Abstract
Cinnamyl alcohol dehydrogenase (CAD) is a key enzyme in the biosynthesis of lignin. We have isolated full length of a cDNA encoding CAD (IbCAD1) that was previously identified as the most abundant gene in an EST library of sweetpotato suspension cells. Phylogenetic analysis revealed that IbCAD1 belongs to the family of defense-related CADs. High levels of IbCAD1 mRNA were found in the roots of sweetpotato, but not in the leaves and petioles. The IbCAD1 gene transcripts were highly induced by cold, wounding, and reactive oxygen species. Analyses of transcriptional regulation of the IbCAD1 gene in transgenic tobacco plants carrying the IbCAD1 promoter–GUS revealed that IbCAD1 promoter expression was strong in the roots, but barely detectable in the cotyledons. IbCAD1 promoter activity increased with increasing root age, and strong promoter expression was observed in the lateral root emergence sites and in root tips. Weak GUS expression was observed in lignified tissues of vascular system of mature leaves and stems. IbCAD1 promoter activity was strongly induced in response to the biotic and abiotic stresses, with the strongest inducer being wounding, and was also induced by salicylic acid (SA) and jasmonic acid (JA) as well as by abscisic acid (ABA) and 6-benzylaminopurine. Taken together, our data suggest that IbCAD1 can be involved in JA- and SA-mediated wounding response and ABA-mediated cold response, respectively. The IbCAD1 gene may play a role in the resistance mechanism to biotic and abiotic stresses as well as in tissue-specific developmental lignification.
Keywords: Cinnamyl alcohol dehydrogenase, Promoter activity, Biotic and abiotic stresses, Hormone response, Lignification
Introduction
Lignin is the second most abundant biopolymers in nature after cellulose. It forms an integral part of the secondary cell walls of specialized conducting and supporting tissues of plants, facilitating water transport and providing mechanical strength, respectively (Boudet 2000). The lignification of tissues is also thought to play a role as a defense barrier to limit pathogen invasion (Boerjan et al. 2003; Goujon et al. 2003). The biosynthesis of lignin proceeds via the general phenylpropanoid pathway, which generates a pool of hydroxycinnamyl-CoA esters. These hydroxycinnamyl-CoA esters are then channeled into the lignin-specific pathway to produce monolignols through the action of two enzymes, cinnamoyl-CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD). CCR catalyzes the conversion of hydroxycinnamyl-CoA esters to their corresponding cinnamaldehydes, and these are subsequently converted to their corresponding alcohols (p-coumaryl, coniferyl, and sinapyl alcohol) by CAD (Walter et al. 1988; Goujon et al. 2003).
In angiosperm species, CAD is encoded by a multigene family. Nine CAD genes (AtCAD1 to AtCAD9) have been identified in Arabidopsis and classified into three groups based on both homology and biochemical properties in vitro (Sibout et al. 2003; Kim et al. 2004, 2007). The first group consists of AtCAD4 (AY302081) and AtCAD5 (AY302082). These genes are associated with the highest CAD enzyme activity and homology (74–83% similarity) with respect to the bona fide CADs, such as Eucalyptus EgCAD2 and rice OsCAD2, and act as primary genes in lignin biosynthesis in Arabidopsis. EgCAD2 is the best characterized CAD involved in developmental lignin synthesis (Goffner et al. 1992). The second group of Arabidopsis CADs consists of AtCAD2 (AY302077), AtCAD3 (AY302078), AtCAD7 (AY302079) and AtCAD8 (AY302080). These all have a relatively lower CAD catalytic activity. AtCAD7 (AY302079) and AtCAD8 (AY302080) show the highest homology (~78%) to poplar sinapyl alcohol dehydrogenase (PtSAD) (Li et al. 2001). PtSAD has been shown to be highly specific for sinapaldehyde substrates and is possibly responsible for syringyl unit deposition in poplar. AtCAD7 and AtCAD8 are induced in response to pathogen infection (Kiedrowski et al. 1992). The members of the last group, AtCAD1 (AY288079), AtCAD6 (AY302075) and AtCAD9 (AY302076), lack detectable CAD catalytic activities in vitro, but they are expressed predominantly in vascular (lignin deposition) tissues. The expression of AtCAD1 was found to be induced by bacterial pathogen (Tronchet et al. 2010). The true biochemical roles of these CAD enzymes still need to be elucidated, particularly so as differences in the lignin composition of tissues in different plant species result from the different activities of several CAD isoenzymes, each with a different specificity.
Detailed knowledge of the role of CADs in relation to the biosynthesis of developmental lignins would contribute extensively to our understanding of the in vivo enzyme activity of CADs and subsequent lignin composition in tissues. In comparison to wild-type Arabidopsis plants, the content of lignin in the stem of double mutant Atcad4/Atcad5 plants is reduced by 60% and the level of coniferyl and sinapyl alcohol is reduced by 94% (Sibout et al. 2003, 2005). The rice gold hull and internode2 (gh2) mutant, which results from a mutation in the OsCAD2 gene, has reduced levels of p-hydroxyphenol, guaiacyl and syringyl monomers—in almost the same ratio—compared to the wild type (Zhang et al. 2006). Transgenic plants having reduced levels of lignins are more readily attacked by fungi and bacteria, as shown by double mutant Atcad4/Atcad5 plants, whose disease symptoms are distinctly more severe than those of the wild type (Tronchet et al. 2010). The relationship between CAD and biotic stress has been well-established. The mRNA level of alfafa MsaCADs and ryegrass LpCADs was observed to increase following wounding treatments (Brill et al. 1999; Lynch et al. 2002), and the addition of fungal elicitor to bean cell cultures leads to the rapid accumulation of CAD transcripts (Walter et al. 1988). In addition, syringyl lignin was found to be accumulated during the hypersensitive resistance response in wheat (Menden et al. 2007). All of these results indicate that the amount and unit composition of lignin is influenced by both developmental and environmental conditions. However, little information is yet available on the role of CAD in relation to abiotic stress.
