Abstract
The Sir2 family proteins (sirtuins) are evolutionally conserved NAD+ (nicotinamide adenine dinucleotide)-dependent protein deacetylases and ADP-ribosylases, which have been shown to play important roles in the regulation of stress response, gene transcription, cellular metabolism and longevity. Recent studies have also suggested that sirtuins are downstream targets of calorie restriction (CR), which mediate CR-induced beneficial effects including life span extension in a NAD+-dependent manner. CR extends life span in many species and has been shown to ameliorate many age-associated disorders such as diabetes and cancers. Understanding the mechanisms of CR as well as the regulation of sirtuins will therefore provide insights into the molecular basis of these age-associated metabolic diseases. This review focuses on discussing advances in studies of sirtuins and NAD+ metabolism in genetically tractable model system, the budding yeast Saccharomyces cerevisiae. These studies have unraveled key metabolic longevity factors in the CR signaling and NAD+ biosynthesis pathways, which may also contribute to the regulation of sirtuin activity. Many components of the NAD+ biosynthesis pathway and CR signaling pathway are conserved in yeast and higher eukaryotes including humans. Therefore, these findings will help elucidate the mechanisms underlying age-associated metabolic disease and perhaps human aging.
Keywords: Sir2, NAD+, calorie restriction, longevity regulation, aging
1. Introduction
The Sir2 family proteins (sirtuins) are evolutionally conserved longevity factors that were originally discovered and studied in yeast. SIR2 and its mammalian orthologs encode NAD+-dependent protein deacetylases and ADP-ribosylases [1]. By coupling the cleavage of NAD+ and the modifications of target proteins, sirtuins serve as a potential molecular link relaying the cellular energy state to the machinery of life span regulation. Recent studies have shown that sirtuins play important roles in CR-induced life span extension [2–8] as well as in regulating stress response, cell survival and energy metabolism [9–11], suggesting a role for sirtuins in age-related metabolic diseases.
In addition to being a co-substrate for sirtuin activities, NAD+ is also an essential pyridine nucleotide functioning in many biological processes including metabolic redox reactions and DNA repair [12]. NAD+ also plays an important role in CR-mediated life span extension by regulating NAD+-dependent longevity factors, including the Sir2 family proteins [2]. Aberrant NAD+ metabolism has also been linked to certain age-associated diseases [13–15]. In eukaryotes, NAD+ is synthesized via the de novo and salvage pathways. Cells can also assimilate extracellular NAD+ precursors into intracellular NAD+ pool via specific transporters and the salvage pathway. In this review, we summarize recent studies on yeast sirtuins and their regulation by NAD+ metabolism and CR as well as discussing possible implications of these findings in the mechanisms underlying age-related metabolic diseases.
2. Sir2 and Sir2 family proteins
Sir2 (silent information regulator 2), the founding member of the sirtuin family (silent mating-type information regulator 2 homolog), was first identified as one of the components of Sir1/2/3/4 silencing complex in yeast [16, 17]. The SIR silencing complexes are required for the maintenance and repression of three repetitive genomic regions: telomeres, the cryptic mating type loci (HML and HMR), and the ribosomal DNA loci (Fig. 1A) [18–21]. Each locus employs a unique set of DNA binding factors to recruit specific Sir proteins to mediate transcriptional silencing. Among these Sir proteins, only Sir2 is required for maintaining the silencing of all three repetitive regions. It is now clear that the repression of these loci is achieved by generating a compact chromatin configuration. The main structure of chromatin is established by packaging DNA around histones. Biochemical analysis of silenced genomic regions reveals that the core histones, mainly H3 and H4, resided within transcriptionally repressed loci are hypoacetylated in their extended N-terminal tails, which results in a compact chromatin structure less accessible for DNA binding proteins including transcription regulators [21, 22]. The fact that Sir2 exhibits a NAD+-dependent histone deacetylase activity in vitro and in vivo provides a molecular mechanistic basis for SIR complex-mediated transcriptional silencing [23–25].
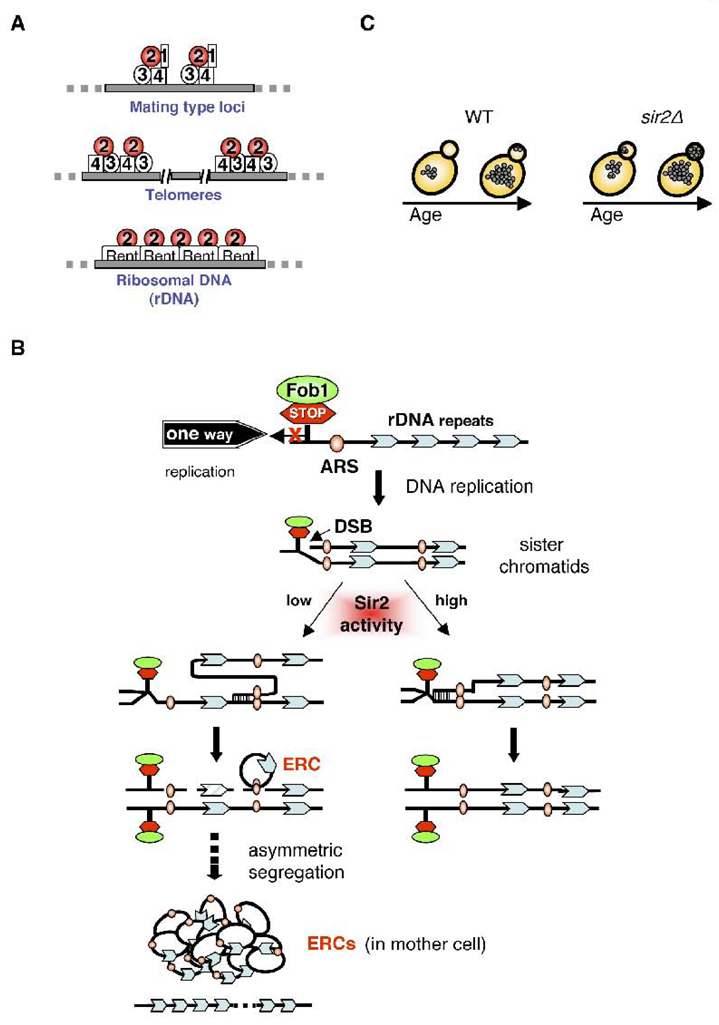
Fig. 1.
