Abstract
Mammals possess an intricate regulatory system for controlling flux through fuel utilization pathways in response to the dietary availability of particular macronutrients. Under fasting conditions, for instance, mammals initiate a whole body metabolic response that limits glucose utilization and favors fatty acid oxidation. Understanding the underlying mechanisms by which this process occurs will facilitate the development of new treatments for metabolic disorders such as type II diabetes and obesity. One of the recently identified components of the signal transduction pathway involved in metabolic reprogramming is PGC-1α. This transcriptional coactivator is able to coordinate the expression of a wide array of genes involved in glucose and fatty acid metabolism. The nutrient-mediated control of PGC-1α activity is tightly correlated with its acetylation state. In this review, we evaluate how the nutrient regulation of PGC-1α activity squares with the regulation of its acetylation state by the deacetylase Sirt1 and the acetyltransferase GCN5. We also propose an outline of additional experimental directives that will help to shed additional light on this very powerful transcriptional coactivator.
1. Introduction
The availability of food is one of the most capricious environmental variables that mammals have been subjected to over the course of their natural history. At first glance, an unreliable supply of food would seem to pose a major dilemma for homeothermic organisms like mammals, which have an intrinsically high metabolic rate. Compounding this problem is the metabolic peculiarity that although most mammalian tissues are able to use glucose, fatty acids, or amino acids as fuel substrates there are some tissues, such as the central nervous system, retina, red blood cells, and renal medulla, that rely almost exclusively upon an uninterruptable supply of glucose as fuel. In spite of these existential problems, however, mammals have evolved a highly regulated and effective system of fuel utilization pathways that manages to accommodate the tissue-specific requirements of fuels over a wide range of food and macronutrient availability.
When a mammal is deprived of food, its body undergoes both quantitative and qualitative changes in the way in which stored macronutrients are mobilized and metabolized as fuel. These changes constitute a homeostatic response that ultimately aids in the preservation of a constant supply of circulating blood glucose for those tissues that exclusively require it and in the reduction of overall energy expenditure. Part of this response includes switching to the utilization of free fatty acids as the major macronutrient fuel in peripheral tissues such as muscle as well as increasing the rate of glucose synthesis in the liver and kidney. The coordinated initiation and subsequent maintenance of this remarkable change in fuel utilization is a highly complicated process that requires the integration of many hormonal, transcriptional, translational, and allosteric signals. Identification of the regulatory components in this response has been an important research endeavor for the purpose of aiding not just our understanding of basic biology but also in the development of treatments for pathophysiological conditions where glucose homeostasis has clearly gone awry, such as in obesity and type II diabetes. Elucidation of this regulatory pathway may also help to shed light on the mechanism by which caloric restriction is able to retard or prevent the age-related deterioration of mammalian tissues. One exciting branch of research into this problem over the past 10 years has indicated that a major participant in the physiological process of glucose maintenance and fuel utilization is the protein Peroxisome Proliferator-Activated Receptor Gamma-Coactivator-1 α (PGC-1α). In this review, we discuss the role of PGC-1α in regulating the central pathways of metabolism. We also discuss how the acetyl transferase GCN5 and the deacetylase Sirt1 are capable of changing the acetylation state of PGC-1α in response to nutrient state and reciprocally alter the transcriptional coactivating properties of PGC-1α.
2. PGC-1α and Its Effects on Fuel Metabolism in Fasting
PGC-1α was first identified as a binding partner and co-activator of the transcriptional activity of PPARγ as part of a study to identify transcriptional components of the pathway responsible for the metabolic changes accompanying adaptive thermogenesis [1]. Since that time, however, the list of transcription factors to which PGC-1α is known to bind to and consequently modulate the activity thereof has expanded considerably. This list includes PPARα, glucocorticoid receptor, hepatic nuclear factor-4α, members of the estrogen related receptor (ERR) family, myocyte enhancer factor 2, nuclear respiratory factors 1 and 2, FoxO1, and YY1 [2–6].
The precise mechanism by which PGC-1α is able to enhance the ability of these proteins to initiate gene transcription is not altogether clear, although it is clear that the enhancement of activity is contingent upon the physical interaction of PGC-1α and its cognate transcription factor [7]. Most transcriptional co-activators possess intrinsic enzymatic activity, such as histone acetyltransferase activity, that enables them to modify chromatin to make the genetic locus at which they are located more amenable to transcription. PGC-1α does not appear to possess any enzymatic activity. Its N-terminus, however, is an effective docking site for two well-established histone acetyltransferases, SRC-1 and CBP/p300, which have been shown to be strong enhancers of PGC-1α activity [7]. Although they enhance the activity of PGC-1α, SRC-1 and CBP/p300 do not acetylate PGC-1α. In fact, acetylation of PGC-1α is correlated with a decrease in PGC-1α activity (see below).
It should be noted that although PGC-1α is a co-activator it is not merely a blunt instrument for effecting a wholesale change in the transcriptional activity of its binding partners—its co-activational properties can apparently be selectively tuned for different promoters even when coupled to the same transcription factor. The UCP-1 and aP2 genes, for instance, contain conserved PPARγ binding sites within their promoters but it is only the gene expression of UCP-1 that changes in response to levels of PGC-1α [1]. The mechanism by which this specificity is mediated has not been fully explored.
From a physiological standpoint, the co-activation of a litany of cognate transcription factors by PGC-1α has important metabolic repercussions; collectively, the set of transcription factors to which PGC-1α binds controls the expression of genes involved in gluconeogenesis, glycolysis, lipogenesis, peroxisomal and mitochondrial fatty acid oxidation, and mitochondrial respiration efficiency. Thus, PGC-1α can single handedly coordinate the gene expression of multiple energy pathways. This point is underscored by the PGC-1α knockout mouse, which shows a reduced respiratory capacity, diminished hepatic TCA cycle flux, reduced rates of hepatic gluconeogenesis and β-oxidation, hepatic steatosis under fasting conditions, and hypoglycemia [8–10].
