm, B–C = 30 μm, other scale bars = 10 μm.
Etymology: Name refers to Colombia, the country this fungus was isolated from.
Telomorpha Calonectriae pauciramosa similis, sed ascosporis brevioribus, (28–)31–36(–40) × 3–5 μm (in medio 34 × 4 μm). Culturae homothallicae. Anamorpha Cylindrocladio pauciramoso simile, sed vesiculis obpyriforme vel fusiforme (8–12 μm diam.) et conidiis maioribus (33–)35–39(–40) × 3–4 μm, in medio 37 × 3 μm.
Perithecia solitary or in groups, orange to red, becoming red-brown with age; in section, apex and body yellow to orange, base red-brown, sub-globose to ovoid, 270–410 μm high, 175–285 μm diam, body turning dark red, and base dark red-brown (KOH+). Perithecial walls rough, consisting of 2 thick-walled layers: outside layer of textura globulosa, 24–90 μm wide; becoming more compressed towards inner layer of textura angularis, 18–22 μm wide; becoming thin-walled and hyaline towards the center, outer cells, 38–55 × 16–40 μm; inner cells, 3–12 × 3–7 μm: perithecial base up to 114 μm wide; consisting of dark red, angular cells; merging with an erumpent stroma, cells of the outer wall layer continuing into the pseudoparenchymatous cells of the erumpent stroma. Asci 8-spored, clavate, 87–162 × 12–18 μm, tapering to a long thin stalk. Ascospores aggregated in the upper third of the ascus, hyaline, gluttulate, fusoid with rounded ends, straight to slightly curved, 1-septate, not or slightly constricted at the septum, (28–)31–36(–40) × 3–5 μm (av. = 34 × 4 μm). Cultures homothallic. Conidiophores with a stipe bearing a penicillate suite of fertile branches, stipe extensions, and terminal vesicles. Stipe septate, hyaline, smooth, 45–126 × 6–9 μm; stipe extensions septate, straight to flexuous, 143–173 μm long, 5–7 μm wide at the apical septum, terminating in an obpyriform to ellipsoid vesicle, 8–12 μm diam. Conidiogenous apparatus 38–115 μm long, and 35–91 μm wide; primary branches aseptate or 1-septate, 19–37 × 5–8 μm; secondary branches aseptate, 9–17 × 4–5 μm; tertiary and additional branches (–4), aseptate, 8–13 × 3–4 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 9–12 × 3–4 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (33–)35–39(–40) × 3–4 μm (av. = 37 × 3 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Specimens examined: Colombia, La Selva, from soil, June 1995, M.J. Wingfield, Herb. PREM 60295, holotype of Calonectria colombiana, cultures ex-type CBS 115127 = CMW 30871 = CPC 1160; La Selva, June 1995, M.J. Wingfield, CBS 111041 = CMW 30767 = CPC 1163; La Selva, June 1995, M.J. Wingfield, CBS 111136 = CMW 30812 = CPC 1151; La Selva, June 1995, M.J. Wingfield, CBS 115638 = CMW 30766 = CPC 1161 (Herb. PREM 60296); La Selva, June 1995, M.J. Wingfield, CBS 115694 = CMW 30813 = CPC 1162, CMW 9058.
Culture characteristics: Colonies fast growing (35–55 mm diam after 7 d) with optimal growth temperature at 25 °C (growth at 10–30 °C) on MEA, reverse amber to sepia-brown after 7 d; abundant white aerial mycelium with sparse sporulation; chlamydospores extensive throughout the medium, forming microsclerotia.
Substrate: Soil.
Distribution: Colombia.
Notes: Isolates of Ca. colombiana were previously regarded as either Ca. pauciramosa or Ca. scoparia (Crous 2002) based on the morphological similarity of the anamorph states of these species. Based on macroconidial dimensions, Ca. colombiana (av. 37 × 3 μm) can be distinguished from Ca. pauciramosa (av. 50 × 4.5 μm) and Ca. scoparia (av. 60 × 4.5 μm) in having smaller, 1-septate macroconidia. Both Ca. pauciramosa and Ca. scoparia have a biallelic, heterothallic mating system (Schoch et al. 1999, 2001), whereas Ca. colombiana is homothallic.