Sir2 functions in yeast. (A) The SIR complexes mediate silencing at the mating type loci, telomeres, and rDNA loci. Sir2 associates with Sir1/3/4 to repress the transcription of cryptic mating type loci (HML and HMR), while Sir2/3/4 are recruited to telomeres by telomere binding proteins to mediate telomere maintenance. In these two loci, Sir3 is suggested to mediate the spreading of SIR complex along the region to accomplish the assembly of silent chromatin. At the rDNA loci, Sir2 is recruited to the nucleolus to form RENT complexes with nucleolar protein Net1 and phosphatase Cdc14 to repress the recombination of rDNA repeats and to maintain genome integrity. (B) A model for ERC formation at the rDNA loci. Fob1-dependent replication fork block (STOP) induces double strain breaks (DBSs) within rDNA repeats during DNA replication. The repair of DBS could be initiated via recombination by using unequal sister chromatid (left) or equal sister chromatid (right) as template. The unequal sister-chromatid recombination results in the excision of rDNA repeats and the formation of extrachromosomal rDNA circle (ERC). ERCs are preferentially retained in the mother cell during asymmetric cytokinesis, and this accumulation of ERCs is suggested to be the primary cause of replicative aging in yeast. Sir2 functions in maintaining rDNA stability by promoting the cohesion of sister chromatids, which favors the proceeding of equal sister-chromatid recombination to fill up the double strand break. (C) Asymmetric segregation of oxidatively damaged proteins in yeast. In wild type cells (WT), oxidatively damaged proteins (shown as grey dots within cells) are asymmetrically retained in the mother cells, which is lost in old cells. In the sir2Δ deletion mutant, this age-dependent asymmetry is abolished, suggesting a role of Sir2 in protecting newborn cells by promoting an asymmetric segregation of damaged proteins.
Comparative genetic analyses have identified Sir2 homologs in a wide variety of organisms, ranging from bacterium to human [26, 27], showing that the Sir2 family proteins are highly conserved throughout evolution. The Sir2 family members harbor a conserved core domain required for enzyme activity, and these proteins are now collectively called sirtuins. Many sirtuins have been biochemically characterized and found to be NAD+-dependent protein deacetylases or ADP-ribosylases [28]. In higher eukaryotes, sirtuins reside in different intracellular compartments (nucleus, cytosol and mitochondria) to regulate a variety of proteins involved in different cellular processes. Examples of sirtuin targets include transcription factors [29–33], cytoskeleton components [34], and metabolic enzymes [35–38].
Sir2 homologs in yeast include Hst1, Hst2, Hst3 and Hst4 (homologous to Sir two) [26]. Among those, Hst1 has highest homology with Sir2 and has been shown to suppress silencing defects at the mating type loci of a sir2Δ deletion mutant when overexpressed [26]. However, it is suggested that Hst1 mediates transcription regulation independent of the SIR silencing complex. By interacting with another transcription regulator Sum1, Hst1 regulates transcription in a gene-specific manner [39, 40]. Hst2 is the most abundantly expressed Sir2 homolog in yeast and accounts for most of the intracellular NAD+-dependent deacetylase activity [24]. The primary subcellular localization of Hst2 is somewhat perplexing. Although Hst2 is reported as a cytoplasmic protein [24, 41], it has also been shown that Hst2 works in concert with Hst1 to down-regulate subtelomeric gene expression [42]. Interestingly, Hst2 also plays a role in regulating rDNA silencing [41] and recombination [5]. It is now believed that Hst2 shuttles between the nucleus and the cytoplasm, however, detailed mechanisms underlying the regulation of Hst2 shuttling remains unclear [43]. Although deleting any HST genes alone does not result in silencing defect, deleting HST3 and HST4 together affect telomeric silencing and cell cycle progression, accompanied by increased mitotic recombination and genome instability [26]. This genome-wide effect is possibly due to continuous acetylation of H3 at lysine 56, which is of importance in silencing and the formation of heterochromatin, further linking the Sir family to regulation of genome stability [44]. In addition, a transcriptome analysis of strains carrying individual deletions of the Sir2 family members reveals a significant overlap in regulation targets among Hst1, Hst2 and Hst3, further complicating the characterization of individual yeast sirtuin [45].
3. Sir2 family members as longevity regulators in yeast
Budding yeast propagates by asymmetric cell division, in that the partitioning between two resultant cells is unequal in morphological and in molecular aspects. The larger one is designated as mother cell, and each yeast cell can only undergo a certain number of divisions, which is known as the replicative life span (RLS). The process of approaching the limit of cell division is recognized as replicative aging. Similar to higher eukaryotic cells, aged yeast cells show declined division potential and reduced fitness. Another type of yeast life span is chronological life span (CLS), which measures the length of time cells remain viable at a non-dividing state. Yeast cells enter a non-dividing stationary phase (or post-diauxic phase) when nutrients are limited. This quiescent state has been suggested to resemble the G0 state in higher eukaryotes [46]. Several longevity factors have been identified through RLS and CLS studies [2, 47–52]. Since the roles of yeast Sir2 family proteins in CLS regulation remain unclear, in this section, we focus on the discussion of the roles of Sir2 family proteins in RLS regulation.
A primary cause of yeast replicative aging stems from structural changes within the nucleolus, a distinct region that is responsible for ribosomal RNA transcription from the rDNA loci and ribosome assembly [53]. The yeast rDNA loci consist of a stretch of ~9 kb rDNA repeats and are inherently recombinagenic. Homologous recombination between adjacent repeats is known to result in the excision of repeat unit and the formation of extrachromosomal rDNA circles, known as ERCs. ERCs are autonomously replicating circles that preferentially accumulate in the ageing mother cells due to asymmetrical segregation [53]. As a result, the abundance of ERCs increases exponentially in mother cells to a level that even exceeds the yeast genome [53]. Given the abundance of ERCs, it is possible that they might cause cell death by titrating away essential transcription and/or replication factors; however, detailed mechanisms remain unclear [54].