2.1. PGC-1α Function in the Liver
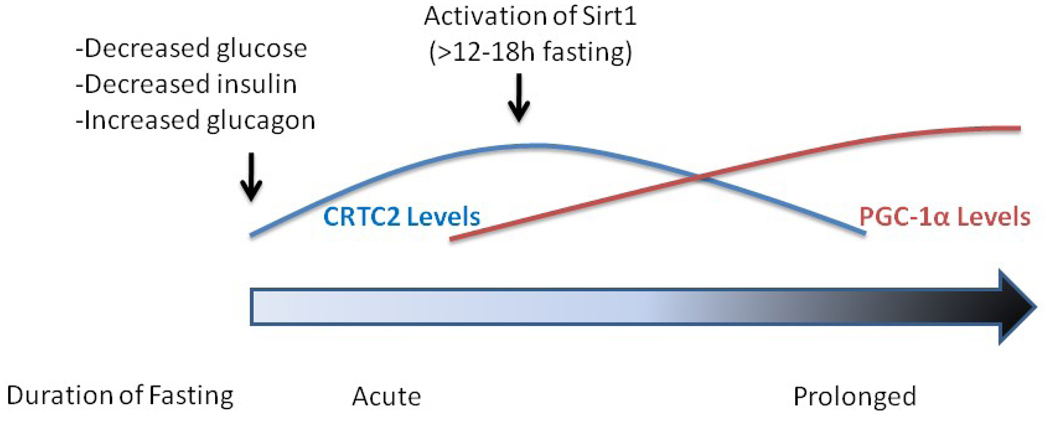
In terms of its contribution to the problem of diet-dependent maintenance of energy homeostasis in mammals, there is a body of evidence to suggest that the co-transcriptional activity of PGC-1α is important for the compensatory metabolic responses that occur during food deprivation. The two organs for which this process is best understood are the liver and the skeletal muscle. In the liver during fasting, gluconeogenesis is profoundly upregulated primarily at the level of transcription. A compelling argument can be made for dissecting the upregulation of gluconeogenesis into two temporally distinct phases. In the first and more acute phase (<12–18h), an increase in the transcription of gluconeogenic enzymes is initiated in part by the activation of cAMP response element binding protein (CREB) and its coactivator CRTC2 (Figure 1). With prolonged fasting (>12–18h), however, CRTC2 protein is degraded [11] and its contribution to the fasting transcriptional response is significantly diminished [12]. The initial signal for CRTC2 degradation appears to be deacetylation by Sirt1 [11]. The maintenance of the gluconeogenic response through prolonged fasting is thought to be mediated by PGC-1α and its cognate transcription factors. Indeed, hepatic levels of PGC-1α protein significantly increase under fasting conditions, driven in part by an increase in transcription caused by CRTC2 activation [13] and in part by an increase in the half-life of the protein induced by an attenuation of insulin signaling [14]. The elevated levels of PGC-1α facilitate increased hepatic glucose output by promoting the expression of gluconeogenic genes such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase, most likely through the co-activation of HNF4α and FoxO1 [15]. Interestingly, whereas the increase in Sirt1 activity that accompanies prolonged fasting attenuates CRTC2 signaling, it actually improves PGC-1α’s ability to increase hepatic gluconeogenesis (see below).
Figure 1.
A proposed theory for the temporal regulation of the two major transcriptional coactivators involved in hepatic gluconeogenesis during fasting. At the onset of fasting, circulating levels of glucose fall. This results in an increase in blood levels of glucagon and a decrease in insulin. This hormonal change produces an increase in both the levels and activity of the transcriptional coactivator CRTC2, which facilitates an increase in the expression of hepatic gluconeogenic genes. CRTC2 also increases the transcription of PGC-1α. With sustained fasting (>12–18h), Sirt1 becomes activated and deacetylates CRTC2. This event permits the ubiquitination of the protein by the E3 ligase COP1 and targets the enzyme for destruction [70]. The activation of Sirt1 also results in the deacetylation of PGC-1α, which is associated with an increase in the activity of this transcriptional coactivator.