Calonectria polizzii L. Lombard, Crous & M.J. Wingf., sp. nov. MycoBank MB515066, Fig. 4.
Fig. 4.
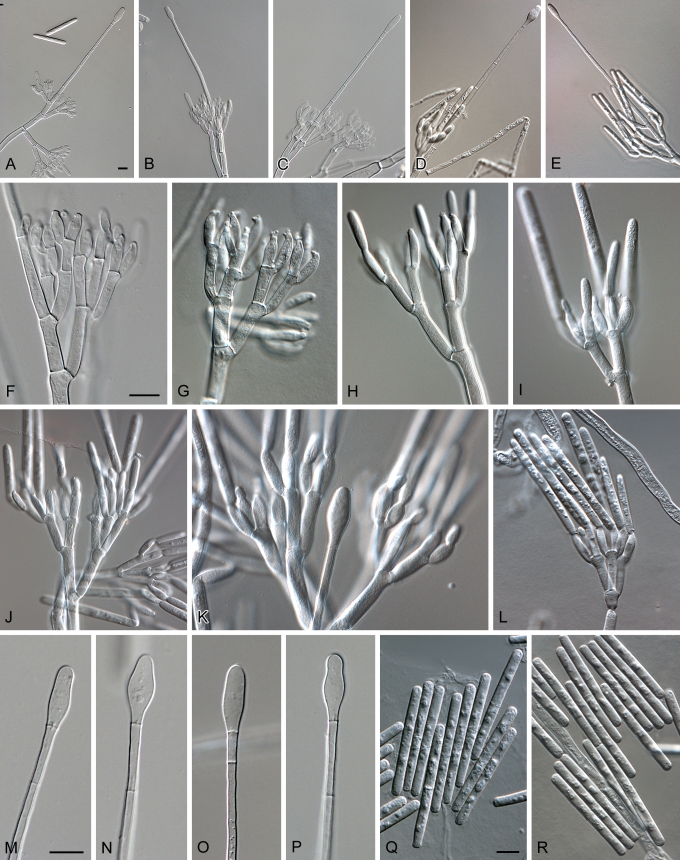
Calonectria polizzii. A–E. Macroconidiophores. F–L. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. M–P. Obpyriform to ellipsoid vesicles. Q–R. One-septate macroconidia. Scale bars = 10 μm.
Etymology: The name honours Prof. dr. Giancarlo Polizzi, who isolated the fungus in Italy.
Teleomorpha ignota. Cylindrocladio pauciramoso simile, sed vesiculis clavato vel obpyriforme (6–9 μm diam.) et conidiis maioribus (31–)32–42(–49) × 3–5 μm, in medio 37 × 4 μm.
Teleomorph unknown. Conidiophores with a stipe bearing a penicillate suite of fertile branches, stipe extensions, and terminal vesicles. Stipe septate, hyaline, smooth, 58–108 × 5–7 μm; stipe extensions septate, straight to flexuous, 111–167 μm long, 5–6 μm wide at the apical septum, terminating in an obpyriform to ellipsoid vesicle, 6–9 μm diam. Conidiogenous apparatus 27–57 μm long, and 28–51 μm wide; primary branches aseptate or 1-septate, 15–35 × 4–6 μm; secondary branches aseptate, 12–26 × 3–5 μm; tertiary branches aseptate, 10–15 × 4–5 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 8–13 × 3–4 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (31–)32–42(–49) × 3–5 μm (av. = 37 × 4 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Specimens examined: Italy, Sicily, Carrubba, on Arbutus unedo, 1997, G. Polizzi, Herb. PREM 60297, holotype of Calonectria polizzii, cultures ex-type CBS 123402 = CMW 30872; Sicily, on Callistemon citrinus, 1997, G. Polizzi, CMW 7804 = CPC 2681 = CBS 125270; Sicily, on Callistemon citrinus, 1997, G. Polizzi, CMW 10151 = CPC 2771 = CBS 125271 (Herb. PREM 60298).