Since ERCs in an old cell could arise from a single initial recombination event, suppression of rDNA recombination should forestall or delay the initiation of aging process in the mother cell. A major mechanism by which yeast cells suppress ERC formation is the packaging of DNA and histones into a “silent structure” analogous to heterochromatin in higher eukaryotes. In yeast, heterochromatin is formed at telomeres, the HM loci, and the rDNA loci [18–21]. The formation of heterochromatin at the HM loci and telomeres is mediated by silencing complex Sir2/3/4 [18, 19], whereas heterochromatin at the rDNA loci is catalyzed by the RENT (regulator of nucleolar silencing and telophase exit) complex, which includes Sir2, Net1 and Cdc14 (Fig. 1A) [55–57]. Interestingly, Sir2 is the only factor that is required for silencing at all three regions [20, 58, 59]. Supporting the ERCs model of yeast replicative aging, expressing one extra copy of Sir2 increases yeast replicative life span, whereas deleting SIR2 accelerates ERC generation and shortens replicative life span [60]. In addition, other factors that can affect homologous recombination between rDNA repeats have also been demonstrated to function as longevity regulators [5, 61–63].
ERC production is likely due to a combination of Fob1-dependent rDNA repeat expansion, contraction, double strand break formation and repair (Fig. 1B)[64]. Fob1 is a nucleolar protein that binds to rDNA at the replication fork block site, creating a replication barrier in the rDNA loci, and may lead to double strand breaks [62, 65, 66]. In addition to modulating chromatin configurations, Sir2 also counteracts ERC formation via the establishment and/or maintenance of sister-chromatid cohesion at the rDNA loci [64]. Increasing the contacts between the two sister chromatids at the rDNA loci during DNA replication may reduce unequal-sister chromatid recombination upon double strand break repair [64]. Therefore, deleting Fob1 in a sir2Δ deletion mutant mitigates the formation of ERCs and rescues replicative life span back to wild type levels [60, 62]. Sir2 also affects general genomic stability. The sir2Δ deletion mutant has an increased mutation frequency compared to wild type during the initial growth phase of yeast cultures [67]. These findings also raise the question of the causal relationship between genomic instability and aging.
In addition to regulating rDNA recombination and ERC formation, Sir2 also plays a role in stress resistance and the regulation of newborn cell fitness (Fig. 1C). It has been shown that oxidatively damaged proteins, namely carbonylated proteins, are retained in mother cells and are not inherited by daughter cells during cytokinesis [68]. Interestingly, the accumulation of damaged proteins in the mother cell is a function of replicative age, and the asymmetrical partitioning of damaged proteins is lost in the sir2Δ deletion mutants [68]. Quantitative comparisons of reactive oxygen species (ROS) and ROS scavenging enzyme catalase in the mother and the daughter cells suggest a Sir2-dependent free-radical defense mechanism that confers a superior ROS management in the daughter and therefore protects the progeny from aging [69]. This rejuvenation process not only functions in asymmetrical cell division system but might also provide beneficial effects in binary division system [70].
Telomere silencing and maintenance also affect the progression of aging in multiple organisms including metazoan. Telomere maintenance in yeast requires multiple telomere binding proteins working in concert to preserve the integrity of chromosome ends, while telomere silencing is mainly achieved by the formation of heterochromatic structure. Sirtuins have been implicated both in telomere silencing and Ku protein-mediated telomere maintenance [71–73]. A very recent study shows that Sir2-mediated deacetylation of histone H4 lysine 16 decreases with age suggesting telomere silencing also plays a critical role in regulating yeast life span [73].
Although it is still unclear whether sirtuin-mediated telomere silencing and ERC-formation also contribute to longevity regulation in higher eukaryotes, accumulating evidence from various model systems suggest that sirtuins play significant roles in mediating life span regulation in complex organisms. For example, over-expressing Sir2 homologs in Drosophila and C. elegans also confers extended life span [3, 74]. In addition, the mammalian Sir2 family proteins (SIRT1-7) have been shown to regulate stress response, cell survival, as well as insulin and fat metabolism [9, 10], suggesting a role for sirtuins in age-related metabolic diseases and perhaps human aging. Future studies are warranted to determine whether over-expressions of sirtuins would indeed extend life span and/or ameliorate age-associated diseases.
4. Regulation of the Sir2 family proteins
4.1. Biochemistry of Sir2 activity
Sir2 exhibits a NAD+-dependent protein deacetylase activity, which is highly conserved among Sir2 family members [23–25] and is required for most sirtuin-mediated regulations. During deacetylation reactions, Sir2 catalyzes the transfer of acetyl group from specific lysine residues on protein substrates to the ADP ribose moiety of NAD+, followed by releasing nicotinamide, O-acetyl-ADP ribose, and deacetylated substrates [1]. In addition to serving as co-substrate in Sir2-mediated deacetylations, NAD+ and its reduced form NADH are essential redox carriers that connect the metabolism of biomolecules to ATP synthesis. The ratio of NAD+ to NADH therefore reflects the intracellular redox state and is considered as readout of metabolic activity [75]. Since Sir2 couples the cleavage of NAD+ to protein modification, it has been suggested that Sir2 functions as a molecular link relaying the signals from energy metabolism to the biological processes that Sir2 regulates.
Since NAD+ is absolutely required for Sir2 activity, deficiencies in the NAD+ synthesis salvage pathway, for example deleting the critical enzyme Npt1 (nicotinic acid phosphoribosyltransferase), abolish Sir2-mediated silencing [24, 76, 77], whereas over-expressing Npt1 confers rDNA stability and life span extension [77]. Sir2 is also subject to negative regulation by its reaction product nicotinamide (Nam). It has been shown that dosing yeast cells with Nam or genetically deleting the nicotinamidase Pnc1 reduces Sir2-mediated silencing and shortens life span [78, 79]. Extensive structural analysis of Sir2 reveals that the presence of Nam could drive Sir2 to undergo a base-exchange reaction and prevent further proceedings of deacetylation [1, 80–82], which mechanistically explains Nam as a natural non-competitive feedback inhibitor of Sir2. On the other hand, the role of another small molecule generated from Sir2-catalyzed reaction, O-acetyl-ADP ribose (AADPR), is relatively unclear yet intriguing. Structural studies show that AADPR forms a complex with Sir2 and could potentially act as an inhibitor [1, 83]. However, studies of the in vitro reconstitution of silencing complexes indicate that AADPR promotes the assembly of silencing complex and potentiates silencing activity [84]. In contrast, another study has shown that AADPR seems to be dispensable for the assembly and spreading of silencing complexes [85]. Finally, although NADH is not derived from Sir2-catalyzed reactions, it has also been demonstrated to function as a competitive inhibitor of Sir2 [75]. In addition to these endogenously produced Sir2 activators/inhibitors, many recent studies have identified small molecules that can modulate Sir2 activity, for example, sirtinol and resveratrol [1, 4, 86, 87], which are also invaluable tools for studying the functions of sirtuins.