2.2 PGC-1α Function in Skeletal Muscle
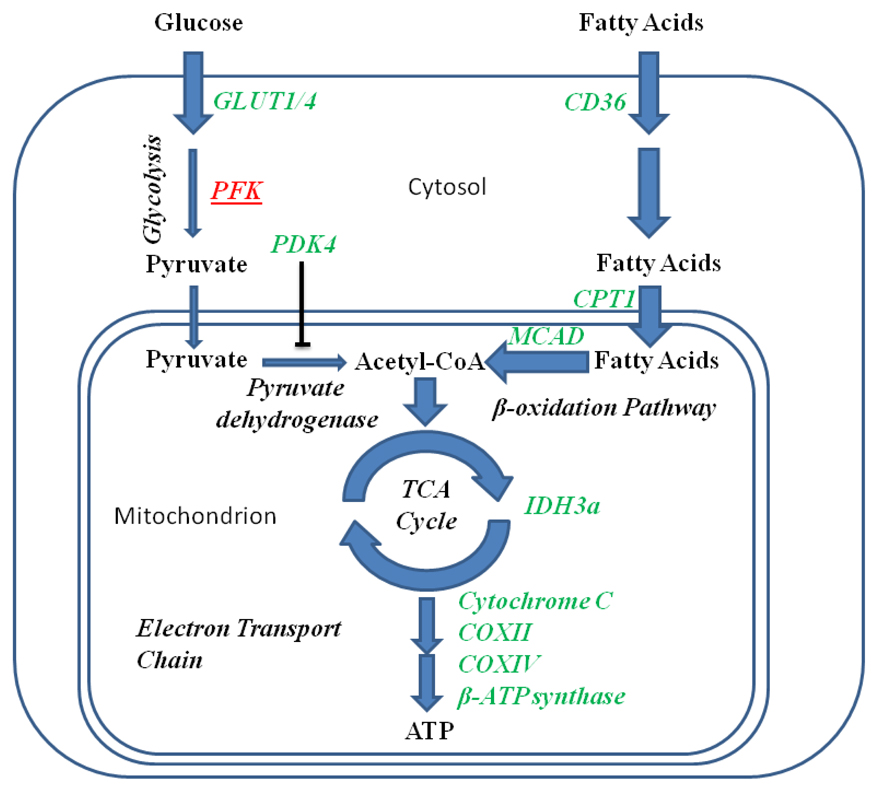
In skeletal muscle during fasting, induction of PGC-1α helps to orchestrate a series of metabolic changes that results in muscle tissue using less glucose and more fatty acids for oxidative phosphorylation (Figure 2). PGC-1α has been shown to increase the expression of the glucose transporters GLUT1 and GLUT4 in muscle tissue [16–18]. In the case of GLUT4, this effect is dependent upon the transcription factor MEF-2c [18]. Although PGC-1α can induce glucose transporter expression, net utilization of glucose by skeletal myocytes is significantly mitigated by an increase in PGC-1α activity. The mechanism by which this occurs involves at least two different processes. The first mechanism is a PGC-1α-induced decrease in the expression of phosphofructokinase and an accompanying diminution in glycolytic flux [16].
Figure 2.
Conditions that increase PGC-1α activity, such as fasting, increase the oxidation of fatty acids and decrease the oxidation of glucose in skeletal muscle. Genes that are upregulated by increases in PGC-1α activity are indicated in green whereas those that are downregulated are indicated in red and are underlined. PGC-1α increases the rates at which both glucose and fatty acids are transported into myocytes. Both the rate of glycolysis and the rate at which glucose-derived pyruvate is converted into acetyl-CoA, however, are significantly reduced by PGC-1α. Fatty acid transport into mitochondria is significantly enhanced, on the other hand, by PGC-1α. The rate at which fatty acids are oxidized in the mitochondrion is similarly increased, resulting in an increase in the acetyl-CoA pool derived from this macronutrient. Genes involved in TCA cycle flux, electron transport, and proton-coupled ATP synthesis are upregulated by PGC-1α, improving the efficiency with which ATP can be generated from fatty acids.
The second proposed mechanism involves a PGC-1α-induced increase in the expression of pyruvate dehydrogenase kinase-4 [16, 19, 20], a negative regulator of pyruvate dehydrogenase, which abates the entry of glucose-derived pyruvate into the TCA cycle. PGC-1α’s coactivation of ERRα is responsible for increased PDK4 expression. Higher levels of PGC-1α also permit muscle cells to oxidize more fatty acids. PGC-1α facilitates the delivery of free fatty acids across the cell membrane by increasing the expression of the integral membrane protein CD36, which is capable of binding to long-chain fatty acids and mediates their internalization into the cytosol [16, 17]. Transport of free fatty acids from the cytoplasm across the outer mitochondrial membrane is also facilitated by an increase in the expression of the free fatty acid transporter carnitine palmitoyltransferase 1 (CPT-1) [16, 20]. The complete oxidation of fatty acids and its coupling to ATP production within the mitochondria are facilitated by an increase in the levels of medium chain acyl CoA dehydrogenase (MCAD), cytochrome oxidases II and IV, isocitrate dehydrogenase 3A, β-ATP synthase, and cytochome C [20].
3. Regulation of PGC-1α Co-transcriptional Acitivity: The Contribution of Acetylation
Given the preponderance of evidence supporting a role for PGC-1α in coordinating the compensatory metabolic responses that accompany food restriction, it is logical to inquire how the biological activity of PGC-1α changes under these circumstances. One conceivable mode of regulation is simply that the absolute concentration of PGC-1α protein is increased under low nutrient conditions and forces, by the principle of mass action, an increase in the activity of its transcription factor binding partners. This is definitely possible within the liver, where the concentration of PGC-1α protein significantly increases during periods of acute fasting [21]. Evaluations of skeletal muscle, on the other hand, have shown that there is no appreciable change in the level of PGC-1α during fasting despite very clear increases in the target genes of PGC-1α [20, 22]. Such data strongly suggest the involvement of post-translational regulation of protein activity by covalent modification, allosteric modulation, or changes in intracellular localization/compartmentation.