Culture characteristics: Colonies fast growing (35–40 mm diam after 7 d) with optimal growth temperature at 25 °C (growth at 10–30 °C) on MEA, reverse amber to sepia-brown after 7 d; abundant white aerial mycelium with sparse sporulation; chlamydospores extensive throughout the medium, forming microsclerotia.
Substrates: Arbutus unedo, Callistemon citrinus.
Distribution: Italy.
Notes: Calonectria polizzii is morphologically similar to Ca. pauciramosa and Ca. zuluensis. The macroconidia of Ca. polizzii (av. 37 × 4 μm) are smaller to those of Ca. pauciramosa (av. 50 × 4.5 μm). Mating tests also showed that Ca. polizzii does not mate with either of the tester strains of Ca. pauciramosa (Schoch et al. 2001) used in this study. However, the isolates of Ca. polizzii tested might represent a single mating type, or might have lost their ability to mate, and further studies incorporating more isolates will be required to confirm this.
Calonectria zuluensis L. Lombard, Crous & M.J. Wingf., sp. nov. MycoBank MB515067, Fig. 5.
Fig. 5.
Calonectria zuluensis. A. Perithecium. B. A vertical section through a perithecium, showing the wall layers. C–D. Asci. E–G. Ascospores. H–L. Macroconidiophores. M–P. Conidiogenous apparatus with conidiophore branches and doliiform to reniform phialides. Q–U. Ellipsoid to obpyriform vesicles. V–W. One-septate macroconidia. Scale bars: A = 70 μm, B = 30 μm, other scale bars = 10 μm.
Etymology: Name refers to KwaZulu-Natal, South Africa, the province were the fungus was isolated.
Telomorpha Calonectriae pauciramosa similis, sed ascosporis brevioribus, (26–)29–34(–38) × 4–5 μm (in medio 32 × 4 μm). Culturae homothallicae. Anamorpha Cylindrocladio pauciramoso simile, sed vesiculis clavato vel obpyriforme (6–10 μm diam) et conidiis maioribus (31–)34–38(–40) × 3–5 μm, in medio 36 × 4 μm.
Perithecia solitary or in groups, orange to red, becoming red-brown with age; in section apex and body yellow to orange, base red-brown, sub-globose to ovoid, 292–394 μm high, 170–285 μm diam, body turning dark red, and base dark red-brown (KOH+). Perithecial walls rough, consisting of 2 thick-walled layers: outside layer of textura globulosa, 30–80 μm wide; becoming more compressed towards inner layer of textura angularis, 20–22 μm wide; becoming thin-walled and hyaline towards the center, outer cells, 40–50 × 18–40 μm; inner cells, 4–12 × 3–5 μm: perithecial base up to 116 μm wide; consisting of dark red, angular cells; merging with an erumpent stroma, cells of the outer wall layer continuing into the pseudoparenchymatous cells of the erumpent stroma. Asci 8-spored, clavate, 92–140 × 10–16 μm, tapering to a long thin stalk. Ascospores aggregate in the upper third of the ascus, hyaline, gluttulate, fusoid with rounded ends, straight to slightly curved, 1-septate, not or slightly constricted at the septum, (26–)29–34(–38) × 4–5 μm (av. = 32 × 4 μm). Cultures homothallic. Conidiophores with a stipe bearing penicillate clusters of fertile branches, stipe extensions, and terminal vesicles. Stipe septate, hyaline, smooth, 57–84 × 6–9 μm; stipe extensions septate, straight to flexuous, 110–171 μm long, 5–8 μm wide at the apical septum, terminating in ellipsoid to obpyriform vesicles, 6–10 μm diam. Conidiogenous apparatus 35–67 μm long, and 37–70 μm wide; primary branches aseptate or 1-septate, 16–28 × 4–6 μm; secondary branches aseptate, 11–20 × 3–5 μm; tertiary branches aseptate, 8–13 × 3–4 μm, each terminal branch producing 2–6 phialides; phialides doliiform to reniform, hyaline, aseptate, 10–13 × 3–4 μm; apex with minute periclinal thickening and inconspicuous collarette. Macroconidia cylindrical, rounded at both ends, straight, (31–)34–38(–40) × 3–5 μm (av. = 36 × 4 μm), 1-septate, lacking a visible abscission scar, held in parallel cylindrical clusters by colourless slime. Megaconidia and microconidia not seen.