We have isolated a cDNA (IbCAD1) encoding CAD and the upstream region of the IbCAD1 gene from sweetpotato and have investigated this IbCAD1 gene transcript and the activity of its promoter in response to various development stages. To gain an understanding of the regulation mechanism of IbCAD1 gene by biotic and abiotic stresses, we subjected wild type and transgenic plants to various stresses and then monitored promoter activity and tissue-specific expression. The effect of various plant hormones on IbCAD1 promoter activity was also examined. We report here the results of these analyses and suggest that the IbCAD1 gene may be involved in lignification induced by both abiotic and biotic stresses and in tissue-specific developmental lignification.
Materials and methods
Plant materials
Sweetpotato plants (Ipomoea batatas cv. Yulmi) and transgenic tobacco plants were maintained in pots in a greenhouse at 25°C under a 16/8-h light/dark photoperiod. They were propagated from cuttings taken from the top growing tip and grown for 3 weeks for the stress treatments. For the Agrobacterium-mediated transformation procedure, tobacco (Nicotiana tabacum cv. Xanthi) seeds were surface sterilized and sown on MS medium (Murashige and Skoog 1962) containing 3% sucrose and 0.8% agar.
Southern blot analysis
Whole cells of sweetpotato cultured in cell suspension were frozen and ground to a fine powder under liquid nitrogen, and genomic DNA was isolated according to the manufacturer’s instructions (QIAGEN, Germany). The genomic DNA (10 μg) was then digested with EcoRI and HindIII independently, followed by electrophoretic separation in a 0.8% agarose gel. These restriction enzymes do not have any recognition sequences within the IbCAD1 cDNA sequence. After a complete denaturation and subsequent renaturation, the gel was blotted onto a positively charged nylon membrane (Hybond-N+, Amersham Biosciences, UK). Biotin (biotin-14-dCTP, Invitrogen, USA) was used as a probe and labeled by PCR amplification using IbCAD1 cDNA as a template. The PCR analysis was performed in a 20 μl volume containing 1.25 U ExTaq DNA polymerase (Takara, Japan), 2 μl of 10× Taq buffer, 4 μl of 5× dNTPs mix (0.25 mM biotin-14-dCTP, 0.25 mM dCTP, 0.5 mM dATP, 0.5 mM dGTP and 0.5 mM dTTP), and 10 pmol of T3 and T7 primers. The PCR cycling conditions consisted of 94°C for 5 min, 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, with a final extension step of 7 min at 72°C. The labeled probe was purified using a PCR Purification kit (QIAGEN). The membrane was hybridized at 65°C for 16 h and then washed twice with 2× SSC/1% SDS at room temperature for 10 min, twice with 0.1× SSC/1% SDS at 65°C for 20 min, and twice with 1× SSC at room temperature for 10 min. Hybridized signals were detected using the Southern-Star™ System (Tropix, USA).
Reverse transcription-PCR analysis
Semi-quantitative reverse transcription (RT)-PCR was performed to analyze the expression pattern of IbCAD1. Total RNA (2 μg) was utilized for the synthesis of first strand cDNA, and the RT was performed using oligo (dT)20 (Invitrogen, USA) as a primer and Superscript™ II reverse transcriptase (Invitrogen). Aliquots consisting of one-tenth of the RT product were used as a template for each of the subsequent PCR amplifications. To amplify the IbCAD1, we designed PCR primers for gene-specific amplification in the 5′- and 3′-untranslated region (UTR) of the IbCAD1 cDNA. Sequences of the forward (F) and reverse (R) primers used to amplify cDNA were: F, 5′-ATCTTGATTGTCTCAATCTA3′, R, 5′-GGACATTATTACATTACAC-3′. Tubulin was used as the internal control (F, 5′-CAACTACCAGCCACCAACTGT-3′; R, 5′-CAAGATCCTCACGAGCTTCAC-3′). PCR amplification was carried out in a 20 μl reaction mixture containing 0.2 mM of each dNTPs, 1 pmol of each forward and reverse primers, and 0.125 U of Taq polymerase (NEB, USA), including cDNA products as a template. After an initial denaturation at 94°C for 5 min, amplification was carried out at 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, with a final incubation for 7 min at 72°C. There were 30 and 20 PCR cycles for IbCAD1 and tubulin, respectively. The amplified PCR products were separated in a 1% agarose gel.
Construction of genomic library and isolation of the IbCAD1 promoter
The 5′-flanking region of IbCAD1 was isolated from the genomic DNA of sweetpotato cells grown in suspension culture following the instructions of the Universal GenomeWalker kit (Clontech, USA), which is a simple method for cloning unknown genomic sequences adjacent to a DNA fragment of known sequence. The first step consisted of constructing adaptor-ligated sweetpotato genomic DNA libraries. This was accomplished by completely digesting separated aliquots of sweetpotato genomic DNA with different restriction enzymes, namely, NaeI, NruI, HpaI, and ScaI. Each batch of digested product was then ligated separately to the GenomeWalker Adaptor. In the primary PCR, the four adaptor-ligated genomic libraries were used as a template. The PCR was performed in 50 μl aliquots, each containing 1 μl of each template, 5 μl of 10× PCR reaction buffer, 4 μl of dNTPs (2.5 mM each), 1 μl of outer adaptor primer (AP1, 5′-GTAATACGACTCACTATAGGGC-3′), 1 μl of gene-specific primer (GSP1, 5′-AAAGGAGAAAGAACTCCAGAGGTGTCC-3′), and 0.5 μl of ExTaq polymerase (Takara, Japan). The PCR cycling conditions consisted of seven cycles at 94°C for 25 s and 72°C for 3 min; 32 cycles of 94°C for 25 s and 67°C for 3 min; one final cycle of 67°C for 7 min. The primary PCR products were then diluted and used as a template in a second PCR using a nested adaptor primer (AP2, 5′-ACTATAGGGCACGCGTGGT-3′) and a nested gene-specific primer (GSP2, 5′-CCACAGGCCTTCACTGGGTGCTCGTTT-3′). After the amplification step, the major PCR product was isolated and cloned into pGEM-T Easy vector (Promega, USA) for sequencing.