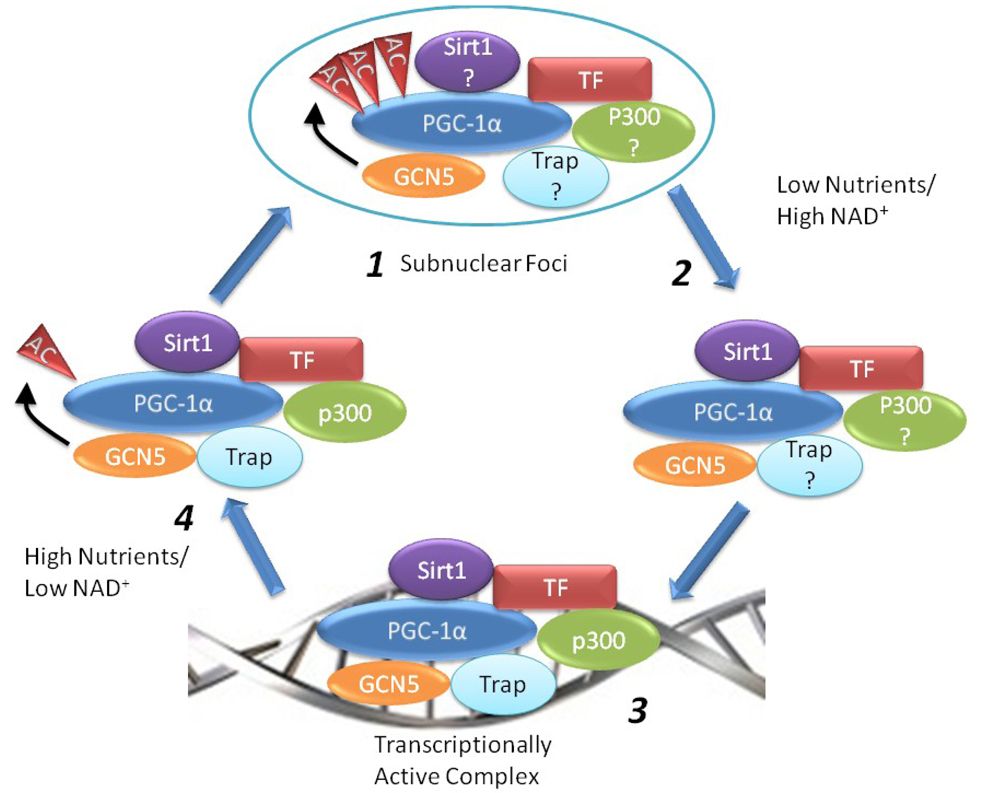
One proposed mechanistic model for explaining changes in PGC-1α activity under fed and fasting conditions that does not require a change in total protein is the alteration of activity by acetylation state [21, 23]. At least 13 lysine acetylation sites have been identified on PGC-1α [21]. Under an acetylation-based control model, it follows that nutrient availability affects the steady-state level of PGC-1α by altering the relative rates at which the protein is either acetylated by GCN5 or deacetylated by Sirt1 and this is subsequently translated into a change in the co-transcriptional activity of the protein (Figure 3). Under high nutrient conditions and low intracellular NAD+ concentrations, PGC-1α is hyperacetylated by GCN5 and located within punctate nuclear bodies along with its transcription factor binding partners. In this state, the PGC-1α complex is effectively transcriptionally inactive. As cells are confronted with low nutrient availability, however, intracellular NAD+ levels increase and lead to an increase in the rate at which PGC-1α is deacetylated by Sirt1. The change in PGC-1 α acetylation coincides with an increased occupancy of PGC-1α at the promoters of its target genes and an increase in transcriptional activation by remodeling of the local chromatin environment, by proteins such as p300, and greater interaction with general transcriptional machinery, facilitated by proteins such as the TRAP/Mediator complex [24].
Figure 3.
A model for the regulation of PGC-1α acetylation in response to nutrient availability. When nutrient availability is high and intracellular NAD+ is low, PGC-1α is acetylated and re-located within punctate nuclear bodies along with its transcription factor partners (TF) (1). As nutrients are exhausted and NAD+ rises, Sirt1 activity is enhanced and results in the deacetylation of PGC-1α (2). This coincides with a greater occupancy of PGC-1α at its target promoters and an increase in the transcription of the target genes (3). When nutrient levels return to higher levels, the activity of Sirt1 relative to GCN5 declines and PGC-1α is acetylated (4) and sequestered within specific nuclear foci (1). See text for additional details. Abbreviations used: TF, transcription factor
Support for this model is readily found in correlative evidence from whole animal studies that have consistently shown, in both liver [21] and skeletal muscle [20, 25], that steady-state levels of acetylated PGC-1α are high when animals are fed food and decline significantly when animals are deprived of food. Biochemical and genetic dissection of the association between nutrient supply and PGC-1α acetylation has revealed that the nutrient status of cells can directly alter both the rate at which PGC-1α is acetylated as well as the rate that it is deacetylated. What has been less well defined, however, is how acetylation is physically communicated into a change in the biological activity of PGC-1α. These aspects and how they pertain to the regulatory model as a whole are discussed below.
3.1. Nutrient Supply and the Regulation of GCN5: an Endogenous Acetyltransferase of PGC-1α
Overexpression experiments with p300 suggested that this enzyme could be the acetyltransferase responsible for acetylating PGC-1α [26]. Subsequent experiments involving the purification of intact PGC-1α protein complexes from cells, however, identified the acetyltransferase General Control Non-repressed Protein 5 (GCN5) as being bound to PGC-1α and showed that this enzyme was a far more efficient acetyltransferase for PGC-1α than p300 in vivo [23].
The interaction of GCN5 and PGC-1α is dependent upon the C-terminal bromodomain of GCN5 and a stretch of amino acids located within the first 200 residues of PGC-1α. This region within PGC-1α, incidentally, is also an effective docking site for other acetyltransferases such as SRC-1 and CBP/p300 which strongly induce PGC-1α activity but do not result in significant increases in PGC-1α acetylation [7]. In contrast to these other acetyltransferases, the binding to and acetylation of PGC-1α by GCN5 is associated with a decrease in PGC-1α co-transcriptional activity. Overexpression of GCN5, for example, was able to effectively suppress the induction of PGC-1 α target genes caused by the overexpression of wild-type PGC-1α in cultured hepatoma cells [23]. Along these same lines, knockdown of GCN5 expression in these cells enhanced the effect of wild-type PGC-1α expression. When extended to whole animal models, the hepatic overexpression of GCN5 during fasting resulted in a suppression of the gluconeogenic genes PEPCK and glucose-6-phosphatase as well as in a reduction of de novo synthesis of glucose from pyruvate [23]. Although the metabolic effects of GCN5 overexpression in vivo are consistent with the PGC-1α acetylation model established in cells, additional experiments in which GCN5 is overexpressed in mice where PGC-1α expression has been knocked down or genetically extirpated are needed to prove that the effects of this acetyltransferase are actually contingent upon PGC-1α.