Specimens examined: South Africa, KwaZulu-Natal, Kwambonambi, from Eucalyptus grandis clonal cutting, Feb. 2001, L. Lombard, Herb. PREM 60292, holotype of Calonectria zuluensis, cultures ex-type CBS 125268 = CMW 9188; KwaZulu-Natal, Kwambonambi, E. grandis × urophylla hybrid cutting, Feb. 2001, L. Lombard, CMW 9115, CMW 9208 (Herb. PREM 60293), CMW 9215, Pietermarizburg, E. grandis × urophylla hybrid cutting, Mar. 2001, L. Lombard, CMW 9896 = CBS 125272.
Culture characteristics: Colonies fast growing (35–40 mm diam after 7 d) with optimal growth temperature at 25 °C (growth at 10–30 °C) on MEA, reverse amber to sepia-brown after 7 d; abundant white aerial mycelium with sparse sporulation; chlamydospores extensive throughout the medium, forming microsclerotia.
Substrate: Eucalyptus grandis and E. grandis × urophylla rooted cuttings.
Distribution: South Africa.
Notes: Calonectria zuluensis can be distinguished from Ca. pauciramosa and Ca. scoparia based on its homothallic mating system. Macroconidia of Ca. zuluensis (av. 36 × 4 μm) are also smaller than those of Ca. pauciramosa (av. 50 × 4.5 μm) and Ca. scoparia (av. 60 × 4.5 μm). This species is morphologically very similar to Ca. colombiana. However, Ca. zuluensis can be distinguished from Ca. colombiana based on the fact that it has broadly clavate to obpyriform vesicles as compared with the obpyriform to fusiform vesicles in Ca. colombiana. Furthermore, Ca. zuluensis can easily be distinguished based on phylogenetic inference.
DISCUSSION
Considerable variation observed amongst isolates of “Ca. pauciramosa” from different geographical localities was illustrated in this study. Morphological characteristics, phylogenetic inference and mating studies revealed the presence of three cryptic species accommodated in cultures that have collectively been treated as Ca. pauciramosa. This is consistent with the results of previous studies (Schoch et al. 1999, 2001), which noted variation within Ca. pauciramosa, although at that time the sample size was inordinately small to consider the matter further. Schoch et al. (2001) also noted a high level of variation among isolates from South America, but concluded that this most likely reflected diversity consistent with an endemic population.
Crous (2002) suggested that mating isolates with recognised mating tester strains represented an important step in identifying isolates of Ca. pauciramosa. Various studies (Crous et al. 1993, Crous & Wingfield 1994, Crous et al. 1998, Schoch et al. 1999, 2001, Crous 2002) have used CLA as standardised medium to study sexual compatibility amongst isolates of Cylindrocladium. However, CLA has its limitations in that carnation leaf pieces are not always available and the present study used both CLA and MSA amended with sterile tooth picks, which proved to be very successful. Effective application of the latter technique to induce teleomorphs in culture has also been achieved for various other plant pathogenic genera, including Glomerella (Geurber & Correll 2001) and Neonectria (Halleen et al. 2006).
