Abstract
Using ammonia monooxygenase α-subunit (amoA) gene and 16S rRNA gene, the community structure and abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in a nitrogen-removing reactor, which was operated for five phases, were characterized and quantified by cloning, terminal restriction fragment length polymorphism (T-RFLP), and quantitative polymerase chain reaction (qPCR). The results suggested that the dominant AOB in the reactor fell to the genus Nitrosomonas, while the dominant AOA belonged to Crenarchaeotal Group I.1a in phylum Crenarchaeota. Real-time PCR results demonstrated that the levels of AOB amoA varied from 2.9 × 103 to 2.3 × 105 copies per nanogram DNA, greatly (about 60 times) higher than those of AOA, which ranged from 1.7 × 102 to 3.8 × 103 copies per nanogram DNA. This indicated the possible leading role of AOB in the nitrification process in this study. T-RFLP results showed that the AOB community structure significantly shifted in different phases while AOA only showed one major peak for all the phases. The analyses also suggested that the AOB community was more sensitive than that of AOA to operational conditions, such as ammonia loading and dissolved oxygen.
Keywords: Ammonia monooxygenase α-subunit (amoA) gene, Ammonia-oxidizing archaea (AOA), Ammonia-oxidizing bacteria (AOB), T-RFLP, qPCR
Introduction
Nitrification, the key and often rate-limiting step in nitrogen removal, includes two steps, i.e., oxidation of ammonia to nitrite catalyzed by ammonia-oxidizing bacteria (AOB), and nitrite to nitrate by nitrite-oxidizing bacteria. AOB are thought to be largely responsible for the oxidation of ammonia to nitrite in wastewater treatment plants (WWTPs) and natural environments. In particular, members of the betaproteobacterial genera Nitrosomonas and Nitrosospira are considered as the most important AOB in activated sludge (Purkhold et al. 2000).
Recently, however, it was discovered that autotrophic oxidation of ammonia is not restricted to the Bacteria domain (Treusch et al. 2005; Könneke et al. 2005). The first cultivated representative of ammonia-oxidizing archaea (AOA), named Nitrosopumilus maritimus, which, like AOB, grows chemolithoautotrophically by oxidizing ammonia to nitrite, was isolated from a marine aquarium tank (Könneke et al. 2005). Both AOA and AOB have ammonia monooxygenase (AMO), one of the key enzymes responsible for the conversion of ammonia to nitrite. AMO is composed of three subunits encoded by genes of amoA, amoB, and amoC (Klotz et al. 1997). Among these three genes, the amoA gene has been much more extensively used for the study of ammonia oxidizers and has a well-established database, compared with amoB and amoC (Francis et al. 2005; Okano et al. 2004).
By using culture-independent DNA-based techniques, AOA has been found quite diverse and abundant in various natural environments, such as marine sediment (Francis et al. 2005; Park et al. 2008), soil (He et al. 2007), estuary (Beman and Francis 2006), and seawater (Coolen et al. 2007). So far, most studies on AOA have been focused on their diversity and quantity in various natural environments, revealing its high diversity and widespread distribution. However, only very limited works on the presence of AOA in wastewater treatment reactors/plants have been done (Park et al. 2006; Zhang et al. 2009). Most studies of nitrifying activated sludge only focused on the presence and abundance of AOB (Dionisi et al. 2002; Geets et al. 2007). So far, there was only one new report to indicate that AOB was predominant rather than AOA in targeted full-scale bioreactors (Wells et al. 2009). Lacking more data, it remains unknown whether it will be the same or not in other bioreactors.
AOB are highly sensitive to several environmental factors, such as temperature, salinity, pH, dissolved oxygen (DO), ammonium concentration, and hydraulic retention time (HRT; Limpiyakorn et al. 2007; Lydmark et al. 2007). Any improper condition may result in nitrification failure in wastewater treatment. Although the study on the factors affecting AOA is quite limited, it is believed that AOA responds to different environmental conditions (Erguder et al. 2009). Therefore, knowledge on the diversity and dynamics of AOA and AOB in wastewater treatment will be helpful to improve the reactor performance.
In this study, a laboratory-scale nitrogen-removing reactor was carefully controlled to gradually change the running conditions (DO, ammonium concentration, and HRT) in five phases in order to investigate the response of AOA and AOB. Using culture-independent DNA-based approaches, the goals of this study are (1) to investigate the diversity of AOA and AOB, as well as the total archaeal and bacterial community based on clone libraries of 16S rDNA and amoA, (2) to monitor changes of composition of AOA/AOB populations under different running conditions using terminal restriction fragment length polymorphism (T-RFLP), and (3) to quantify the AOA/AOB amoA gene copies using quantitative polymerase chain reaction (qPCR).