GCN5 also affects the subcellular localization of PGC-1α [23]. Assessment of ectopically expressed PGC-1α has revealed that most of the intracellular pool of PGC-1α is localized to the nucleus. Within this organelle, PGC-1α resides in a largely homogenously diffuse distribution. In the presence of ectopically overexpressed GCN5, however, the majority of nuclear PGC-1α becomes concentrated into numerous punctate speckles. The exact composition of these speckles has yet to be adequately resolved, although it is known that they co-localize with the nuclear receptor co-repressor RIP140 [23].
The relocalization of PGC-1α to nuclear speckles is dependent upon the acetyltransferase activity of GCN5: a catalytically incompetent mutant of GCN5 does not cause PGC-1α to relocalize to speckles [23]. Despite the fact that PGC-1α relocalization is dependent upon GCN5 activity, it has yet to be directly demonstrated that the PGC-1α in nuclear speckles is actually acetylated. The possibility thus remains that relocalization is a consequence of GCN5 activity that is independent of PGC-1α acetylation. Additional experiments with mutant isoforms of PGC-1α that cannot be acetylated by GCN5 are needed to clarify this point. It is known, however, that relocalization is strictly dependent upon an intact C-terminus of PGC-1α, a region that is not involved in GCN5 binding but does contain a RNA binding domain as well as an arginine/serine (RS)-enriched domain that has been identified in splicing complex proteins [27]. This domain also contains two known acetylation sites [21], raising the possibility that these acetylations are important in the relocalization signaling PGC-1α. From a functional standpoint, the GCN5-induced movement of PGC-1α to nuclear speckles correlates with a significant decrease in the binding of PGC-1α to the promoters of its target genes [23]. This could be interpreted to mean that the reduction in PGC-1α activity by GCN5 is operating via a spatial discrimination mechanism whereby PGC-1α is recompartmentalized away from its transcriptional targets and rendered functionally inactive.
There are some data to suggest that macronutrient availability could affect the ability of GCN5 to acetylate PGC-1α through changes in steroid receptor coactivator protein (SRC-3). SRC-3 positively regulates the activity of GCN5 in mammalian cells. SRC-3 has been shown to bind to GCN5 and this interaction is crucial to ability of SRC-3 to coactivate retinoic acid receptor [28]. Knockout of SRC-3 in mice has been shown to significantly reduce the expression of GCN5 in muscle and brown adipose tissue [22]. In SRC-3 knockout mice, PGC-1α acetylation levels were found to be similarly reduced in both muscle and brown adipose tissue. Correlating with the change in PGC-1α acetylation levels, many mitochondrial gene targets of PGC-1α, such as CPT-1, UCP-1 through 3, cytochrome C, and ATP synthase, were significantly elevated relative to wild-type controls [22]. In wild-type mice, both SRC-3 and GCN5 expression positively correlated with nutrient availability. Following a short-term fast (6 h), levels of SRC-3 in gastrocnemius in wild-type animals were found to decline along with the level of GCN5 [22]. Concomitant with the decrease in GCN5, the total acetylation state of PGC-1α fell. When wild-type animals were fed a high fat diet for 4 weeks, SRC-3 protein in gastrocnemius was elevated as was GCN5 expression. According to the model proposed by the authors, diet composition/nutrient availability directly regulates the level of SRC-3 which subsequently influences the expression of GCN5 which, in turn, alters the acetylation state/co-transcriptional activity of PGC-1α. More detailed molecular analysis of this model, however, has raised additional questions over how SRC-3 is able to alter the expression and/or activity of GCN5. Knockdown of SRC-3 in C2C12 cells significantly reduced the expression of GCN5, reduced the acetylation of PGC-1α, and increased the expression of PGC-1α regulated mitochondrial genes [22]. Overexpression of SRC-3 by itself, however, increased endogenous GCN5 transcription but did not elevate the amount of PGC-1α acetylation in cultured C2C12 myotubes. Only when SRC-3 was overexpressed in conjunction with GCN5 overexpression was there a significant increase in PGC-1α acetylation. The disparity in results between the whole animal and cell culture experiments warrants further pursuit of the mechanism that connects changes in SRC-3 levels with changes in PGC-1α acetylation and target gene expression. Of particular interest would be an evaluation of the effects of SRC-3 on PGC-1α gene targets in cells which have had either GCN5 or PGC-1α knocked down.
Although abundant evidence suggests that GCN5 is able to acetylate PGC-1α and suppress its activity, evidence for the dietary control of this process is somewhat limited. Far more evidence exists, however, in support of the hypothesis that the changes in both acetylation state and co-transcriptional activity of PGC-1α accompanying dietary intake are principally due to fluctuations in the rate at which it is deacetylated by the protein Sirtuin 1.
3.2. Nutrient Supply and the Regulation of Sirt1: an Endogenous Deacetylase of PGC-1α
Sirtuin 1 (Sirt1) is a NAD+-dependent protein deacetylase that has been implicated in a panoply of physiological processes in mammals, including control of lipolytic rates in white adipose tissue [29], modulation of insulin secretion from pancreatic β-cells [30], control of cytoplasmic and mitochondrial acetyl-CoA synthetase activity [31], regulation of the circadian clock [32–34], and regulation of the genetic response to various stressors such as heat shock [35], genotoxicity [36], and hypoxia [37]. To this long list of biological functions, one can also add the regulation of PGC-1α acetylation state. Sirt1 has thus far been the only identified protein capable of binding to PGC-1α and deacetylating it both in vivo and in vitro [21, 26]. Sirt1 binds to a region of PGC-1α that is contained within amino acid residues 200–400 and deacetylates the protein in a NAD+-dependent manner [21, 26].