The descriptions of Ca. colombiana, Ca. zuluensis and Ca. polizzii add three new species to the Ca. scoparia species complex. This complex is characterised by species having ellipsoidal to obpyriform vesicles and producing 1-septate macroconidia (Schoch et al. 1999, Crous 2002). The complex was previously regarded as having a biallelic, heterothallic mating system (Schoch et al. 1999, 2001). However, both the newly described Ca. colombiana and Ca. zuluensis are homothallic. The occurrence of both heterothallic and homothallic Calonectria species in a single complex is not unique, having previously been found in the Ca. kyotensis species complex (Crous et al. 2004b).
Schoch et al. (2001) considered female fertility of Ca. pauciramosa, and found variation in BT sequence data for isolates from Italy. This variation has most likely been captured in the description of Ca. polizzii in the present study. This new species has thus been shown as unique based on morphological, phylogenetic inference and biological characteristics, separating it from Ca. pauciramosa. Morphologically, Ca. polizzii can be distinguished from Ca. pauciramosa by its smaller 1-septate macroconidia. Isolates of Ca. polizzii were also not capable of mating with the Ca. pauciramosa mating-tester strains or other Ca. pauciramosa isolates from different geographic regions.
Schoch et al. (2001), noted variation amongst isolates of Ca. pauciramosa from South America, and suggested that the fungus could be native to that continent. Results of the present study, including isolates from Colombia, led to the description of Ca. colombiana. This fungus is distinct from Ca. pauciramosa in having a homothallic mating system, smaller macroconidia and quaternary branches on the conidiophores. Although Ca. insularis also forms conidiophores with quaternary branches (Schoch et al. 1999), Ca. colombiana can easily be distinguished from it based on DNA sequence comparisons and its homothallic mating system.
More than eight species of Calonectria have been recorded from South Africa (Crous et al. 1991, Crous et al. 1993, Schoch et al. 1999, Crous 2002) and the description of Ca. zuluensis adds another species to those already reported from the country. Calonectria zuluensis has a homothallic mating system, which is different from Ca. pauciramosa with a biallelic, heterothallic mating system (Schoch et al. 2001). The two species can also easily be distinguished from each other based on DNA sequence comparisons.
In the analyses of the SNP's for the three gene regions used in this study, several fixed and shared SNP alleles were found for Ca. colombiana, Ca. polizzii and Ca. zuluensis. The majority of the fixed SNPs are shared between Ca. polizzii and Ca. zuluensis, indicating that these are sibling species, and that genetic isolation between them occurred recently (Taylor et al. 2000). For Ca. colombiana, fewer of the fixed SNPs are shared with Ca. polizzii and Ca. zuluensis, indicating that speciation occurred less recently than that of Ca. polizzii and Ca. zuluensis. These three species do not share the same alleles with Ca. pauciramosa, clearly distinguishing it from them.
Calonectria brasiliensis has been elevated to species level based on phylogenetic inference. Although Peerally (1974) indicated that the macroconidia of Ca. brasiliensis (24–38 × 2–3 μm) are smaller than those of Ca. morganii (av. 45 × 4 μm), Crous & Wingfield (1994) reduced Ca. brasiliensis to synonymy under Ca. morganii, based on similar conidial dimensions and vesicle morphology observed in culture. It is possible, however, that the original ex-type strain of Ca. brasiliensis was in fact morphologically degenerated, appearing atypical for the species. Several isolates from Brazil, previously identified as Ca. pauciramosa, grouped with the ex-type strain of Ca. brasiliensis (CBS 230.51). Previous DNA sequence comparisons and mating studies with Ca. morganii (Crous et al. 1993, Overmeyer et al. 1996, Schoch et al. 2000, 2001) failed to include the ex-type strain CBS 230.51 of Ca. brasiliensis, as this species was seen as a synonym of Ca. morganii (Crous 2002).
This study has shown the importance of combining morphological, biological and phylogenetic data to identify cryptic species of Calonectria. Although the biological species concept is regarded as insufficient for this purpose and needs to be clearly defined in Calonectria (Crous 2002), this study has shown that it has some use in identifying cryptic species within Ca. pauciramosa. The presence of homothallic and heterothallic mating strategies in closely related fungi is interesting and could well provide another opportunity to analyse the genetics of mating systems in ascomycetes. This study has shown, however, that morphology in combination with phylogenetic inference provides the most useful approach to identify cryptic species in Calonectria (Lombard et al. 2009). The present study has also shown the importance of the multi-gene approach in studying the phylogenetic relationships of phenotypic closely related Calonectria spp.