Materials and methods
Reactor operation
Nitrifying activated sludge was collected from Hong Kong Shatin sewage treatment plant, which treats 150,000 m3 saline sewage containing an average of 6,000 mg Cl−/L, 300 mg COD/L, and 20 mg NH+4–N/L, and used as the sludge seed for an automatically controlled continuous stirred-tank reactor (CSTR) of 2.6 L (working volume) followed by a cylindered sedimentation separation unit of 300 ml (length 15 cm and diameter 9 cm). The influent of the reactor was synthesized by deionized water (67%) and seawater (33%) to simulate the salinity of sewage in Hong Kong. NH4Cl was added at different concentrations in the five phases. For each gram of NH+4-N in the synthetic wastewater, 7 g of NaHCO3 and 0.07 g of phosphorus were added. Table 1 shows the detailed operational conditions and the wastewater compositions in five different phases. The pH was kept at 7.0 by adding NaHCO3 solution. DO was controlled by the combination of aeration and stirring. Temperature was 23 ± 2°C throughout the whole operation. Mixed liquor suspended solid (MLSS), nitrate, nitrite, and ammonium were measured accordingly to the standard method (APHA 2005).
Table 1.
Operational conditions for nitrogen-removing CSTR
| phase I | phase II | phase III | phase IV | phase V | |
|---|---|---|---|---|---|
| Time range (days) | 1–45 | 46–86 | 87–142 | 143–204 | 205–286 |
| Feed flow rate (L day−1) | 5 | 5 | 5 | 5 | 7.5 |
| HRT (h) | 12.5 | 12.5 | 12.5 | 12.5 | 8.3 |
| Feed nitrogen concentration (mg NH+4–N L−1) | 100 | 100 | 100 | 200 | 200 |
| Feed COD concentration (mg COD L−1) | 100 | 100 | 0 | 0 | 0 |
| DO (mg L−1) | 1.0 | 0.5 | 0.5 | 0.5 | 0.5 |
| Nitrogen loading rate (mg NH+4–N L−1 day−1) | 192 | 192 | 192 | 384 | 576 |
| Ammonium-oxidizing rate (g N g SS−1 day−1) | No data | No data | 0.13 | 0.21 | 0.26 |
| Nitrogen removal rate (g N g SS−1 day−1) | No data | No data | 0.09 | 0.17 | 0.20 |
| MLSS concentration (g L−1) | 1.1 | 1.7 | 1.5 | 1.8 | 2.2 |
DNA extraction and PCR
Genomic DNA was extracted from 1.5 ml of sludge sample using a FastDNA® SPIN Kit for Soil (MP Biomedicals, LLC, Illkirch, France) following the instruction of the producer. The amount of DNA was determined by NanoDrop® Spectrophotometer ND-1000 (Thermo Fisher Scientific, USA). The primers used were listed in Table 2. The PCR was performed in a total volume of 25 µl containing 20 ng of DNA template, 1 U of AmpliTaq® DNA polymerase (Applied Biosystem, Branchburg, USA), 0.1 µM of each primer, and 12.5 µl FailSafe™ PCR Premix F (EPICENTRE Biotechnologies, Madison, USA). Thermal cycling was carried out by an initial denaturation step at 94°C for 4 min. The major cycling program for each primer set was listed in Table 2. The presence and sizes of the PCR amplification products were determined by agarose (1%) gel electrophoresis.
Table 2.
Primers used for PCR amplification
| Target gene | Primer | Sequence (5′–3′) | PCR program | Reference | |
|---|---|---|---|---|---|
| 16S rRNA | Bacterial | EBU8F | AGAGTTTGATCMTGGCTCAG | (94°C, 60 s; 55°C, 45 s; 72°C, 60 s) × 30 | Heuer et al. 1997 |
| UNIV1392R | ACGGGCGGTGTGTRC | Ferris et al. 1996 | |||
| Archaeal | ARC23F | TGCAGAYCTGGTYGATYCTGCC | (94°C, 45 s; 56°C, 45 s; 72°C, 60 s) × 35 | Burggraf et al. 1991 | |
| ARC934R | GTGCTCCCCGCCAATTCCT | Giovannoni et al. 1998 | |||
| amoA | AOA | cren amo_F | ATGGTCTGGCTAAGACGMTGTA | (94°C, 45 s; 55°C, 45 s; 72°C, 45 s) × 35 | Hallam et al. 2006 |
| amoARa | GCGGCCATCCATCTGTATGT | Francis et al. 2005 | |||
| AOB | amoA-1F | GGGGTTTCTACTGGTGGT | (94°C, 60 s; 55°C, 45 s; 72°C, 60 s) × 40 | Rotthauwe et al. 1997 | |
| amoA-2Ra | CCCCTCKGSAAAGCCTTCTTC | Rotthauwe et al. 1997 | |||
aPrimers used for T-RFLP analysis were labeled by Hex
Cloning and sequencing
The PCR products were purified by using PCRquick-spin™ PCR Products Purification Kit (iNtRON Biotechnology, Seongnam-Si, Korea). Then the PCR products were cloned using InsT/Aclone™ PCR Cloning Kit (Fermentas, Vilnius, Lithuania). White colonies were picked for insert screening using PCR with M13F and M13R primers, and then grouped by RFLP with restriction enzymes RsaI and/or MspI. For each RFLP group, representatives were selected and sequenced. The sequencing was performed using M13F primer on ABI 3730xl capillary sequencers (PE Applied Biosystems, Foster City, USA).