Construction of the IbCAD1 promoter–GUS fusion and plant transformation
To generate a binary vector for the transformation of tobacco plants, the IbCAD1 promoter region was amplified with gene-specific primers (F, 5′-ACGCGTCGACCTGGTAACAAAACTATTGGAT-3′; R, 5′-CGGGATCCTTTCCTTGTTGCAGGGGGAT-3′). The reverse primers were designed in front of the ATG start codon of the IbCAD1 gene. The underlined sequences, which are SalI and BamHI sites, were introduced at the end of primers in order to facilitate subcloning. The PCR product was then inserted into the SalI/BamHI sites of pBI101 carrying the GUS reporter gene, and the resulting construct was utilized to transform tobacco by Agrobacterium tumefaciens (GV3101) infection (Martinez-Trujillo et al. 2004). The transformants were identified by genomic DNA PCR. Primary transformants were grown to maturity in soil in a greenhouse to produce seeds. The seeds were germinated on MS medium after sterilization in distilled water:chlorox:0.5% Tween 20 (3:2:1). The IbCAD1 promoter activity in transgenic tobacco was analyzed using T2 plants.
Stress and hormone treatments
Sweetpotato plants were grown in a greenhouse at 25°C for 3 weeks and subjected to several stresses. As cold stress, sweetpotato plants in a pot were exposed to 15°C for 24, 48, and 72 h, respectively. For the methyl viologen (MV) treatment, 50 μM of a solution containing 0.125% Triton X-100 was sprayed onto whole plants and the plants incubated for 6, 12, and 24 h, respectively. For the H2O2 treatment, the second and third leaves from the top were removed from plants and incubated in 440 mM hydrogen peroxide (H2O2) solution at 25°C for 12 and 24 h, respectively; sterile water was used as a control for the H2O2 treatment. Wounding stress was performed according to Sasaki et al. (2002). Fully expanded leaves were detached from the plant and immediately cut into pieces with a razor blade. After the midrib had been removed, six pieces from each leaf, from six different leaves (n = 36 leaf pieces), were placed on filter paper moistened with distilled water and incubated for 1, 4, 8, 12, and 24 h, respectively, at 25°C under continuous illumination. Untreated and stress-treated plant samples were collected, frozen in liquid nitrogen, and stored at −70°C for RNA purification.
Four-day-old seedlings were used for the stress treatments of transgenic tobacco plants containing the IbCAD1 promoter. For the cold stress, 4-day-old seedlings in MS medium were exposed to 4°C for 48 h. For the H2O2 stress, transgenic seedlings were transferred to MS medium containing 0.1 M H2O2 at 25°C for 24 h. For the wounding treatment, detached leaves and stems of mature plants were cut into slices and incubated for 24 h at 25°C. For the hormone treatments, 4-day-old tobacco seedlings were transferred to MS medium containing 100 μM abscisic acid (ABA), 1 mM salicylic acid (SA), or 50 μM jasmonic acid (JA) for 24 h or to MS medium containing 10 μM 6-benzylaminopurine (BA) for 72 h. Untreated and hormone-treated plant samples were collected, and crude extracts were prepared for the analysis of GUS activity.
Fluorometric and histochemical GUS assay
The crude plant extract was prepared and used for the quantitative fluorometric assays of GUS activity according to Jefferson et al. (1987) using 4-methylumbelliferyl glucuronide (MUG; Sigma, USA). GUS values were expressed as nmoles of MUG per minute per milligram protein. Protein concentrations were determined by the method of Bradford (1976). The histochemical analysis of GUS activity was performed by incubating plant tissues with 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc, Gold Biotechnology, USA) for 16–24 h at 37°C (Jefferson et al. 1987). Chlorophyll was removed from the tissues by immersion in 70% ethanol. The samples were observed and photographed under a SZ61 zoom stereo microscope (Olympus, Japan) equipped with digital camera system (Canon, USA).
Results
Isolation and sequence analysis of IbCAD1 cDNA
In an earlier study, we constructed an expressed sequence tag (EST) library from sweetpotato cells grown in suspension culture and identified highly abundant and cell culture-specific genes, of which the most abundant and unique gene was CAD (Kim et al. 2006). Here, we have isolated and characterized the full length of the cDNA clone (IbCAD1, GU380306). The IbCAD1 cDNA consisted of 1,355 nucleotides, and the largest open reading frame (ORF) encoded a polypeptide of 357 amino acids. It encoded a 38.5-kDa protein with an isoelectric point of 6.26. The putative polyadenylation signal AATAAT was located 94 bases upstream from the start of the poly(A)+ tail. We found no clear targeting domains within the sequence, suggesting a cytoplasmic location of the protein (WoLF PSORT Prediction; probability cytoplasm 12.0, chloroplast 1.0).
The deduced protein sequence of IbCAD1 was highly homologous to that of other plant CADs identified to date, including strawberry FxaCAD (AF320110, 73.5% identity), parsley ELI-3 (X67817, 71.2% identity), poplar PtSAD (AF273256, 70.2% identity), alfafa MsaCAD1 (AF083332, 67.8% identity), Arabidopsis AtCAD8 (AY302080, 67.6% identity) and AtCAD7 (AY302079, 66.9% identity). All of these CADs clustered with defense-related proteins compared to classical development-related lignifications CADs. The sequence alignment indicated that the IbCAD1 protein possessed all of the structural characteristics of CADs, including the catalytic Zn1 binding sequence [GHE(X)2G(X)5G(X)2V], the Zn2 binding sequence [GD(X)9,10C(X)2C(X)2C(X)7C], and the coenzyme (NADPH) binding domain GXG(X)2G (McKie et al. 1993) (Fig. 1). The Zn1 and Zn2 binding sequences in IbCAD1 were located at amino acid residues 72–86 and 92–118, respectively. A coenzyme binding site located at residues 188–193 and structural zinc ion coordinating residues, Cys 104, Cys 107, Cys 110, and Cys 118, have also been identified (Vallee and Aulds 1990; Grima-Pettenati et al. 1993). His 73 and Cys 163 may be involved as a catalytic zinc ion coordinating residue and thus play an important role in active zinc binding (Vallee and Aulds 1990; Knight et al. 1992). Gly 299 is a key determinant residue of substrate specificity (Bomati and Noel 2005).