4.2. Regulation of Sir2 by CR and environmental stresses
Calorie restriction, CR, is the most effective intervention examined to extend life span in a wide spectrum of species including mammals [88, 89]. Moderate CR can be imposed in yeast by reducing the glucose concentration from 2% to 0.5% in rich media [2, 5, 90–94]. Under this CR condition, the growth rate remains robust and both replicative and chronological life spans are extended. Variations in CR protocols have also been described in which limitation of amino acids and/or further reduction in carbon sources are employed [91, 95–97]. In yeast, moderate CR is suggested to function through reducing the activities of conserved nutrient-sensing pathways to extend life span. Decreasing the activity of the Ras-cAMP/PKA (cyclic-AMP activated protein kinase A) pathway, which regulates cell growth and stress response, extends life span [2, 48]. Deleting the nutrient responsive Sch9 (homolog of mammalian S6K kinases) or Tor1 (target of rapamycin) kinases also promotes longevity [48, 50]. Both tor1Δ and sch9Δ mutants have been suggested to be genetic mimics of CR [48, 50].
Life span extension induced by low-glucose mediated CR has been shown to require both NAD+ and Sir2 [2]. Two major models have been proposed to explain how CR-induced signals are translated to Sir2-dependent life span extension (Fig. 2). The first model centers on the metabolic shift in response to CR. CR induces a shunting of carbon metabolism from fermentation towards the mitochondrial TCA cycle [98]. The concomitant increase in respiration is necessary and sufficient for the activation of Sir2-mediated silencing and life span extension [98]. One important aspect of mitochondrial respiration is to couple ATP synthesis, a process fueled by electron transport chain resided in the inner membrane of mitochondrium, to the regeneration of NAD+ via NADH oxidation. In yeast, CR induces an increase in intracellular NAD+/NADH ratio by decreasing the NADH level [75]. Since NADH can function as a competitive inhibitor of Sir2 activity [75], it is suggested that the increase of NAD+/NADH ratio induced by CR activates Sir2. In support for this model, genetic manipulations that cause a decrease in NADH levels are shown to increase Sir2 activity and extend life span [75, 98]. However, since NADH is a weak inhibitor of Sir2, it has been questioned that reported in vivo NADH levels are too low to inhibit Sir2 activity [99]. It is possible that intracellular compartmentalization of NAD+ and NADH and/or specific protein-protein interactions create local high NAD+/NADH ratios thereby activating Sir2 in vivo. It has also been suggested that the affinity/sensitivity of Sir2 towards its substrates and inhibitors varied when Sir2 was in complex with different interacting partners [100].
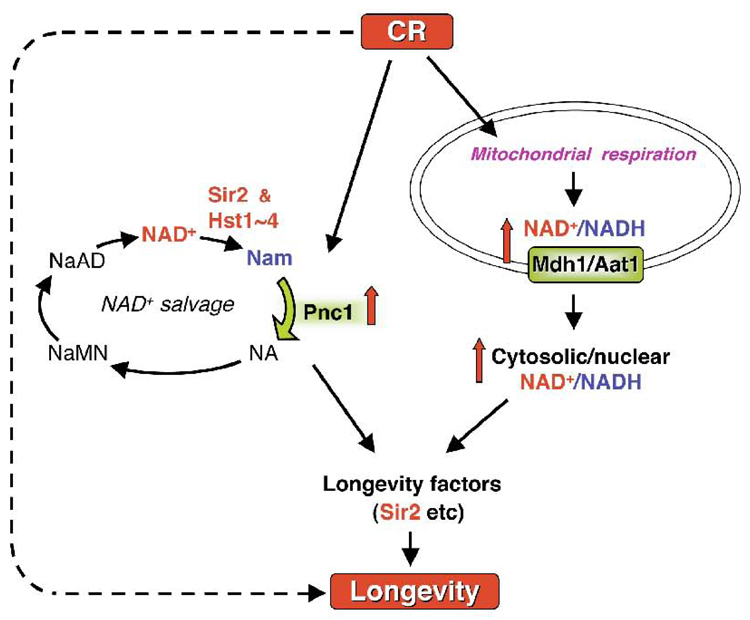
Fig. 2.
A model for CR-mediated life span extension in yeast. Multiple pathways mediate CR-induced life span extension. CR increases mitochondrial respiration and the concomitant elevation in NAD+/NADH ratio. The malate-aspartate NADH shuttle components (Mdh1/Aat1) function to balance redox equivalents between the mitochondrial and the cytosolic/nuclear pools, and are essential for relaying the metabolic signals in the mitochondria to downstream longevity factors, such as Sir2. CR also increases protein level of nicotinamidase Pnc1 in NAD+ salvage pathway. Up-regulation of Pnc1 facilitates the clearance of Nam and enhances Sir2 activity. Other Sir2-independent pathways also exist to mediate CR-induced beneficial effects including life span extension.
Supporting the NAD+/NADH ratio model, the NADH shuttle system has been shown to play important roles in CR [94]. The mitochondrial inner membrane is impermeable to NAD+ and NADH, therefore, the NADH shuttle system is required to move small permeable redox equivalents of NAD+ and NADH across the mitochondrial membrane to balance the NAD+/NADH ratio between the mitochondrial and the cytosolic/nuclear pools (Fig. 3) [101]. It has been shown that deleting NADH shuttle components abolishes CR-mediated life span extension whereas over-expressing NADH shuttle components extends life span [94]. In mammals, impairments of mitochondrial metabolism and NADH shuttles have also been implicated in age-associated diseases such as diabetes [38, 102]. The NADH shuttles thus play important roles in mitochondrial metabolism, metabolic fitness and life span regulation. In line with this model, CR has also been reported to increase mitochondrial respiration in multiple model organisms and humans [6, 103–107].