Phylogenetic analyses
The nucleotide sequences were compared with those from the GenBank using BLASTn in the National Center for the Biotechnology Information server (http://www.ncbi.nlm.nih.gov). The sequences in this study and the reference sequences retrieved from the GenBank were aligned by ARB (http://www.arb-home.de; Ludwig et al. 2004) to construct phylogenetic trees using the neighbor-joining method (based on Jukes–Cantor corrected distance). Bootstrap value was calculated based on 1,000 replications.
Quantitative PCR
qPCR was performed using an iCycler IQ System (Bio-Rad, Hercules, USA), two replicates for each sample. For AOA, the PCR was performed in a total volume of 30 µl containing 15 µl of FailSafe™ PCR Premix F, 5 µl of DNA template, 0.1 µM of each primer, 1.5 U of AmpliTaq® DNA polymerase, 15 mM MgCl2, and 0.5× SYBR® Green I (Invitrogen, Eugene, USA). For AOB, the PCR was performed in a total volume of 30 µl containing 15 µl of iQ™ SYBR® Green Super Mix (Bio-Rad, Hercules, USA), 5 µl of DNA template, and 0.1 µM of each primer. The real-time PCR thermocycling steps were set as follows: 95°C for 4 min and 45 cycles at 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s. Cycling was completed by a final elongation step at 72°C for 10 min. The negative control containing no DNA was subjected to the same procedure to exclude or detect any possible contamination. After real-time PCR assay, the specificity of amplification was verified by generation of melting curves and agarose gel electrophoresis.
T-RFLP analysis
PCR was performed by using fluorescently labeled primers amoAR-Hex and amoA-2R-Hex for AOA and AOB, respectively. PCR conditions were the same as those for clone library construction. RsaI and HhaI were selected to digest PCR products of AOA and AOB amoA gene purified with PCRquick-spin™ PCR Products Purification Kit, respectively, as these two enzymes may classify the different operational taxonomic units (OTUs) of AOA or AOB into unique terminal restriction fragments (T-RFs) as shown in the Table 3.
Table 3.
amoA OTUs and clones of AOA and AOB and their corresponding T-RFs
| T-RF | OTU | Clones | |
|---|---|---|---|
| AOA | 247 | AOA-OTU-1, 4 | D30-C-1, D30-C-8, D30-C-33 |
| 56 | AOA-OTU-3 | D30-C-3 | |
| AOB | 214 | AOB-OTU-1 | F-AOB-5, F-AOB-19, F-AOB-40 |
| 371 | AOB-OTU-3 | AS-1 | |
| 421 | AOB-OTU-2 | F-AOB-1, F-AOB-25 | |
| 486 | AOB-OTU-4 | AS-60 |
The restriction–digestion mixture, containing 17 µl of purified PCR product (about 200 ng DNA), 2 µl of buffer, and 1 µl of restriction enzyme (10 U/µl), was incubated at 37°C for 5 h. The digested DNA was directly precipitated with 50 µl 100% ethanol by centrifugation at 14,000×g for 15 min at 4°C. The DNA pellets were washed with 100 µl 70% (v/v) ethanol and then air dried. The precipitate (about 70–100 ng) was dissolved in 15 µl of distilled water. The fluorescently labeled T-RFs were run through an ABI 3730xl capillary sequencer in the GeneScan mode. T-RFLP data was analyzed using GeneMarker V1.6 (SoftGenetics LLC, Pennsylvania, USA). Because of the detection range of internal marker GS500, T-RFs smaller than 50 bp and larger than 500 bp were excluded from further analysis. The peaks were first selected by the default parameters setting of the software GeneMarker with the threshold cutoff of peak intensity of 100. The relative abundance of each T-RF was determined by calculating the ratio between the area of each peak and the total area of all peaks in one sample. The peaks with relative abundance <1% were neglected in this study.