Acknowledgments
We thank members of the Tree Protection Co-operative Programme (TPCP), the Centraalbureau voor Schimmelcultures (CBS) and the University of Pretoria for financial and technical support to undertake the study. We also thank Dr H. Glen, South African National Botanical Institute (SANBI), for the Latin descriptions and for providing names for the new species.
Taxonomic novelties: Calonectria brasiliensis (Bat. & Cif.) L. Lombard, M.J. Wingf. & Crous, comb. nov., Calonectria colombiana L. Lombard, Crous & M.J. Wingf., sp. nov., Calonectria polizzii L. Lombard, Crous & M.J. Wingf., sp. nov., Calonectria zuluensis L. Lombard, Crous & M.J. Wingf., sp. nov.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic Local Alignment Search Tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Crous PW (2002). Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera. APS Press, St. Paul, Minnesota, U.S.A.
- Crous PW, Alfenas AC, Junghans TG (1998). Variability within Calonectria ovata and its anamorph Cylindrocladium ovatum from Brazil. Sydowia 50: 1–13. [Google Scholar]
- Crous PW, Alfenas AC, Wingfield MJ (1993). Calonectria scoparia and Calonectria morganii sp. nov., and variation among isolates of their Cylindrocladium anamorphs. Mycological Research 97: 701–708. [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004a). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hyde KD (2006). Calonectria species and their Cylindrocladium anamorphs: species with clavate vesicles. Studies in Mycology 55: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hywel-Jones N (2004b). Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Phillips AJL, Wingfield MJ (1991). The genera Cylindrocladium and Cylindrocladiella in South Africa, with special reference to forest nurseries. South African Forestry Journal 157: 69–89. [Google Scholar]
- Crous PW, Wingfield MJ (1994). A monograph of Cylindrocladium, including anamorphs of Calonectria. Mycotaxon 51: 341–435. [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, Nelson PE (1982). Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Geurber JC, Correll JC (2001). Characterization of Glomerella acutata, the teleomorph of Colletotrichum acutatum. Mycologia 93: 216–229. [Google Scholar]
- Gueidan C, Roux C, Lutzoni F (2007). Using multigene phylogeny analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycological Research 111: 1145–1168. [DOI] [PubMed] [Google Scholar]
- Halleen F, Schroers H-J, Groenewald JZ, Rego C, Oliveira H, Crous PW (2006). Neonectria liriodendra sp. nov., the main causal agent of black foot disease of grapevine. Studies in Mycology 55: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acid Research 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioke ST, Crous PW (2001). First report of root and crown rot of myrtle in California caused by Cylindrocladium pauciramosum. Plant Disease 85: 448. [DOI] [PubMed] [Google Scholar]
- Kioke ST, Henderson DM, Crous PW, Tjosvold SA (1999). A new root and crown rot disease of heath in California caused by Cylindrocladium pauciramosum. Plant Disease 83: 589. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J (2009). DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lombard L, Bogale M, Montenegro F, Wingfield BD, Wingfield MJ (2008). A new bark canker disease of the tropical hardwood tree Cedrelinga cateniformis in Ecuador. Fungal Diversity 31: 73–81. [Google Scholar]
- Lombard L, Rodas CA, Crous PW, Wingfield BD, Wingfield MJ (2009). Cylindrocladium species associated with dying Pinus cuttings. Persoonia 23: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Zhou XD, Crous PW, Wingfield BD, Wingfield MJ (2010). Calonectria species associated with cutting rot of Eucalyptus. Persoonia 24: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Gamer R, Kellogg E (1996). Testing for phylogenetic conflict among molecular datasets in the tribe Tiriceae (Graminae). Systematic Biology 45: 524–545. [Google Scholar]
- Nirenburg HI (1981). A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany 59: 1599–1609. [Google Scholar]
- Nylander JAA (2004). MrModeltest v. 2. Programme distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O'Donnell K, Cigelnik E (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeyer C, Lünnemann S, Wallburnn C von, Meinhardt F (1996). Genetic variability among isolates and sexual offspring of the plant pathogenic fungus Calonectria morganii on the basis of random amplification of polymorphic DNA (RAPD) and restriction fragment length polymorphism (RFLP). Current Microbiology 33: 249–255. [DOI] [PubMed] [Google Scholar]
- Peerally A (1974). Cylindrocladium brasiliense. CMI Descriptions of Pathogenic Fungi and Bacteria no. 427.