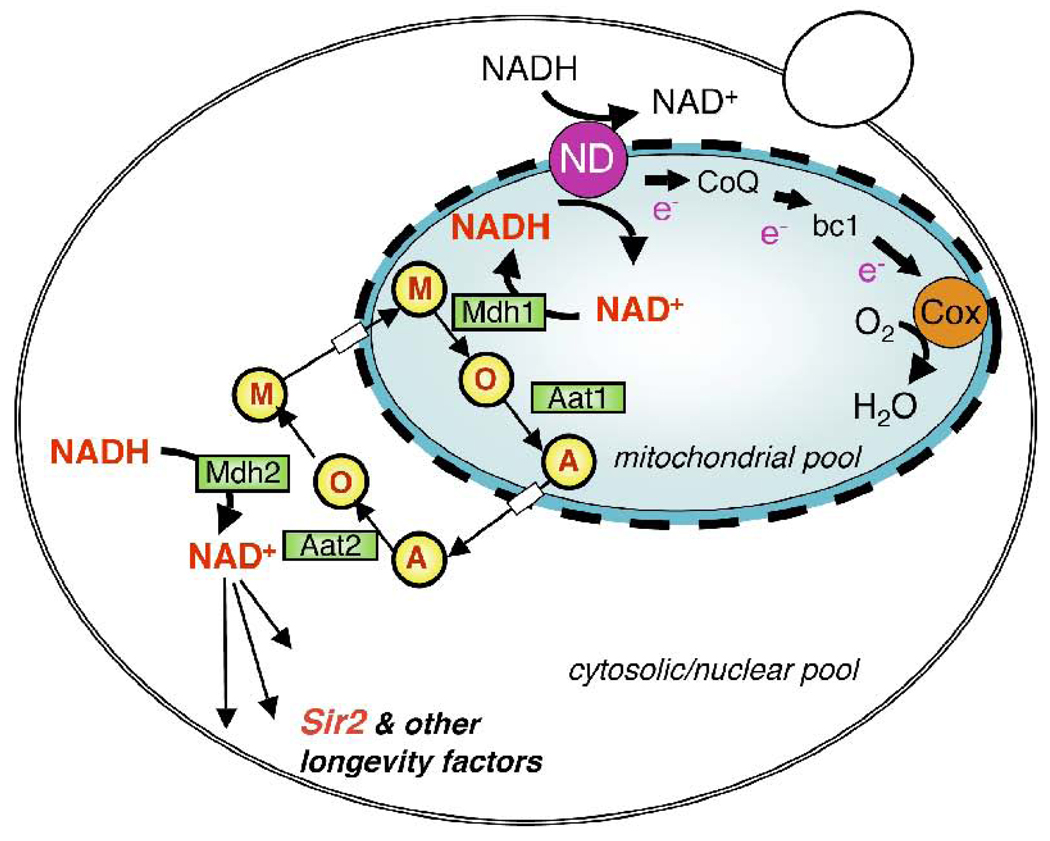
Fig. 3.
Schematic presentations of the yeast mitochondrial respiratory chain and the NADH shuttle system. Mitochondrial NADH dehydrogenases (ND) oxidize NADH from both the cytosolic and mitochondrial pools to regenerate NAD+. The electrons acquired are then passed down the electron transport chain to the terminal acceptor oxygen (O2). CR activates mitochondrial activity, which results in an increase in NAD+/NADH ratio. Since the mitochondrial inner membrane is impermeable to NAD+ and NADH, this increase in NAD+/NADH ratio is transmitted into the cytosolic/nuclear pools via the NADH shuttle system. The NADH shuttle system affects the NAD/NADH ratio indirectly by coupling the redox reaction of NAD+ and NADH with the redox reactions of permeable redox equivalents of NAD+ and NADH. In the example shown here, cytosolic malate dehydrogenase (Mdh2) couples the oxidation of NADH with the reduction of oxaloacetate (O). The resulting reduced product malate (M) enters the mitochondria via specific carriers and thereby shuttling redox equivalents into the mitochondrial matrix. In the mitochondrial matrix, mitochondrial Mdh1 catalyzes the generation of NADH from NAD+ and electrons carried by malate (M) are concurrently oxidized to oxaloacetate (O). Mitochondrial aspartate aminase (Aat1) then converts oxaloacetate to aspartate (A), which is subsequently transported to the cytosol via other carriers. Cytosolic Aat2 then regenerates oxaloacetate from aspartate, and another round of NADH shuttling is ready to resume. With the NADH shuttle system, alterations of the mitochondrial NAD+/NADH ratio resulted from the increase of mitochondrial respiration could be conveyed to the cytosolic/nuclear compartments.
The other model is derived from biochemical studies of Sir2 inhibition. As mentioned above, nicotinamide (Nam) is a by-product generated during Sir2-mediated deacetylation, which is also a potent non-competitive inhibitor of Sir2 [78, 80]. The nicotinamidase Pnc1 in yeast is responsible for the clearance of Nam by converting it to nicotinic acid (NA) and ammonia [108]. In vitro experiments show that recombinant Pnc1 stimulates Sir2 activity [79]. Moreover, over-expression of Pnc1 increases life span [109] and prevents the inhibition of Nam towards rDNA and telomeric silencing [79]. Along with the enzyme activity of Pnc1, the finding that Pnc1 protein level is up-regulated under CR conditions places Pnc1 to the CR pathway [109]. In addition, several types of environmental mild stresses, such as increased osmolarity, salinity, and amino acid starvation, as well as internal stresses also profoundly induce the elevation of Pnc1 protein [79, 108–110], a response primarily mediated by stress responsive transcription factors Msn2 and Msn4 [108, 111]. Given that many mild stresses have been shown to extend life span [90, 109, 112] and that the reduction in calorie intake might also be sensed by cells as mild stress signals, it has been proposed that Pnc1 is the key factor that translates stress signals (including CR) to Sir2 activation [113]. Interestingly, Nampt (nicotinamide phosphoribosyltransferase), the functional homolog of Pnc1 in higher eukaryotes, is also responsive to stresses and suggested to be the mediator of CR-induced benefit [114, 115].