Accession number
The sequences reported in this study were deposited in the GenBank under accession number FJ917560–FJ917595.
Results
Reactor performance
As shown in Table 1, the ammonium-oxidizing rate was increased from 0.13 to 0.21 and 0.26 mg N g SS−1 day−1 from phase III to phase V. Correspondingly, the nitrogen removal rates were 0.09, 0.17, and 0.20 mg N g SS−1 day−1 and the removal percentages were 64%, 70%, and 71%, respectively.
Bacterial community
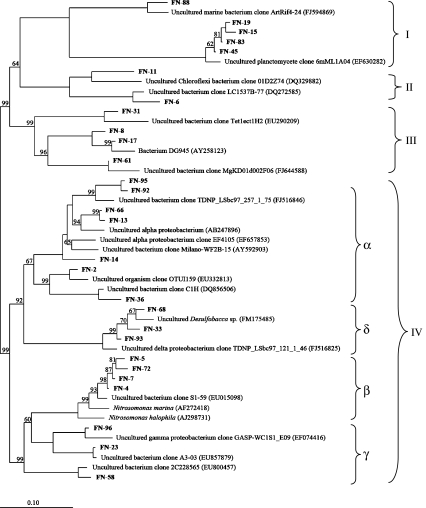
Eighteen OTUs were recovered from 75 clones of a library based on bacterial 16S rRNA gene using 3% nucleotide cutoff. Most of these sequences were closely related to unidentified clones from environmental samples. Bacteria in the sludge of this study belonged to four divisions: Proteobacteria (11 OTUs), Chloroflexi (two OTUs), Bacteroidetes (three OTUs), and Planctomycetales (two OTUs; Fig. 1). In the subdivision of betaproteobacteria, the OTUs FN-4, 5, 7, and 72 were affiliated with bacteria Nitrosomonas spp. at similarities of 97% to 99%.
Fig. 1.
Neighbor-joining tree based on Jukes–Cantor corrected DNA distances showing the phylogenetic affiliation of bacterial 16S rRNA gene sequences from activated sludge of CSTR and reference sequences from other environmental samples or pure stains. The sequences obtained in this study are printed in bold. All the sequences were grouped as I Planctomycetales, II Chloroflexi, III Bacteroidetes, and IV Proteobacteria
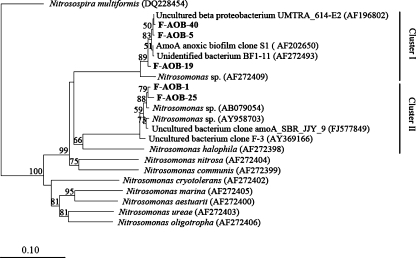
AOB community diversity based on amoA gene
As shown in Fig. 2 and Table 3, two AOB amoA OTUs (based on the 3% nucleotide cutoff), i.e., AOB-OTU-1 and AOB-OTU-2, obtained from the sludge formed two clusters. The sequences of all these OTUs were closely related to those of Nitrosomonas spp., instead of Nitrosospira spp., in agreement with results of bacterial 16S rRNA gene clone library. Most of the sequences in cluster II were from activated sludges of different WWTPs. Especially, Nitrosomonas sp. ML1 (AY958703) was isolated from a bioreactor operated under low DO (0.12–0.24 mg L−1; Park and Noguera 2007), which was even lower than DO levels of 0.5–1.0 mg L−1 in this study.
Fig. 2.
Neighbor-joining tree based on Jukes–Cantor corrected DNA distances showing the phylogenetic affiliation of AOB amoA gene sequences from activated sludge of CSTR and other environmental samples or pure strains. The sequences obtained in this study are printed in bold
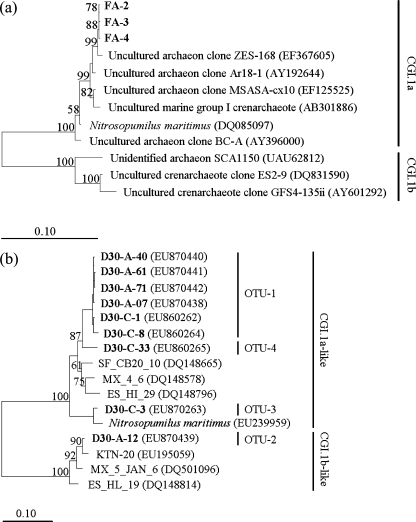
Archaeal and AOA community
The results of the archaeal 16S rRNA gene clone library showed very low diversity of archaeal community. RFLP analysis of 16 clones showed only one OTU. Three representatives were sequenced, as shown in Fig. 3a. It was affiliated with an archaeon clone ZES-168 (EF367605) from a tropical estuary, belonging to Crenarchaeotal Group I.1a (CGI.1a) in phylum Crenarchaeota. As reported before (Zhang et al. 2009), there were four AOA amoA OTUs (on the basis of the 5% nucleotide cutoff) recovered by using two primer sets in this sludge. As shown in Fig. 3b, they were dominantly distributed in the CGI.1a cluster and only one OTU fell in the CGI.1b cluster, according to the classification of Park et al. (2008), in agreement with the results of the archaeal 16S rRNA gene clone library.