Alterations in the level of Sirt1 activity are closely associated with changes in the expression of PGC-1α targets in liver and muscle tissue. In hepatoma cultures, knockdown of Sirt1 significantly increased the acetylation of PGC-1α, and reduced both the induction of gluconeogenic genes and the corresponding rate at which glucose was produced following PGC-1α overexpression [21]. In myotube cultures, Sirt1 was found to be bound to the promoters of PGC-1α target genes and occupancy at these sites could be significantly enhanced by Sirt1 overexpression [20]. Overexpression of Sirt1 was also accompanied by a significant decrease in PGC-1α acetylation and an increase in the expression of PGC-1α targets; knockdown, on the other hand, produced a corresponding decrease in the expression of PGC-1α targets. Activation of endogenous Sirt1 activity by resveratrol in cultures of C2C12 myotubes was also able to induce the deacetylation of overexpressed PGC-1α protein and potentiate the effects of PGC-1α on MCAD, ERRα, and cytochrome C [38]. Resveratrol, however, was not able to enhance the activity of PGC-1α when all 13 of the known acetylated lysine residues were mutated to an arginine.
Whole animal studies have largely supported the mechanistic associations borne out by the cell culture experiments. In agreement with the cell culture studies, overexpression of Sirt1 in the liver significantly increased the expression of PEPCK and glucose-6-phosphatase in a PGC-1α-dependent manner [39]. Similarly, knockdown of Sirt1 attenuated hepatic glucose output induced by PGC-1α overexpression in a pyruvate tolerance test in mice [39] and reduced both fasting glucose levels and hepatic gluconeogenesis in rats [40]. Pharmacological activation of Sirt1 in animals has also provided corroborating evidence for the models of PGC-1α regulation that were developed in cell culture systems, with the caveat that there is currently controversy over whether these activators are actually directly modulating Sirt1 activity in vivo [41, 42]. The Sirt1 activator SRT1720 was associated with a metabolic change in mouse skeletal muscle to an oxidative fiber type, a decrease in PGC-1α acetylation, and an increase in the expression of PGC-1α targets involved in fatty acid oxidation [43]. Treatment of animals with resveratrol increased the expression of a much larger number of PGC-1α targets in skeletal muscle [38]. Interestingly, resveratrol administration also appeared to be activating AMP kinase in these animals. Evidence that PGC-1α is phosphorylated by AMP kinase has been shown to increase the biological activity of PGC-1α therefore warrants consideration as an alternate explanation for these findings [25, 44].
It should be noted that the findings of two whole animal studies have somewhat contradicted the activational effects of Sirt1 on PGC-1α activity observed in cell culture. In one of these studies [45], transgenic mice overexpressing a single copy of Sirt1 showed significantly reduced glucose output by the liver under fed conditions. Nevertheless, isolated hepatocytes from these mice did show increased PGC-1α activity in the form of elevated expression of gluconeogenic genes. This suggests that the whole animal metabolic phenotype could be due to counter regulatory non-cell autonomous factors such hormones. In the second study [46], electrotransfection of Sirt1 into rat hindlimb skeletal muscle increased total Sirt1 expression but decreased the level of PGC-1α protein. Concomitant with the decrease in PGC-1α protein was a decrease in proteins associated with mitochondrial biogenesis including cytochrome oxidase IV and mitochondrial transcription factor A.
One major issue for forwarding our understanding of the control of steady-state levels of PGC-1α acetylation by Sirt1 is identification of the potential mechanisms by which changes in nutrient availability are translated into changes in Sirt1 activity. Although there is evidence to suggest that post-translational modifications such as phosphorylation [47] and sumoylation [48] can modulate Sirt1 activity as well as some interacting proteins such as AROS [49] and DBC1 [50, 51], most proposals for how Sirt1 activity is controlled by nutrient availability in vivo involve either changes in the total level of Sirt1 protein or with the availability of NAD+.
Levels of Sirt1 have been reported to change in response to nutrients. Some investigators have found that Sirt1 transcription is inversely responsive to changes in nutrient load by a mechanism that is dependent upon either the transcription factor Foxo3a [52] or the transcriptional co-repressor CtBp and its binding partner Hypermethylated In Cancer [53]. Others, however, report that there are increases in Sirt1 protein caused by nutrient deprivation in both cultured cells and mice, but are not accompanied by changes in the rates of transcription or in steady-state mRNA levels [21, 54]. Among these reports there is disagreement as to whether Sirt1 actually increases in the liver. From this body of work, it is clear that additional studies need to be done to establish the exact mechanism by which Sirt1 protein is controlled by nutriture and if this control is cell autonomous or tissue specific.
Levels of NAD+ are also known to change in response to nutrient availability. NAD+ is a necessary co-substrate for the deacetylase activity of sirtuin proteins. The strict requirement of this substrate is one of the two linchpins for the hypothesis that the major regulator of sirtuin proteins is the concentration of NAD+. The second is that the concentration of NAD+ is ordinarily at or below the sirtuins’ Km for NAD+ (100–300 µM) [55, 56]. Cell culture experiments in which NAD+ biosynthesis from nicotinamide was artificially manipulated by varying the expression level of nicotinamide phosphoribosyl transferase (NAMPT)—an enzyme involved in the salvaging of NAD+ from nicotinamide—have shown that it is possible to produce significant changes in Sirt1 activity by altering NAD+ levels [57]. Interestingly, an ancillary effect of NAMPT activity is to reduce intracellular nicotinamide levels—a product of the sirtuin deacetylase reaction and an inhibitor of their activity—and raises the possibility that NAMPT increases sirtuin activity not only by increasing the concentration of the co-substrate but also by alleviating product inhibition. It has also been recently shown that nicotinamide monoadeninenucleotide transferase, the other enzyme involved in the NAD+ salvage pathway positioned immediately downstream of NAMPT, is physically complexed with Sirt1 at the promoters of certain genes and plays a supporting role in allowing Sirt1 to activate gene transcription [58]. In light of these observations, physiological states that result in a perturbation of steady-state NAD+ levels may very well alter Sirt1 activity and PGC-1α acetylation.