It is noteworthy that in addition to these models, other schemes for the mechanisms of CR are highly possible and remain to be explored. For example, the requirement of Sir2 as well as mitochondrial respiration in CR-induced replicative life span (RLS) extension has remained controversial [116]. It has been shown that under extreme CR conditions or in certain genetic backgrounds, neither functional mitochondria nor Sir2 are required for RLS extension [95–97, 117]. In addition, Sir2 also appears to be dispensable for CR-induced chronological life span extension [67, 92]. Identification of additional CR downstream factors and understanding their relationships with Sir2 activity and NAD+ metabolism will help elucidate the molecular aspects of these CR models. It is, however, generally believed that CR modulates a complex metabolic network and multiple longevity factors functioning in accordance to improve overall fitness and to ensure optimal survival of organisms.
4.3. NAD+ metabolism and the regulation of sirtuins
The function of NAD+ in carrying electrons from one pathway to another does not directly affect the size of the pyridine nucleotide pool. In contrast, the participation of NAD+ as a co-substrate in protein modifications catalyzed by sirtuins, for example, results in consumption of NAD+ accompanied by the generation of Nam. It has been suggested that the basal levels of intracellular NAD+ are accounted by those bound to redox enzymes to fulfill the function in delivery of redox equivalents, and are kept relatively constant [118]. On the other hand, those involved in sirtuin-mediated reactions are free-form NAD+ and rely on salvage pathway for regeneration [118]. The best example to explicate the significance of NAD+ salvaging is the finding that deleting the critical salvage enzyme Npt1 results in a significant decrease in total cellular NAD+ level [75, 76], and abolishes Sir2-dependent benefits induced by CR [2]. On the other hand, over-expression of Npt1 is sufficient to extend life span, although the steady-state level of NAD+ remains unchanged [77]. It is therefore suggested that increased flux through the salvage pathway might also help increase the availability of NAD+ to Sir2 [77].
With the significance of NAD+ for cellular metabolism and sirtuin activities in response to environmental cues and internal signals, it is essential to understand NAD+ biosynthetic pathways. There are two major strategies for cells to generate NAD+ [119] (Fig. 4A). In yeast, the de novo synthesis uses tryptophan to make intermediate NaMN (nicotinic acid mononucleotide) through the oxygen-dependent kynurenine pathway, which consists of 6 enzymatic steps and one spontaneous reaction [119]. The other strategy of making NaMN involves the salvage of Nam and NA generated intracellularly or taken from the environment via the classical Preiss-Handler pathway, which is featured by Pnc1, Npt1 and Tna1 (transporter of nicotinic acid) [120–122]. NaMN produced from both pathways are first adenylated by adenylyltransferases Nma1/Nma2 to nicotinic acid adenine dinucleotide (NaAD), followed by amidation via the NAD+ synthetase Qns1 to form NAD+ [121, 122]. Cells also synthesize NAD+ from nicotinamide riboside (NmR) and nicotinic acid riboside (NaR), another salvage pathway conserved from bacterium to human. Assimilation of NmR/NaR could be initiated via phosphorylation to form mononucleotide by NmR kinase 1 (Nrk1) [123, 124] or via hydrolysis/phosphorolysis to form Nam/NA by three nucleoside-splitting enzymes (Urh1/Pnp1/Meu1) to enter the Preiss-Handler pathway [118]. Since Nrk1-dependent NmR assimilation replenishes NAD+ pool without producing Nam (Fig. 4A), it is possible that increasing the activity of this NmR salvage branch might also stimulate sirtuin activity.
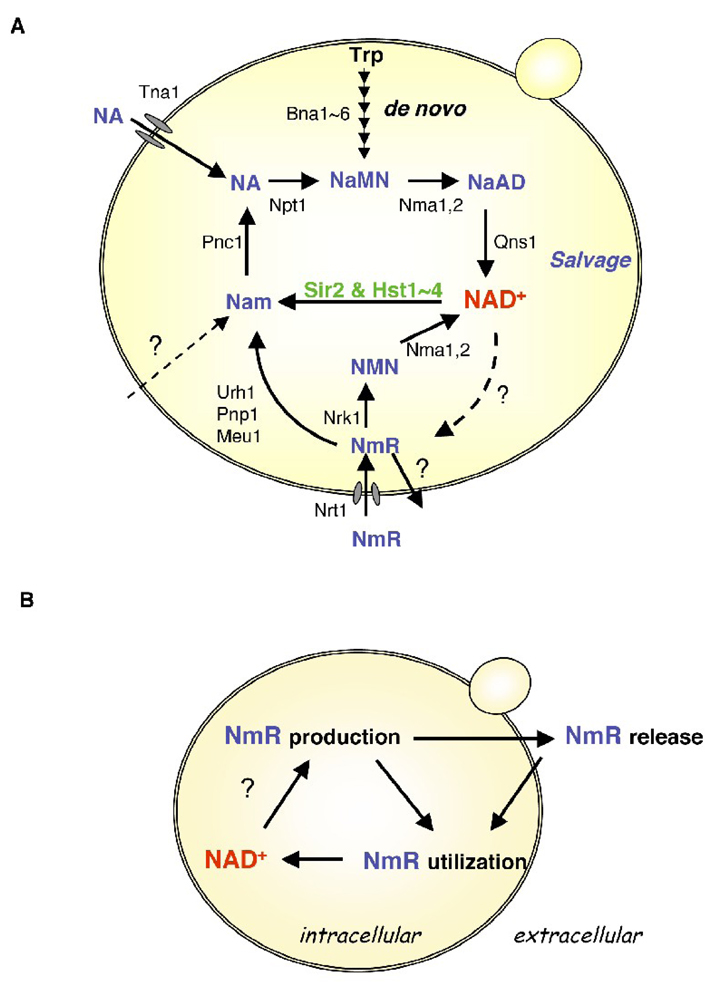
Fig. 4.
NAD+ biosynthesis and homeostasis in yeast. (A) The model of NAD+ biosynthesis pathways in yeast. NAD+ is produced via de novo synthesis from tryptophan or the salvage of NAD+ metabolites from NAD+-consuming reactions and from environment. See text for detail steps. (B) A schematic model of extended NAD+ pool. Yeast cells constitutively produce, release, and retrieve nicotinamide riboside (NmR). This extended cycle encompasses intracellular and extracellular compartments and might functions as a flexible pyridine nucleotide reservoir.