Fig. 1.

Alignment of amino acid sequences of IbCAD1 and other plant CADs. Conserved important regions identified previously are marked as follows: inverted filled triangle denotes catalytic zinc ion coordinating residue, filled circle denotes structural Zn ion coordinating residue, star denotes key residues for substrate specificity. Locations of the Zn1, Zn2, and NADPH binding domains are shown in boxes. The alignment was performed with the ClustalX and GENEDOC software programs
To estimate the genomic constitution of the IbCAD1 gene, we analyzed genomic DNA isolated from sweetpotato cells grown in suspension culture by Southern blot analysis (Fig. 2). Three major bands and several minor ones were identified on the Southern blots, indicating that IbCAD1 belongs to a member of a multigenic family. This was an expected result as, with the exception of gymnosperm species, all of the CAD genes isolated from plant species to date have been present as members of a multigene family (MacKay et al. 1995).
Fig. 2.
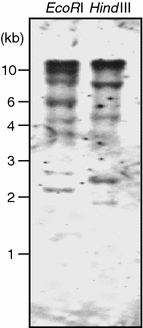
Southern hybridization analysis of the IbCAD1 gene. Genomic DNA (10 μg) purified from sweetpotato suspension cells was digested with EcoRI and HindIII, respectively, and then separated in a 0.8% agarose gel. The full length of the IbCAD1 cDNA was used as a probe. Migration position of the DNA size standards are shown at left
Effects of environmental stresses on the transcription levels of the IbCAD1 gene
In an earlier publication, we reported that IbCAD1 was highly expressed in the log and stationary phases of sweetpotato suspension-cultured cells (Kim et al. 2006). Here, we investigated the tissue specificity and stress response of IbCAD1 in sweetpotato plants using a comparative quantitative RT-PCR analysis. The IbCAD1 gene was most strongly expressed in the roots, less actively in the petioles, and only rarely expressed in the leaves of sweetpotato plants (Fig. 3a). To study the response to environmental stresses, we subjected 3-week-old sweetpotato plants to cold, MV of a superoxide-generating herbicide, H2O2, and wounding and then analyzed the time-course accumulations of the IbCAD1 transcript in sweetpotato leaves. The IbCAD1 transcript was highly induced at 72 and 12 h after the start of the cold stress and MV treatments, respectively, and at 12 and 24 h after the start of the H2O2 treatment. The response of the IbCAD1 gene to wounding was very fast (1 h after wounding), and the transcription levels remained high until 24 h post-treatment initiation (Fig. 3b). Taken together, these results show a root-specific expression of the IbCAD1 gene and that its transcription can be highly induced by various stress treatments.
Fig. 3.

Expression of the IbCAD1 gene in sweetpotato. a Tissue-specific expression of IbCAD1. Total RNA was isolated from leaves (L), petioles (P) and roots (R) of sweetpotato, and RT-PCR was performed. Tubulin was used as an internal control. b IbCAD1 expression in response to various stresses. Sweetpotato plants were subjected to cold, methyl viologen (MV), wounding, and H2O2 treatments, respectively. The cold treatment consisted of exposing the plants to 15°C. For the MV treatment, a 50 μM solution containing 0.125% Triton X-100 was sprayed onto whole plants. The wounding treatment consisted of cutting sweetpotato leaves into small pieces and incubating these at 25°C. For the H2O2 treatment, two or three leaves from the top of the growing tip were removed from sweetpotato plants and incubated in a 440 mM H2O2 solution. RNA was isolated from leaves and subjected to RT-PCR analysis. Tubulin was used as an internal control
Nucleotide sequence analysis of the IbCAD1 promoter
To isolate a 5′ flanking region of the IbCAD1 gene, we designed specific primers in the ORF region of the IbCAD1 cDNA and performed PCR-based genome walking. A 1,260-bp product was obtained, and the nucleotide sequence of the product was compared with that of the IbCAD1 cDNA. This PCR product encoded 16 amino acids, including an ATG start codon, and corresponded exactly with the 5′-UTR and N-terminal region of the IbCAD1 cDNA. The nucleotide site that corresponded in sequence to the end of 5′-UTR of the IbCAD1 cDNA was designated as +1. A putative TATA box (−65 to −58) and CAAT box (−1043 to −1040, −831 to −828, −138 to −135, and −27 to −24), which act as general signals for eukaryotic gene expression, were observed in IbCAD1 promoter.
A promoter motif search carried out using the software program PLACE to define putative cis-elements (Higo et al. 1999) (Fig. 4) revealed that the IbCAD1 promoter contained putative regulatory elements corresponding to known cis-elements of eukaryotic genes. One of the most abundant motifs detected was the nuclear response regulator (ARR) binding site (NGATT), which is involved in cytokinin-mediated regulation during root differentiation (Yokoyama et al. 2007). Many DOF (AAAG) binding sites were also found. DOF has been found to influence phenylpropanoid metabolism in an environmental and tissue-specific manner (Skirycz et al. 2007). ACGT (ACGT) and GT-1 (GAAAAA/GRWAAW) motifs were also present, both of which are required for gene expression during dehydration stress (Simpson et al. 2003). The stress-responsive MYB and MYC elements (Abe et al. 1997) and the W box (TGACY) and WRKY binding sites (TGAC), all of which are involved in pathogen and wounding, were also present. These results indicate that the transcriptional regulation of IbCAD1 gene may be involved in root development and in the response to various stresses.