One of the outstanding mysteries in mammalian NAD+ biology is how changes in dietary intake cause the intracellular NAD+ concentration of tissues to change. Intracellular concentrations of liver and muscle NAD+ have been shown to increase in rodents during fasting [21, 25, 59]. Nevertheless, under these circumstances, it is not clear if cells are responding to a change in a particular macronutrient such as glucose or fatty acids, one of their respective downstream metabolites, or perhaps a hormonal signal. Because cultured cells show an increase in intracellular NAD+ following the withdrawal of glucose in culture medium [20, 21], however, it is possible that tissues in vivo are also altering intracellular NAD+ based on glucose availability. Research with cell culture models has led to the hypothesis that a reduction in glucose availability causes a decrease in the synthesis of ATP and an increase in AMP levels, which results in an activation of AMP-activated protein kinase (AMPK). The activation of AMPK, in turn, produces an increase in the rate of NAD+ biosynthesis and the resulting elevation in NAD+ stimulates sirtuin activity. Experimental dissection of the proposed pathway, however, has yielded some conflicting results about how changes in AMPK activity are eventually translated into an increase in NAD+ biosynthesis. Some researchers have posited that an activation of AMPK causes an increase in the expression of NAMPT and increases the rate at which NAD+ is synthesized from nicotinamide—indeed, knockdown of NAMPT was able to prevent NAD+ levels from rising in C2C12 cells cultured in low glucose medium [60]. Another study employing C2C12 cultures, however, has show that low glucose increases both AMPK activity and NAD+ levels but does so by a mechanism that is independent of changes in NAMPT expression [25]. The significant time lag between the onset of AMPK activation and when changes in intracellular NAD+ levels were observed in the latter study, however, suggests that some as yet unidentified intermediary transcriptional response may be involved in initiating the change in NAD+. Although these cell culture studies have formulated an intriguing connection between the activities of AMPK and Sirt1, any generalization of the involvement of AMPK as a proximate sensor for dietary glucose/energy availability and master regulator of Sirt1 activity in vivo is tempered by the fact that many tissues in rodents fail to show substantial increases in AMPK activity following short-term fast or long-term caloric restriction [61, 62]—dietary conditions known to significantly increase Sirt1 activity. Empirical validation for the applicability of the hypothesis in vivo could be obtained by seeing what effect inhibition of AMPK and/or NAMPT has on the ability of fasting/caloric restriction to induce Sirt1 activity and PGC-1α deacetylation in living animals.
4. Overall Regulatory Model for PGC-1α Acetylation
A synthesis of the current data for the regulation of PGC-1α acetylation in response to nutrient availability is presented in Figure 3. Under conditions of high nutrients/low NAD+ levels, PGC-1α is heavily acetylated and biologically inactive (1). It is sequestered within subnuclear foci and away from its target genes. PGC-1α is physically associated with GCN5 in these bodies and it is this acetyltransferase that maintains the overall acetylation state of PGC-1α. Unpublished data from our lab suggests that acetylated PGC-1α is able to bind to its cognate transcription factors with the same relative affinity as unacetylated PGC-1, and thus we have assumed in this model that they are also complexed with PGC-1α in the subnuclear foci (1). We have also presumed that Sirt1 is associated with PGC-1α in these bodies, but its catalytic activity relative to that of GCN5 is insufficient to result in a net deacetylation of PGC-1α (1). Additional experiments are needed to verify this as well as the binding state of other accessory proteins such as SRC-1, p300, and the TRAP complex.
An increase in intracellular NAD+ levels, as would occur following fasting, causes an acute increase in Sirt1 activity and a net deacetylation of PGC-1α protein (2). Under conditions of high Sirt1 activity, such as that achieved by Sirt1 overexpression, it is known that the occupancy of PGC-1α at its target genes is enhanced [20]. Because of this, along with unpublished data showing that overexpression of Sirt1 is able to overcome GCN5-induced inhibition of PGC-1 activity, we have assumed that deacetylation somehow releases PGC-1α from the inactive nuclear foci and permits the protein to be directed to its target genes coupled with the aid of a bound transcription factor (3). Nevertheless, there have not been any studies to directly confirm that activation of Sirt1 causes a release of PGC-1α from GCN5-induced sequestration in nuclear foci. This is a salient experiment for establishing mechanistic causality in light of a report showing that overexpression of the transcription factor ERRα is able to result in a relocalization of PGC-1α from discrete foci within the nucleus to a more diffuse pattern of distribution [63]. Such a result suggests that there may be more than one mechanism of inducing a change in PGC-1α localization.
Once located at the promoter of a target gene, the PGC-1α-transcription factor complex is able to induce RNA polymerase II (3). This is accomplished by the recruitment of a coterie of histone remodeling proteins including p300, Src-1, and the TRAP complex. Again, the binding sequence of these proteins to PGC-1α is not known. It is equally unknown as to whether the binding of these protein factors to PGC-1α is sensitive to acetylation state. This latter point is important given the observation that the docking of PGC-1α to its cognate transcription factors is not strictly sufficient to induce co-transcriptional activity. An intact N-terminal domain is needed to induce transcriptional co-activation [7, 64]. Interestingly, several of the identified acetylation sites of PGC-1a fall within this region and could alter PGC-1α’s affinity for specific factors needed for full co-activational activity.