The biochemistry of NAD+ biosynthesis has been intensively studied; however, the regulation of NAD+ metabolism remains largely unclear. When grown in regular rich medium, cellular NAD+ level of yeast is significantly lowered in the mutant deficient in Nam/NA salvage but unchanged in the mutants defective in the de novo pathway [76], suggesting that salvaging NAD+ metabolites is the primary means to make NAD+. It has been reported that the de novo NAD+ synthesis is regulated by Hst1-mediated repression and that deletion of HST1 raises the steady-state level of NAD+ [45]. However, it remains to be determined whether this elevation in NAD+ production could be delivered into the nucleus where Sir2-dependent regulation of life span takes place. Paradoxically, over-expression of Npt1 extends Sir2-mediated life span without altering the steady-state total NAD+ level [77]. Moreover, studies using a NAD+ reporter system to detect the level of free NAD+ in the nucleus show that CR does not increase nuclear NAD+ level [125], which argues that Sir2 might not be regulated by NAD+ availability under CR [125]. To reconcile all these seemingly conflicting observations, the development of a real-time detection tool with the ability to distinguish NAD+ from the reduced form (NADH) and phosphorylated form (NADP+/NADPH) is essential to reveal how NAD+ residing in spatially separating compartments are modulated under various conditions.
It has been shown in various systems that exogenous NmR is a salvageable form of NAD+ precursors, and total cellular NAD+ level of yeast cells could be effectively replenished with NmR supplement [118, 126, 127]. NmR has also been shown to rescue the silencing and life span defects in cells grown in NA-free media in a Sir2-dependent manner [118]. In our earlier studies, we observed that yeast cells defective in both de novo synthesis and Nam/NA salvage were able to survive when inoculated next to NAD+ prototrophic cells [2]. This fortuitous finding leads to the discovery that NmR is a bona fide endogenous NAD+ metabolite and is constitutively produced, released and retrieved by yeast cells [128], suggesting an extended NAD+ pool that encompasses both the intracellular and extracellular compartments (Fig. 4B). The NmR assimilation pathways that consist of the Nrk1-dependent route and Urh1/Pnp1/Meu1-mediated catabolism are necessary not only for utilizing NmR supplement [118] but also for salvaging endogenously produced NmR [118, 128]. It is still unclear why cells allow NAD+ metabolites to traffic between intracellular and extracellular compartments, which poses the potential risk of losing NAD+. One intriguing possibility is that keeping a flexible NmR/NAD+ pool facilitates prompt adjustments of the activities of NAD+-dependent enzymes [128]. In line with this idea, Nrk2, the isoform of Nrk1 identified in mammals, has been shown to be up-regulated by ~20-fold in protein level in response to stresses [129]. It is suggested that this induction of NAD+ metabolizing enzyme is to support sirtuin-dependent protection of neurons [129]. In addition, it has also been reported that NmR circulates in peripheral bloodstream in mammals [130], supporting the possible function of extracellualr NmR as a pyridine nucleotide reservoir. These observations also raise another interesting speculation that factors regulating NmR pool could also be CR targets. As mentioned above, it has been suggested that Pnc1-mediated Nam salvage increases in response to mild stresses and CR to eliminate sirtuin inhibitor, Nam, and/or to increase NAD+ availability. Before the Pnc1/Nam route is activated, the NmR pool might serve as a prompt supply of NAD+ precursors since NmR can be assimilated into NAD+ via multiple paths. Alternatively, NmR production/release could also be used as a means by which cells down-regulate NAD+ levels to slow down growth rate in response to stresses. Interestingly, it is also found that SIR2 deletion enhances NmR release, which suggests an active role of Sir2 in NmR/NAD+ metabolism [128]. While we are still searching for the answers to how and where NmR is produced, the investigation of the regulation of NmR metaboizing enzymes under various growth conditions as well as the identification of additional factors that modulate NmR/NAD+ metabolism might provide clues to the physiological roles of this extended NAD+ pool and its relationship to NAD+ and central metabolism.
5. Conclusions and Perspective
Despite the simplicity as a unicellular organism, these studies have demonstrated that the budding yeast is a useful model system for studying the conserved longevity regulating factors/pathways. Although these pathways are conserved among different organisms, significant discrepancies also exist due to various degree of complexity. For example, in contrast to yeast Sir2, which mainly functions in the nucleus, mammalian sirtuins are found in different intracellular compartments. The enzymatic substrates of mammalian sirtuins encompass diverse protein targets in addition to histones. These sirtuins may also play different roles in different tissues during development, which further complicates sirtuin functions and elicits concerns about systemic manipulation of sirtuin activities. Furthermore, there are dissimilarities in NAD+ biosynthesis between yeast and higher eukaryotes, although the role of NAD+ in regulating sirtuin activities is highly conserved.
Sirtuins and NAD+ homeostasis factors have emerged as putative metabolic regulators of longevity and have been implicated in CR and many age-associated diseases. With this molecular basis, interventions to modulate sirtuin activity and NAD+ metabolism are now under intense study. For example, administrations of NAD+ precursors such as nicotinamide riboside (NmR) and nicotinamide mononucleotide (NMN) ameliorate deficiencies related to aberrant NAD+ metabolism in yeast, mouse and human cells [129, 131, 132] and therefore appear to be a promising strategy for medical and nutritional purposes. Unraveling novel components of NAD+ metabolic pathways will further our understanding of intracellular NAD+ homeostasis, the regulation of sirtuin activity as well as the mechanisms of CR-induced beneficial effects. These findings will also serve as an invaluable paradigm to facilitate the development of preventative and therapeutic reagents for age-associated diseases and metabolic disorders in human.