Fig. 4.

Nucleotide sequence of the promoter region of IbCAD1 gene. The nucleotide site that corresponded in sequence to the end of 5′-UTR of the IbCAD1 cDNA was designated as +1. The TATA box (TATATAAA) is underlined and the start codon is double underlined. The dotted underline indicates the CAAT box. Several regulatory cis-elements are represented. GSP1 and GSP2 were primers used for PCR-based genome walking
Spatial and temporal regulation of the IbCAD1 gene during development
Transgenic tobacco plants harboring the GUS reporter gene fused to the IbCAD1 promoter were constructed and three independent transgenic lines analyzed. IbCAD1 promoter activity in various tissues during different growth stages was investigated in in vitro-grown seedlings and in mature plants grown in soil using the quantitative GUS assay (Fig. 5A) and by histochemical GUS staining (Fig. 5B). These tests showed that the activity of the IbCAD1 promoter did not change significantly in the cotyledons during seedling development, while that in the roots increased gradually with root maturation (Fig. 5A). Promoter activity was much higher in the roots than in the cotyledons. The results of the histochemical assay revealed that GUS staining was absent in the cotyledons but that it became stronger in the root tips with advancing seedling development (Fig. 5Ba–d). Strong GUS staining was observed around the sites where lateral roots were initiated from the primary roots (Fig. 5Be).
Fig. 5.

GUS activity and histochemical localization of the IbCAD1 promoter at different developmental stages of transgenic tobacco plants. A Transgenic tobacco seedlings were germinated on MS medium and collected at 5, 8, and 12 days after germination (DAG). GUS activity was measured in the cotyledons and roots, respectively. The data represent the average value ± SD from independent experiments (n = 40). Fully developed leaves, petioles, upper stems, and root materials were collected from 4- to 8-week-old mature plants grown in soil and analyzed for GUS activity. The data represent the average value ± SD from independent experiments (n = 6). B (a) Seedling with fully developed cotyledon at 5 DAG; (b) seedling with fully matured cotyledons at 8 DAG; (c) seedling with expanding secondary leaves and initiating lateral roots at 12 DAG; (d) root tip of seedling at 12 DAG; (e) initiation regions of lateral roots at 12 DAG; (f, g) root of 4-week-old plant; (h) root of 8-week-old plant; (i) transverse section of stem; (j) longitudinal section of stem; (k) leaf of 4-week-old plant; (l) stigma; (m) anther; (n) filament; (o) pollen grains
IbCAD1 promoter activity was investigated in the leaves, petioles, stems, and roots of mature plants grown in soil. It was relatively low in 4-week-old mature plants, except for the roots. GUS activity increased only slightly in the leaves, petioles, and stems with increasing plant age. In contrast, there was a dramatic increase in GUS activity in the roots as the plant matured (Fig. 5A), with GUS activity in the roots of 8-week-old plant roots being 2.9- to 6.8-fold higher than that of 4-week-old plant roots. The lateral root initiation site was strongly stained in 4-week-old plants (Fig. 5Bf–g), while much stronger staining was observed in lateral roots and primary roots of 8-week-old plants (Fig. 5Bh). Weak GUS staining was observed in the fibers, phloem, and xylem of the stem and in the midrib and veins of leaves (Fig. 5Bi–k). In the reproductive tissues, the anther, trichomes in the filament, and pollen grains all showed strong GUS staining, while the stigma showed weak GUS staining (Fig. 5Bl–o). Based on these results, the IbCAD1 promoter directs strong expression in the meristematic tissue of the root, but weak expression in the lignified tissue of the leaves and stem.
Up-regulation of IbCAD1 gene expression by both biotic and abiotic stresses
The IbCAD1 promoter has Myb and Myc responsive elements that respond to dehydration stress as well as the W box and WRKY recognition sequence of the wounding responsive element. To investigate the transcriptional regulation of the IbCAD1 gene in response to various stresses, we examined the expression of the GUS gene driven by the IbCAD1 promoter under conditions of cold (abiotic stress), wounding (biotic stress), and H2O2 (reactive oxygen species, ROS). The production of ROS is a common signal which triggers the downstream responses of plants to biotic and abiotic stresses. The cold treatment (4°C) induced an increase in promoter activity of the cotyledons (2.2- to 13.2-fold) and roots (1.2- to 3.8-fold) compared with control plants (Fig. 6A). Although GUS activity was higher in the cotyledons than in the roots following the cold treatment, the GUS activity value in the cotyledons was below the detection limit of the analysis for GUS staining. Thus, following the cold stress treatment, strong GUS staining was only observed in the roots. When seedlings were treated with H2O2, IbCAD1 promoter activity was highly induced compared to control plants—very strongly in the cotyledons (about 15.3- to 47.4-fold) and significantly less in the roots (2.5- to 4.2-fold) (Fig. 6B). Histochemical analysis showed strong GUS staining in cotyledons. These results show that the IbCAD1 promoter was highly activated under these stress conditions, with the induction of promoter activity being much higher in the cotyledons than in the roots.
Fig. 6.