Fig. 3.
Neighbor-joining tree based on Jukes–Cantor corrected DNA distances showing the phylogenetic affiliation of sequences from activated sludge of CSTR and reference sequences from other environmental samples or pure stains: a archaeal 16S rRNA gene; b AOA amoA gene. The sequences obtained in this study are printed in bold
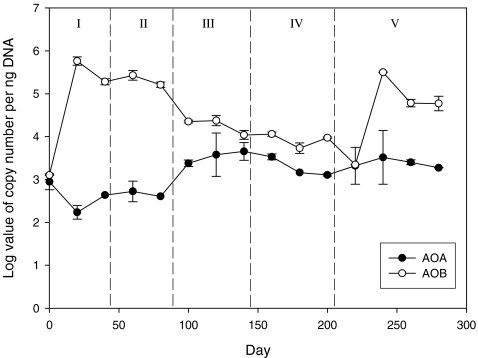
AOA and AOB abundance
Figure 4 shows the change of AOA and AOB population abundance in the whole microbial community of the CSTR in terms of the amoA gene copy number per nanogram DNA in the extract. AOA amoA gene copy number ranged from 1.7 × 102 to 3.8 × 103 copies per nanogram DNA in the whole process. It was firstly down from 8.9 × 102 to 1.7 × 102 copies per nanogram DNA in the initial 20 days (phase I), recovered slightly in phase II, subsequently increased up to reach the highest 3.8 × 103 copies per nanogram DNA in phase III, and finally decreased slightly in last two phases. The average abundance for AOA was 1.9 × 103 copies per nanogram DNA for five phases.
Fig. 4.
AOB (open circles) and AOA (solid circles) amoA gene abundance over five phases, as measured by qPCR. Standard deviations (n = 2) are indicated by error bars. The five phases were separated by vertical dashed lines
The levels of AOB amoA varied from 2.9 × 103 to 2.3 × 105 copies per nanogram DNA during the five phases, fluctuating more significantly than that of AOA. In phases I, II, and V, AOB abundance was over 1,000-fold higher than that of AOA. In phases III and IV, the abundances of AOA and AOB were in the same magnitude. Overall, the average abundance for AOB was 1.2 × 105 copies per nanogram DNA for five phases. Thus, AOB were about 60 times more abundant than AOA in terms of amoA copy number. Assuming 2.5 copies of amoA gene per AOB cell (Norton et al. 2002) and one copy per AOA cell (Hallam et al. 2006), AOB cell numbers were about 24 times higher than that of AOA, indicating their dominance in the nitrifying community and possible leading role in the nitrification process of the CSTR in this study.
T-RFLP analysis of AOA and AOB
The AOA T-RFLP profiles showed that there was only one major T-RF of 247 bp, corresponding to the dominant AOA-OTU-1 and another AOA-OTU-4 (Table 3), which were also derived from this bioreactor (Zhang et al. 2009). This result indicated that AOA-OTU-1 was the most dominant AOA in activated sludge for all phases, and the AOA community was stable and had relatively low diversity in this CSTR (Fig. 5).
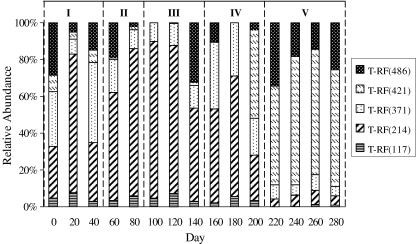
Fig. 5.