Acknowledgments
We are grateful to the researchers whose work provided the basis for this review. The Lin laboratory is supported by the National Institute on Aging (RO1-AG24351).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 2.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 3.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 5.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science (New York, N.Y. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes & development. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 10.Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell Mol Life Sci. 2007;64:752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 13.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nature structural & molecular biology. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 14.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends in biochemical sciences. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhai RG, Zhang F, Hiesinger PR, Cao Y, Haueter CM, Bellen HJ. NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature. 2008;452:887–891. doi: 10.1038/nature06721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 19.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & development. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 20.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & development. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 21.Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 22.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & development. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 23.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 24.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & development. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 27.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 28.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 30.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 31.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, N.Y. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 33.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 34.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 35.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 36.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. From the Cover: Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 39.Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 43.Wilson JM, Le VQ, Zimmerman C, Marmorstein R, Pillus L. Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 2006;7:1247–1251. doi: 10.1038/sj.embor.7400829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 45.Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. Divergent roles of RAS1 and RAS2 in yeast longevity. The Journal of biological chemistry. 1994;269:18638–18645. [PubMed] [Google Scholar]
- 48.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science (New York, N.Y. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 49.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends life span by shifting carbon toward respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 50.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science (New York, N.Y. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 51.Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fabrizio P, Longo VD. Chronological aging-induced apoptosis in yeast. Biochimica et biophysica acta. 2008;1783:1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 54.Sinclair DA, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 55.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 56.Ghidelli S, Donze D, Dhillon N, Kamakaka RT. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 2001;20:4522–4535. doi: 10.1093/emboj/20.16.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shou W, Sakamoto KM, Keener J, Morimoto KW, Traverso EE, Azzam R, Hoppe GJ, Feldman RM, DeModena J, Moazed D, Charbonneau H, Nomura M, Deshaies RJ. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- 58.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & development. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 59.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 60.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 62.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 63.Jazwinski SM. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene. 2005;354:22–27. doi: 10.1016/j.gene.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992;233:355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- 66.Burkhalter MD, Sogo JM. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell. 2004;15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 67.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 68.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 69.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erjavec N, Cvijovic M, Klipp E, Nystrom T. Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proc Natl Acad Sci U S A. 2008;105:18764–18769. doi: 10.1073/pnas.0804550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nature genetics. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 72.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 73.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 75.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes & development. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 78.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 79.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 81.Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 82.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 83.Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J Biol Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- 84.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 85.Yang B, Kirchmaier AL. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Molecular biology of the cell. 2006;17:5287–5297. doi: 10.1091/mbc.E06-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 87.Denu JM. The Sir2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 88.Weindruch W, Walford RL. The retardation of aging and diseases by dietary restriction. Springfield, Illinois, USA: Charles C. Thomas; 1998. [Google Scholar]
- 89.Roth GS, Lane MA, Ingram DK, Mattison JA, Elahi D, Tobin JD, Muller D, Metter EJ. Biomarkers of caloric restriction may predict longevity in humans. Science (New York, N.Y. 2002;297:811. doi: 10.1126/science.1071851. [DOI] [PubMed] [Google Scholar]
- 90.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Easlon E, Tsang F, Dilova I, Wang C, Lu SP, Skinner C, Lin SJ. The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. The Journal of biological chemistry. 2007;282:6161–6171. doi: 10.1074/jbc.M607661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 93.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. Faseb J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 96.Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- 97.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:1381–1387. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 99.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 100.Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol Cell Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bakker BM, Overkamp KM, van Maris AJ, Kotter P, Luttik MA, van Dijken JP, Pronk JT. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 102.Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, Yamauchi N, Kubota N, Murayama S, Aizawa T, Akanuma Y, Aizawa S, Kasai H, Yazaki Y, Kadowaki T. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science (New York, N.Y. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- 103.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 104.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 105.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 106.Zarse K, Schulz TJ, Birringer M, Ristow M. Impaired respiration is positively correlated with decreased life span in Caenorhabditis elegans models of Friedreich Ataxia. Faseb J. 2007;21:1271–1275. doi: 10.1096/fj.06-6994com. [DOI] [PubMed] [Google Scholar]
- 107.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene. PNC1, Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 109.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Silva RM, Duarte IC, Paredes JA, Lima-Costa T, Perrot M, Boucherie H, Goodfellow BJ, Gomes AC, Mateus DD, Moura GR, Santos MA. The yeast PNC1 longevity gene is up-regulated by mRNA mistranslation. PLoS One. 2009;4:e5212. doi: 10.1371/journal.pone.0005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shama S, Lai CY, Antoniazzi JM, Jiang JC, Jazwinski SM. Heat stress-induced life span extension in yeast. Exp Cell Res. 1998;245:379–388. doi: 10.1006/excr.1998.4279. [DOI] [PubMed] [Google Scholar]
- 113.Lamming DW, Wood JG, Sinclair DA. Small molecules that regulate lifespan: evidence for xenohormesis. Mol Microbiol. 2004;53:1003–1009. doi: 10.1111/j.1365-2958.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- 114.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- 116.Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing research reviews. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 117.Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased Life Span due to Calorie Restriction in Respiratory-Deficient Yeast. PLoS Genet. 2005;1:614–621. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 119.Panozzo C, Nawara M, Suski C, Kucharczyka R, Skoneczny M, Becam AM, Rytka J, Herbert CJ. Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 2002;517:97–102. doi: 10.1016/s0014-5793(02)02585-1. [DOI] [PubMed] [Google Scholar]
- 120.Llorente B, Dujon B. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w) FEBS Lett. 2000;475:237–241. doi: 10.1016/s0014-5793(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 121.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958;233:488–492. [PubMed] [Google Scholar]
- 122.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem. 1958;233:493–500. [PubMed] [Google Scholar]
- 123.Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 124.Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS Biol. 2007;5:e263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anderson RM, Latorre-Esteves M, Neves AR, Lavu S, Medvedik O, Taylor C, Howitz KT, Santos H, Sinclair DA. Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science. 2003;302:2124–2126. doi: 10.1126/science.1088697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grose JH, Bergthorsson U, Xu Y, Sterneckert J, Khodaverdian B, Roth JR. Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J Bacteriol. 2005;187:4521–4530. doi: 10.1128/JB.187.13.4521-4530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Merdanovic M, Sauer E, Reidl J. Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae. J Bacteriol. 2005;187:4410–4420. doi: 10.1128/JB.187.13.4410-4420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu SP, Kato M, Lin SJ. Assimilation of Endogenous Nicotinamide Riboside Is Essential for Calorie Restriction-mediated Life Span Extension in Saccharomyces cerevisiae. J Biol Chem. 2009;284:17110–17119. doi: 10.1074/jbc.M109.004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sasaki Y, Araki T, Milbrandt J. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J Neurosci. 2006;26:8484–8491. doi: 10.1523/JNEUROSCI.2320-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmidt-Brauns J, Herbert M, Kemmer G, Kraiss A, Schlor S, Reidl J. Is a NAD pyrophosphatase activity necessary for Haemophilus influenzae type b multiplication in the blood stream? Int J Med Microbiol. 2001;291:219–225. doi: 10.1078/1438-4221-00122. [DOI] [PubMed] [Google Scholar]
- 131.Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annual review of nutrition. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]