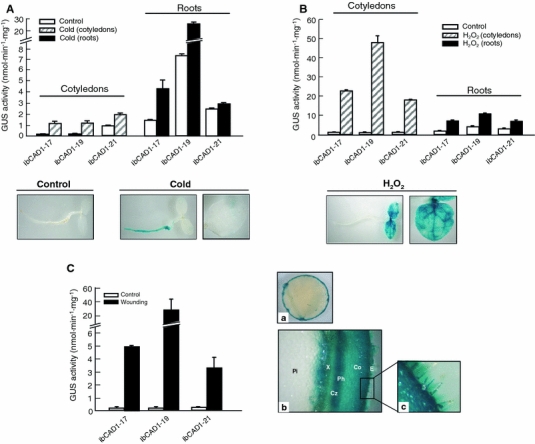
IbCAD1 promoter activity in transgenic tobacco plants following treatment with cold, H2O2, and wounding stresses. Four-day-old seedlings were exposed to 4°C for 48 h in the cold treatment (A) or transferred to MS medium containing 0.1 M H2O2 for 24 h (B). Cotyledons and roots of each transgenic line were collected separately. After the respective stress treatment, the tissues were homogenized and measured for GUS activity. The data represent the average ± SD from independent experiments (n = 40). C For the wounding stress, the leaves of tobacco plantlets were injured by punching and incubated on moistened filter paper. After 24 h, the tissues were homogenized and the GUS activity measured. The data represent the average ± SD of six transgenic plants per line. For the histochemical analysis after the wounding stress, the leaves and stems of transgenic plants were cut and placed on moistened filter paper for 30 and 4 h, respectively, and then stained. (a) Wounded leaves; (b) wounded stem (hand-sectioned); (c) trichomes of stem. Pi Pith, X xylem, Cz cambial zone, Ph phloem, Co cortex, E epidermis
To provoke a wounding response, we wounded detached leaves and stems from 6-week-old mature plants grown in soil by punching or cutting and then incubated the leaf pieces on moistened filter paper. Relative to intact tissues, the GUS activity directed by the IbCAD1 promoter was dramatically increased (15.2- to 174.1-fold) in wounded tissues (Fig. 6C). As shown in Fig. 6Ca, the wounding response was highly localized. Strong GUS staining was also observed at wounded site in the stem, and a histochemical study of cross-sections of the stem revealed that the IbCAD1 promoter was strongly expressed in the xylem, phloem, cortex, and epidermis as well as in the trichomes (Fig. 6Cb, c). Taken together, these results show that the IbCAD1 promoter was highly activated by both biotic and abiotic stresses and that the stress response was tissue-specific.
Effects of hormones on the IbCAD1 promoter activity
The presence of several hormone-responsive cis-regulatory elements suggested the possibility that IbCAD1 gene expression may be controlled by hormones. Many wound-induced genes have been reported to be JA-inducible (Sasaki et al. 2002). To determine whether the IbCAD1 gene mediates hormone-responsive expression, we investigated IbCAD1 promoter activity in 4-day-old transgenic tobacco seedlings treated with ABA, BA, JA and SA by measuring the levels of GUS activity (Fig. 7). Following the ABA and BA treatments, IbCAD1 promoter activity increased by approximately 1.1- to 2.6-fold and 1.1- to 2.5-fold, respectively (Fig. 7a, b); in comparison, treatments with SA and JA separately resulted in a dramatic increase in IbCAD1 promoter activity, 9.1- to 15.4-fold and 6.4- to 8.5-fold, respectively (Fig. 7c, d). These results clearly demonstrate that the expression of the IbCAD1 gene is induced by various hormones. Among them, JA and SA are the strongest inducer of IbCAD1 gene at the seedling stage.
Fig. 7.
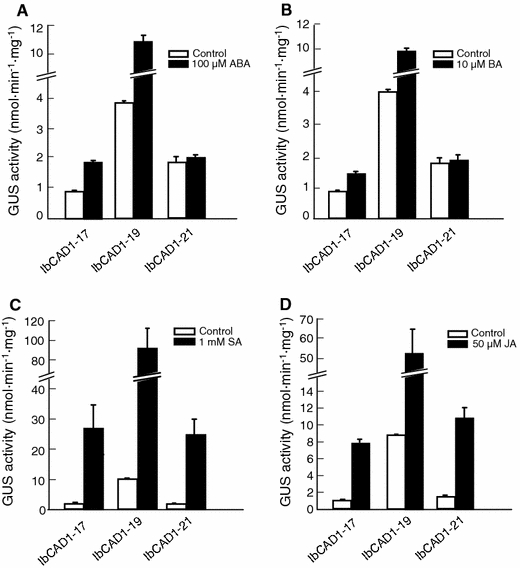
IbCAD1 promoter activity in transgenic tobacco plants treated with different hormones. Four-day-old seedlings were treated with 100 μM abscisic acid (ABA) (a), 1 mM salicylic acid (SA) (c), and 50 μM jasmonic acid (JA) (d) for 24 h, or with 10 μM 6-benzylaminopurine (BA) (b) for 72 h. The data represent the average ± SD from independent experiments (n = 40)
Discussion
Cinnamyl alcohol dehydrogenase (EC 1.1.1.195) is the last enzyme in the lignin biosynthetic pathway, and it mediates the reduction of coniferaldehyde, p-coumaraldehyde, and sinapaldehyde into guaiacyl, p-coumaryl, and syringyl monolignols, respectively. Lignification is a tightly regulated and dynamic process subject to modulation at different levels during normal development and in response to different stresses (Boudet 2000). In this study, we isolated the CAD cDNA (IbCAD1) from sweetpotato, which can be phylogenetically categorized as belonging to the family of defense-related CADs. The Gly 299 residue in IbCAD1 is a key determinant of substrate specificity and is a sinapaldehyde-specific residue according to Bomati and Noel (2005). These researchers reported that the Gly residue at the base of the active site of PtSAD and FxaCAD is responsible for the high specificity of these proteins for the substrate sinapaldehyde. PtSAD is required for the biosynthesis of syringyl lignin (Li et al. 2001). Classical CAD enzymes involved in developmental lignification, such as AtCAD4, AtCAD5, and EgCAD2, have Phe instead of Gly in the active site for substrate specificity. AtCAD5 is able to use all substrates effectively, while AtCAD4 shows a very poor affinity for sinapaldehyde (Kim et al. 2004). Thus, IbCAD1 may have high affinity for the substrate sinapaldehyde based on its structure and, consequently, be involved in the defense-related process.