Temporal dynamics of five AOB OTUs detected from the bioreactor in the five phases (total 280 days). Relative abundance (in percent) of each AOB OTU was determined by the corresponding normalized T-RFLP peak area
In contrast, the five major T-RFs in AOB T-RFLP profiles varied significantly in the five phases in terms of the percentage of each T-RF, indicating the dynamic shift of the AOB community subject to change of operation conditions. As shown in Table 3, two of five T-RFs (214 and 421 bp) may correspond to AOB-OTU-1 and AOB-OTU-2, respectively, which were recovered from the activated sludge in this CSTR. The other two T-RFs (371 and 486 bp) may correspond to AOB-OTU-3 and AOB-OTU-4, respectively, which were recovered from the seed sludge of the CSTR. For T-RF of 117 bp, there was no OTU related to it. From 20 to 180 days, T-RF of 214 bp, i.e., AOB-OTU-1, was the most dominant AOB, accounting for 32–85% of the total AOB, but showed low abundance (4–25%) in the following 100 days (Fig. 5). Oppositely, AOB-OTU-2 (i.e., T-RF of 421 bp) was the most dominant (48–70%) AOB after 200 days although its relative abundance was lower than 10% in previous phases. The other three T-RFs (117, 371, and 486 bp) also showed fluctuation in different phases, but not as significantly as AOB-OTU-1 and AOB-OTU-2, and not the dominant AOB in the sludge.
Discussion
Phylogenic analysis of microbial community
The reactor in this study was operated to conduct nitrification of saline (about 1% salinity) wastewater. The bacterial 16S rRNA gene clone library revealed that sequences of many clones, including OTUs FN-8, 11, 13, 15, 23, 58, 61, 66 and 88, were closely related to bacteria from seawater and marine sediment, indicating that salinity was a key factor shaping the bacterial community in this study. Additionally, both 16S rRNA gene clone library and AOB amoA gene clone library confirmed the presence of the nitrifying genus Nitrosomonas, which had been identified as the major AOB in many nitrifying bioreactors (Egli et al. 2003; Hallin et al. 2005; LaPara and Ghosh 2006; Limpiyakorn et al. 2007; Park and Noguera 2004; Persson et al. 2002; Wells et al. 2009). However, the sludge in this study did not contain another AOB group, the genus Nitrosospira, which was rarely found in nitrogen-removing bioreactors (Schramm et al. 1998; Rowan et al. 2003). Nitrosospira spp. preferred low temperatures of 4–10°C and grew slowly (Avrahami et al. 2003; Siripong and Rittmann 2007) and may not be able to dominate in the reactor operated at 23°C in this study.
Previous studies have suggested that most of AOA belonged to clusters of CGI.1a and CGI.1b in the phylum Crenarchaeota (Hatzenpichler et al. 2008). The clone library of archaeal 16S rRNA gene in this study recovered only one dominant OTU in CGI.1a, indicating the low archaeal diversity. Although four OTUs were recovered from clone libraries of the AOA amoA gene, the most dominant AOA-OTU-1 also belonged to the CGI.1a cluster, showing that the dominant OTU of 16S rRNA gene and amoA gene might represent the same archaeal species. T-RFLP of AOA amoA also showed only one major peak, confirming the results of the two clone libraries. Possibly due to the low abundances, the counterparts of the other three OTUs of AOA amoA were not detected in the T-RFLP profile and the clone library of 16S rRNA gene.
AOA and AOB abundances
It was reported that AOA abundance was remarkably 1–3 orders of magnitude higher than AOB in soil and marine environments (Leininger et al. 2006; Wuchter et al. 2006). Oppositely, AOB was dominant than AOA in estuarine sediment (Mosier and Francis 2008; Santoro et al. 2008). However, little is known about AOA in biological nitrogen removal reactors.
So far, only limited AOA-containing activated sludge were reported for a few WWTPs operated with aerated-anoxic processes in which extremely low DO concentrations were maintained, enabling simultaneous nitrification and denitrification (Park et al. 2006; Zhang et al. 2009). Additionally, AOA-positive samples were collected from WWTPs operating with sludge retention times longer than 15 days and HRT longer than 24 h. It seems that these features (low DO levels and long retention times) might facilitate the growth of AOA (Park et al. 2006).
This study determined the AOA/AOB abundance and ratio of AOA/AOB in activated sludge. The range of AOA amoA gene copy number was from 102 to 103 copies per nanogram DNA at the high end of the levels for marine sediments which was 10−1 to 103 copies per nanogram DNA (Mosier and Francis 2008). The fluctuation of AOA abundance in five phases was <10-fold. However, the AOB amoA copy number fluctuated from 103 to 105 copies per nanogram DNA, much larger than that of AOA. Overall, the abundance of AOA was relative stable, indicating that the change of reactor conditions did not significantly affect AOA, in good agreement with the conclusion by Wells et al. (2009). In contrast, the great change of AOB indicated that AOB might be more sensitive to bioreactor conditions than AOA. It was suggested that AOA are adapted to a broad range of growth conditions and, therefore, have a more versatile metabolism than AOB (Leininger et al. 2006).