The termination of PGC-1α activity occurs under nutrient replete conditions. When nutrient availability increases, there is an accompanying decrease in NAD+ levels. As a consequence, the activity of Sirt1 relative to GCN5 falls and PGC-1α becomes acetylated (4). This results in the transcriptional inactivation of the PGC-1α protein complex and a relocalization of PGC-1α to punctuate nuclear bodies (1).
5. Additional Avenues of Research
Again, we acknowledge that the regulatory model for PGC-1α acetylation and its relationship to the biological activity of the protein may be far more baroque than the simple model presented here. It is our hope, however, that the model will serve as a useful platform for the design of additional experiments that may shed much needed light on this highly important transcriptional coactivator. Some other interesting issues related to the biology of PGC-1α that we believe are worth pursuing are:
Why are there 13 acetylated residues in PGC-1α? Are these sites differentially regulated in response to various stimuli? Which of these residues are primarily responsible for altering the activity of PGC-1α?
Is it possible that it acetylated form of PGC-1α possesses an activity that is antagonistic to its deacetylated form? In our proposed model, we have assumed that the acetylated form has no biological activity. It is also possible that the acetylated form of PGC-1α may have a biological activity that is completely divorced from metabolic regulation.
PGC-1α undergoes other forms of post-translational modifications such as phosphorylation [14, 65, 66], ubiquitination [67], O-linked β-N-acetylglucosamination [68], and methylation [69]. How do these modifications interact with the acetylation state of PGC-1α and modify the protein’s intrinsic activity?
Why is the C-terminal RNA binding domain of PGC-1α important for the GCN5-induced relocalization of PGC-1α? Does relocalization of PGC-1α affect its RNA-binding properties?
Addressing these issues will help to settle some of the key questions surrounding the role of PGC-1α acetylation in the control of this protein’s biological activity. From a much broader perspective, however, the work will no doubt widen our understanding of how environmental cues, such as diet, are able to regulate the activity of mammalian transcriptional machinery. Thorough elucidation of the mechanistic underpinnings behind such external stimulus-transcriptional response pathways will improve our ability to design specific therapeutic interventions to alter the nature of the response for the betterment of disorders such as obesity, type II diabetes, and metabolic syndrome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 2.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 4.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 5.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 7.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 8.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess SC, Leone TC, Wende AR, Croce MA, Chen Z, Sherry AD, Malloy CR, Finck BN. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J Biol Chem. 2006;281:19000–19008. doi: 10.1074/jbc.M600050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Lay J, Tuteja G, White P, Dhir R, Ahima R, Kaestner KH. CRTC2 (TORC2) contributes to the transcriptional response to fasting in the liver but is not required for the maintenance of glucose homeostasis. Cell Metab. 2009;10:55–62. doi: 10.1016/j.cmet.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, Holloszy JO, Kelly DP. A role for the transcriptional coactivator PGC-1alpha in muscle refueling. J Biol Chem. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- 17.Benton CR, Nickerson JG, Lally J, Han XX, Holloway GP, Glatz JF, Luiken JJ, Graham TE, Heikkila JJ, Bonen A. Modest PGC-1alpha overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem. 2008;283:4228–4240. doi: 10.1074/jbc.M704332200. [DOI] [PubMed] [Google Scholar]
- 18.Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 22.Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 25.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 27.Longman D, Johnstone IL, Caceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown K, Chen Y, Underhill TM, Mymryk JS, Torchia J. The coactivator p/CIP/SRC-3 facilitates retinoic acid receptor signaling via recruitment of GCN5. J Biol Chem. 2003;278:39402–39412. doi: 10.1074/jbc.M307832200. [DOI] [PubMed] [Google Scholar]
- 29.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 37.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 38.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Horvath TL, Gao Q, Samuel VT, Shulman GI. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci U S A. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 42.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Resveratrol is Not a Direct Activator of SIRT1 Enzyme Activity. Chem Biol Drug Des. 2009 doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 43.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurd BJ, Yoshida Y, Lally J, Holloway GP, Bonen A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. J Physiol. 2009;587:1817–1828. doi: 10.1113/jphysiol.2008.168096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 50.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 52.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–2423. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 56.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 57.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 58.Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, Dumond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009 doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez AA, Kumar R, Mulligan JD, Davis AJ, Weindruch R, Saupe KW. Metabolic adaptations to fasting and chronic caloric restriction in heart, muscle, and liver do not include changes in AMPK activity. Am J Physiol Endocrinol Metab. 2004;287:E1032–E1037. doi: 10.1152/ajpendo.00172.2004. [DOI] [PubMed] [Google Scholar]
- 62.Kajita K, Mune T, Ikeda T, Matsumoto M, Uno Y, Sugiyama C, Matsubara K, Morita H, Takemura M, Seishima M, Takeda J, Ishizuka T. Effect of fasting on PPARgamma and AMPK activity in adipocytes. Diabetes Res Clin Pract. 2008;81:144–149. doi: 10.1016/j.diabres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Ichida M, Nemoto S, Finkel T. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha) J Biol Chem. 2002;277:50991–50995. doi: 10.1074/jbc.M210262200. [DOI] [PubMed] [Google Scholar]
- 64.Sadana P, Park EA. Characterization of the transactivation domain in the peroxisome-proliferator-activated receptor gamma co-activator (PGC-1) Biochem J. 2007;403:511–518. doi: 10.1042/BJ20061526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 66.Anderson RM, Barger JL, Edwards MG, Braun KH, O'Connor CE, Prolla TA, Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]