Angiosperm CAD is encoded by a multigenic family consisting of members thought to have distinct roles, while gymnosperm CAD is encoded by a single gene (MacKay et al. 1995; Zinser et al. 1998). Different CAD genes in angiosperms may be expressed in different tissues or at different stages during growth and development (Kim et al. 2007; Barakat et al. 2009) in response to the type of lignin required. Several authors have suggested that the chemical composition of defense lignin is different to that of developmental lignin. For example, syringyl lignin is accumulated in the plant cell wall during the hypersensitive resistance response in wheat (Menden et al. 2007) and sinapaldehyde-specific CAD activity is activated in F. oxysporum-treated flax cells (Hano et al. 2006). While much is known on how CAD is induced by biotic stress, such as fungal elicitor (Dixon and Paiva 1995; Brill et al. 1999), relatively little information is available on the role of CAD in relation to lignification under conditions of abiotic stress. We have investigated the transcriptional regulation of the IbCAD1 gene in response to various stresses. RT-PCR analysis shows that IbCAD1 gene expression in sweetpotato was strongly induced by both abiotic stress (cold) and biotic stress (wounding), including ROS (MV and H2O2). ROS is a common signal that acts as a trigger of the downstream stress response. Fujita et al. (2006) reported that ROS may mediate crosstalk between biotic and abiotic stress-responsive gene-expression networks. Our RT-PCR results are consistent with those of our promoter analysis of IbCAD1. The activity of the IbCAD1 promoter was induced by the cold and H2O2 treatments in this study, but the stress response was quite different in different tissues; for example, at the seedling stage, the cotyledons were more sensitive than the roots to IbCAD1 promoter induction. Among the stressors tested, wounding was found to be the strongest inducer of IbCAD1, and the wounding response was highly localized. These data suggest that the expression of the IbCAD1 gene is regulated in a tissue-specific manner under conditions of biotic and abiotic stresses.
Hormone signaling pathways regulated by ABA, SA, JA, and ethylene as well as the ROS signaling pathway all play key roles in the crosstalk between biotic and abiotic stress responses (Fujita et al. 2006). ABA is extensively involved in the plant’s response to abiotic stresses, such as drought, low temperature and osmotic stress, and also regulates a variety of growth and developmental processes. In contrast, SA and JA play central roles in biotic stress signaling following pathogen infection and wounding. We identified several hormone-responsive cis-regulatory elements in the IbCAD1 promoter region, such as ABRE (ABA) (Yamaguchi-Shinozaki and Shinozaki 2005), ARR binding site (cytokinin) (Yokoyama et al. 2007), as-1 element (auxin, SA) (Niggeweg et al. 2000), MYB responsive element, MYC responsive element (Yamaguchi-Shinozaki and Shinozaki 2005), and GARE (gibberellin) (Sutoh and Yamaguchi 2003). AtMYC2, which was identified as an activator involved in the ABA-mediated drought stress signaling pathway, upregulates the expression of genes that are involved in the JA-mediated wounding response stress signaling pathway (Abe et al. 2003; Boter et al. 2004; Lorenzo et al. 2004). Transgenic plants that overexpress both AtMYC2 and AtMYB2 show a greater sensitivity to ABA and enhanced osmotic stress tolerance (Abe et al. 2003). When we tested the GUS activity driven by the IbCAD1 promoter, we found that IbCAD1 promoter activity was enhanced when the plants were treated with various plant hormones, being most strongly induced by JA and SA, respectively. The IbCAD1 promoter was also induced by ABA and cytokinin (BA), but to a lesser degree. Taken together, these results indicate that IbCAD1 may function through a JA- and SA-mediated wounding response and an ABA-mediated cold response.
The expression of the IbCAD1 gene was regulated during growth and development in a tissue-specific manner. Our histochemical analysis showed that weak GUS expression was observed in the xylem, phloem, and fiber of stems, and in the midrib and veins of mature leaves, which are lignified tissues of the plant vascular system. However, the IbCAD1 gene was strongly expressed in the area of the initiation sites for lateral roots developing from primary roots as well as in root tips—but not in the vascular cylinder of lignified root tissue. It has been reported that there is no lignification in root meristem cells, which is primarily involved in the constant progression of cell division (de Obeso et al. 2003). GUS expression was also strong in anther and pollen. We have previously shown that IbCAD1 is the most abundant gene in the EST library of cells grown in suspension culture and that it is highly expressed at the actively dividing-log phase. In light of these results, the expression of the IbCAD1 promoter indicates a new role of CAD protein in lateral root formation, cell-wall synthesis, and the promotion of cell division that is unrelated to constitutive lignin formation. We suggest that IbCAD1 may also be involved in lignifications induced by both abiotic and abiotic stresses and in tissue-specific developmental lignification. Further analysis of the IbCAD1 gene will provide an insight into the signaling pathway involved in the regulation of biotic and abiotic stresses in lignin biosynthesis as well as into the role of CAD protein during cell division.