The results of phases I to II indicated that lower DO possibly decreased the AOB abundance, while the results of phases IV to V showed that higher ammonia concentration and feeding rate were beneficial for AOB. Similarly, a previous study also suggested that AOB abundance was higher at an ammonia load of 250 mg N–NH4 day−1 than that of 130 mg N–NH4 day−1 (Cydzik-Kwiatkowska and Wojnowska-Baryła 2008).
The average abundance of AOB in five phases was much higher (averagely 24 times) than AOA, indicating that AOB played more important roles than AOA in this study. However, in phases III and IV, AOA and AOB were almost equally important, judging from their amoA abundance. Compared to the initial activated sludge (day 0), AOB was enriched more significantly than AOA, suggesting that the overall conditions in this study were more favorable for AOB than AOA.
AOA and AOB community shift
Generally, the AOA community was relatively less diversified and stable than AOB in this reactor. The T-RFLP profile showed that only one AOA amoA OTU remained dominant in activated sludge during the five phases. Another OTU, AOA-OTU-3, only appeared in the clone library with relatively low abundance (Zhang et al. 2009).
In contrast, the AOB community showed obvious shift through the five phases, indicating that AOB were more sensitive to bioreactor conditions, i.e., ammonium concentration and loading rate, than AOA. Previous studies suggested that bioreactors with low or high ammonia loadings enriched different species of Nitrosomonas (Limpiyakorn et al. 2007). The same phenomenon was observed in this study although the specific species identities of these two Nitrosomonas spp., corresponding to T-RFs of 214 and 421 bp, respectively, were still unknown. The AOB-OTU-1 (T-RF of 214 bp) might prefer the lower ammonium loading, while the higher ammonium loading was favorable to AOB-OTU-2 (T-RF of 421 bp).
The DO level may have also significantly affected the AOB community (Wen et al. 2008). However, in this study, when DO was decreased in the first two phases, the AOB community did not show obvious shift although the total abundance of AOB decreased. Wells et al. (2009) reported that the Nitrosospira lineage showed strong negative correlations to DO while the Nitrosomonas-like phylotype showed no significant correlation to DO. In this study, the dominant AOB belonged to genus Nitrosomonas. Thus, the results were consistent with their finding, indicating that various AOB might have different responses to DO change.
Acknowledgements
The authors wish to thank the Hong Kong General Research Fund (HKU7197/08E) for the financial support of this study, and Qingmei Yan wish to thank HKU for the postgraduate studentship.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Tao Jin, Email: jintao@hkucc.hku.hk.
Tong Zhang, Phone: +852-28578551, FAX: +852-25595337, Email: zhangt@hkucc.hku.hk.
Qingmei Yan, Email: h0695089@hkusua.hku.hk.
References
- APHA . Standard methods for the examination of water and wastewater. 21. Washington: American Public Health Association; 2005. [Google Scholar]
- Avrahami S, Liesack W, Conrad R. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol. 2003;5:691–705. doi: 10.1046/j.1462-2920.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- Beman JM, Francis CA. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol. 2006;72:7767–7777. doi: 10.1128/AEM.00946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S, Stetter KO, Rouviere P, Woese CR. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst Appl Microbiol. 1991;14:346–351. doi: 10.1016/s0723-2020(11)80308-5. [DOI] [PubMed] [Google Scholar]
- Coolen MJL, Abbas B, Van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG, Damsté JSS. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- Cydzik-Kwiatkowska A, Wojnowska-Baryła I. The impact of organic carbon and ammonia load in wastewater on ammonia-oxidizing bacteria community in activated sludge. Pol J Microbiol. 2008;57:241–248. [PubMed] [Google Scholar]
- Dionisi HM, Layton AC, Harms G, Gregory IR, Robinson KG, Sayler GS. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl Environ Microbiol. 2002;68:245–253. doi: 10.1128/AEM.68.1.245-253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli K, Langer C, Siegrist HR, Zehnder AJB, Wagner M, van der Meer JR. Community analysis of ammonia and nitrite oxidizers during start-up of nitrification reactors. Appl Environ Microbiol. 2003;69:3213–3222. doi: 10.1128/AEM.69.6.3213-3222.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geets J, de Cooman M, Wittebolle L, Heylen K, Vanparys B, De Vos P, Verstraete W, Boon N. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl Microbiol Biotechnol. 2007;75:211–221. doi: 10.1007/s00253-006-0805-8. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligonucleotide probes for identification of single microbial cells. J Bacteriol. 