- Polizzi G (2000). Prime esperience di lotta chimica nei confrontidel marciume del colletto e delle radici di Polygala myrtifolia causato da Cylindrocladium pauciramosum. Informatore Fitopatologico 11: 39–47. [Google Scholar]
- Polizzi G, Catara V (2001). First report of leaf spot caused by Cylindrocladium pauciramosum on Acacia retinodes, Arbutus unedo, Feijoa sellowiana and Dodonaea viscosa in Southern Italy. Plant Disease 85: 803. [DOI] [PubMed] [Google Scholar]
- Polizzi G, Crous PW (1999). Root and collar of milkwort caused by Cylindrocladium pauciramosum, a new record for Europe. European Journal of Plant Pathology 105: 407–411. [Google Scholar]
- Polizzi G, Grasso FM, Vitale A, Aiello D (2007). First occurrence of Calonectria leaf spot on Mexican blue palm in Italy. Plant Disease 91: 1057. [DOI] [PubMed] [Google Scholar]
- Polizzi G, Vitale A (2001). First report of the prevalence of benzimidazole-resistant isolates in a population of Cylindrocladium pauciramosum in Italy. Plant Disease 85: 1210. [DOI] [PubMed] [Google Scholar]
- Polizzi G, Vitale A, Aiello D, Castello I, Guarnaccia V, Parlavecchio G (2009). First record of crown and root rot caused by Cylindrocladium pauciramosum on brush cherry in Italy. Plant Disease 93: 547. [DOI] [PubMed] [Google Scholar]
- Polizzi G, Vitale A, Aiello D, Parlavecchio G (2006). First record of crown and root rot caused by Cylindrocladium pauciramosum on California lilac in Italy. Plant Disease 90: 1459. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Rayner RW (1970). A mycological colour chart. Commonwealth Mycological Institute, Kew, Surrey. British Mycological Society.
- Ronquist F, Heulsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R (1999). Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248. [Google Scholar]
- Schoch CL, Crous PW, Cronwright G, Witthuhn RC, El-Gholl NE, Wingfield BD (2000). Recombination in Calonectria morganii and phylogeny with other heterothallic small-spored Calonectria species. Mycologia 92: 665–673. [Google Scholar]
- Schoch CL, Crous PW, Polizzi G, Koike ST (2001). Female fertility and single nucleotide polymorphism comparisons in Cylindrocladium pauciramosum. Plant Disease 85: 941–946. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Crous PW, Wingfield BD, Wingfield MJ (1999). The Cylindrocladium candelabrum species complex includes four distinct mating populations. Mycologia 91: 286–298. [Google Scholar]
- Swofford DL (2002). PAUP*. Phylogenetic analysis using parsimony (* and other methods), v. 4.0b10. Computer programme. Sunderland, Massachusetts, USA: Sinauer Associates.
- Taylor JW, Jacobson DJ, Kroken SM, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Vitale A, Aiello D, Castello I, Dimartino MA, Parlavecchio G, Polizzi G (2009). Severe outbreak of crown rot and root rot caused by Cylindrocladium pauciramosum on strawberry tree in Italy. Plant Disease 93: 842. [DOI] [PubMed] [Google Scholar]