Acknowledgments
This work was supported by grants (No. 20080401034022 and 20100301061032) from Biogreen 21 Program funded by the Rural Development Administration, Republic of Korea, and a grant from the 2006 Inje University research grant.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadmorna A, Choi A, Plakkat U, Diloreto DS, Yellanki P, Carlson JE. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization and expression. Plant Biol. 2009;9:26–40. doi: 10.1186/1471-2229-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bomati EK, Noel JP. Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell. 2005;17:1598–1611. doi: 10.1105/tpc.104.029983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruiz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004;18:1577–1591. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet AM. Lignins and lignification: selected issues. Plant Physiol Biochem. 2000;38:81–96. doi: 10.1016/S0981-9428(00)00166-2. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brill EM, Abrahams S, Hayes CM, Jenkins CLD, Watson JM. Molecular characterization and expression of a wound-inducible cDNA encoding a novel cinnamyl alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.) Plant Mol Biol. 1999;41:279–291. doi: 10.1023/A:1006381630494. [DOI] [PubMed] [Google Scholar]
- de Obeso M, Caparrós-Ruiz D, Vignols F, Puigdomènech P, Rigau J. Characterization of maize peroxidases having differential patterns of mRNA accumulation in relation to lignifying tissues. Gene. 2003;309:23–33. doi: 10.1016/S0378-1119(03)00462-1. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Goffner D, Joffroy I, Grima-Pettenati J, Halpin C, Knight ME, Schuch W, Boudet AM. Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase from Eucalyptus xylem. Planta. 1992;188:48–53. doi: 10.1007/BF01160711. [DOI] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Eudes A, MacKay J, Jouanin L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol Biochem. 2003;41:677–687. doi: 10.1016/S0981-9428(03)00095-0. [DOI] [Google Scholar]
- Grima-Pettenati J, Feuillet C, Goffner D, Borderies G, Boudet AM. Molecular cloning and expression of a Eucalyptus gunnii cDNA clone encoding cinnamyl alcohol dehydrogenase. Plant Mol Bol. 1993;221:1085–1095. doi: 10.1007/BF00023605. [DOI] [PubMed] [Google Scholar]
- Hano C, Addi M, Bensaddek L, Crônier D, Baltora-Rosset S, Doussot J, Maury S, Mesnard F, Chabbert B, Hawkins S, Lainé E, Lamblin F. Differential accumulation of monolignol-derived compounds in elicited flax (Linum usitatissimum) cell suspension cultures. Planta. 2006;223:975–989. doi: 10.1007/s00425-005-0156-1. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acid Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl E. Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar KL, Moinuddin SGA, Cardenas CL, Davin LB, Kang CH, Lewis NG. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1455–1460. doi: 10.1073/pnas.0307987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Hur CG, Shin YH, Bae JM, Song YS, Huh GH. Identification and characterization of highly expressed genes in suspension-cultured cells of sweetpotato. J Plant Biol. 2006;49:364–370. doi: 10.1007/BF03178813. [DOI] [Google Scholar]
- Kim SJ, Kim KW, Cho MH, Franceschi VR, Davin LB, Lewis NG. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry. 2007;68:1957–1974. doi: 10.1016/j.phytochem.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Knight ME, Halpin C, Schuch W. Identification and characterization of cDNA clones encoding cinnamyl alcohol dehydrogenase from tobacco. Plant Mol Biol. 1992;19:793–801. doi: 10.1007/BF00027075. [DOI] [PubMed] [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL. The last step of syringyl monolignol biosynthesis in angiosperm is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell. 2001;13:1567–1585. doi: 10.1105/tpc.13.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D, Lidgett A, Mclnnes R, Huxley H, Jones E, Mahoney N. Isolation and characterization of three cinnamyl alcohol dehydrogenase homologue cDNAs from perennial ryegrass (Lolium perenne L.) J Plant Physiol. 2002;159:653–660. doi: 10.1078/0176-1617-0776. [DOI] [Google Scholar]
- MacKay JJ, Liu W, Whetten R, Sederoff RR, O’Malley DM. Genetic analysis of cinnamyl alcohol dehydrogenase in loblolly pine: single gene inheritance, molecular characterization and evolution. Mol Gen Genet. 1995;247:537–545. doi: 10.1007/BF00290344. [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce JL, Herrera-Estrella L. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol Biol Report. 2004;22:63–70. doi: 10.1007/BF02773350. [DOI] [Google Scholar]
- McKie JH, Jaouhari R, Douglas KT, Goffner D, Feuillet C, Grima-Pettenati J, Boudet AM, Baltas M, Gorrichon L. A molecular model for cinnamyl alcohol dehydrogenase, a plant aromatic alcohol dehydrogenase involved in lignification. Biochim Biophys Acta. 1993;1202:61–69. doi: 10.1016/0167-4838(93)90063-w. [DOI] [PubMed] [Google Scholar]
- Menden B, Kohlhoff M, Moerschbacher BM. Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry. 2007;68:513–520. doi: 10.1016/j.phytochem.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assay with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Niggeweg R, Thurow C, Weigel R, Pfitzer U, Gats C. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Hiraga S, Ito H, Seo S, Matsui H, Ohashi Y. A wound-inducible tobacco peroxidase gene expresses preferentially in the vascular system. Plant Cell Physiol. 2002;43:108–117. doi: 10.1093/pcp/pcf013. [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Seguin A, Lapierre C, Jouanin L. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 2003;132:848–860. doi: 10.1104/pp.103.021048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A. Cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell. 2005;17:2059–2076. doi: 10.1105/tpc.105.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Two different novel cis-acting elements of erd1, a clpA homologues Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003;33:259–270. doi: 10.1046/j.1365-313X.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Jozeczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-Roeber B. Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol. 2007;175:425–438. doi: 10.1111/j.1469-8137.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Sutoh K, Yamaguchi D. Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 2003;34:635–645. doi: 10.1046/j.1365-313X.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- Tronchet M, Balagué C, Kroj T, Jouanin L, Roby D. Cinnamyl alcohol dehydrogenase-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol Plant Pathol. 2010;11:83–92. doi: 10.1111/j.1364-3703.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee BL, Aulds DS. Zinc coordination, function and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Walter MH, Grima-Pettenati J, Grand C, Boudet AM, Lamb C. Cinnamyl alcohol dehydrogenase, a molecular marker specific for lignin biosynthesis: cDNA cloning and mRNA induction by fungal elicitor. Proc Natl Acad Sci USA. 1988;85:5546–5550. doi: 10.1073/pnas.85.15.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano YI, Tajima Y, Imamura A, Sakakibara H, Mizuno T. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z. GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl alcohol dehydrogenase in rice. Plant Physiol. 2006;140:972–983. doi: 10.1104/pp.105.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser C, Ernst D, Sandermann H. Induction of stilbene synthase and cinnamyl alcohol dehydrogenase mRNAs in Scots pine (Pinus sylvestris L.) seedlings. Planta. 1998;204:169–176. doi: 10.1007/s004250050243. [DOI] [Google Scholar]