1998;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci U S A. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S, Lydmark P, Kokalj S, Hermansson M, Sörensson F, Jarvis A, Lindgren PE. Community survey of ammonia oxidizing bacteria in full-scale activated sludge processes with different solids retention time. J Appl Microbiol. 2005;99:629–640. doi: 10.1111/j.1365-2672.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di HJ. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz MG, Alzerreca J, Norton JM. A gene encoding a membrane protein exists upstream of the amoA/amoB genes in ammonia-oxidizing bacteria; a third member of the amo operon? FEMS Microbiol Lett. 1997;150:65–73. doi: 10.1111/j.1574-6968.1997.tb10351.x. [DOI] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- LaPara TM, Ghosh S. Population dynamics of the ammonia-oxidizing bacteria in a full-scale municipal wastewater treatment facility. Environ Eng Sci. 2006;23:309–319. doi: 10.1089/ees.2006.23.309. [DOI] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Limpiyakorn T, Kurisu F, Sakamoto Y, Yagi O. Effects of ammonium and nitrite on communities and populations of ammonia-oxidizing bacteria in laboratory-scale continuous-flow reactors. FEMS Microbiol Ecol. 2007;60:501–512. doi: 10.1111/j.1574-6941.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar BA, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydmark P, Almstrand R, Samuelsson K, Mattsson A, Sorensson F, Lindgren PE, Hermansson M. Effects of environmental conditions on the nitrifying population dynamics in a pilot wastewater treatment plant. Environ Microbiol. 2007;9:2220–2233. doi: 10.1111/j.1462-2920.2007.01336.x. [DOI] [PubMed] [Google Scholar]
- Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Norton JM, Alzerreca JJ, Suwa Y, Klotz MG. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch Microbiol. 2002;177:139–149. doi: 10.1007/s00203-001-0369-z. [DOI] [PubMed] [Google Scholar]
- Okano Y, Hristova KR, Leutenegger CM, Jackson LE, Denison RF, Gebreyesus B, Lebauer D, Scow KM. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol. 2004;70:1008–1016. doi: 10.1128/AEM.70.2.1008-1016.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HD, Noguera DR. Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge. Water Res. 2004;38:3275–3286. doi: 10.1016/j.watres.2004.04.047. [DOI] [PubMed] [Google Scholar]
- Park HD, Noguera DR. Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J Appl Microbiol. 2007;102:1401–1417. doi: 10.1111/j.1365-2672.2006.03176.x. [DOI] [PubMed] [Google Scholar]
- Park HD, Wells GF, Bae H, Criddle CS, Francis CA. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Park BJ, Rhee SK. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles. 2008;12:605–615. doi: 10.1007/s00792-008-0165-7. [DOI] [PubMed] [Google Scholar]
- Persson F, Wik T, Sörensson F, Hermansson M. Distribution and activity of ammonia oxidizing bacteria in a large full-scale tricking filter. Water Res. 2002;36:1439–1448. doi: 10.1016/S0043-1354(01)00345-1. [DOI] [PubMed] [Google Scholar]
- Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops HP, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/AEM.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan AK, Moser G, Gray N, Snape JR, Fearnside D, Curtis TP, Barer MR, Head IM. A comparative study of ammonia-oxidizing bacteria in lab-scale industrial wastewater treatment reactors. Water Sci Technol. 2003;48:17–24. [PubMed] [Google Scholar]
- Santoro AE, Francis CA, de Sieyes NR, Boehm AB. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradient in a subterranean estuary. Environ Microbiol. 2008;10:1068–1079. doi: 10.1111/j.1462-2920.2007.01547.x. [DOI] [PubMed] [Google Scholar]
- Schramm A, de Beer D, Wagner M, Amann R. Identification and activity in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripong S, Rittmann BE. Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res. 2007;41:1110–1120. doi: 10.1016/j.watres.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Wells GF, Park HD, Yeung CH, Eggleston B, Francis CA, Criddle CS. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol. 2009;11:2310–2328. doi: 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- Wen QX, Chen ZQ, Shi HC. T-RFLP detection of nitrifying bacteria in a fluidized bed reactor of achieving simultaneous nitrification–denitrification. Chemosphere. 2008;71:1683–1692. doi: 10.1016/j.chemosphere.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GL, Middelburg JJ, Schouten S, Sinninghe Damsté JS. Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Jin T, Yan QM, Shao MF, Wells G, Criddle C, Fang HHP. Occurrence of ammonia-oxidizing archaea (AOA) in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol. 2009;107:970–977. doi: 10.1111/j.1365-2672.2009.04283.x. [DOI] [PubMed] [Google Scholar]