Abstract
Multi-dimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) is a well-developed technology for global lipid analysis, which identifies and quantifies individual lipid molecular species directly from lipid extracts of biological samples. By using this technology, we have revealed three marked changes of lipids in brain samples of subjects with mild cognitive impairment of Alzheimer’s disease including sulfatides, ceramides, and plasmalogens. Further studies using MDMS-SL lead us to the identification of the potential biochemical mechanisms responsible for the altered lipids at the disease state, which are thoroughly discussed in this mini review. Specifically, in studies to identify the causes responsible for sulfatide depletion at the mild cognitive impairment stage of Alzheimer’s disease, we have found that apolipoprotein E is associated with sulfatide transport and mediates sulfatide homeostasis in the nervous system through lipoprotein metabolism pathways and that alterations in apolipoprotein E-mediated sulfatide trafficking can lead to sulfatide depletion in the brain. Collectively, the results obtained from lipidomic analyses of brain samples provide important insights into the biochemical mechanisms underlying the pathogenesis of Alzheimer’s disease.
Keywords: Alzheimer’s disease, apolipoprotein E, ceramide, electrospray ionization-mass spectrometry, plasmalogen, shotgun lipidomics, sulfatides
Introduction to lipidomics
Lipidomics, which studies cellular lipids in a large-scale is a newly emerged research discipline [1, 2]. Research in lipidomics has been focused on determination of the altered levels of lipids induced by a disease state, a gene mutation (knockout, or over-expression), a therapeutic treatment, or other perturbations, and identification of the lipid metabolic pathways and networks responsible for the altered lipids under the pathological and pathophysiological conditions. Attention is also paid to the temporal and spatial changes of cellular lipidomes as well as interactions of lipids with lipids, proteins, and other cellular moieties.
Mass spectrometry (MS), particularly electrospray ionization (ESI) MS, has played an essential role in characterization, identification, and quantitation of lipid species from biological samples [1, 3–8]. Accordingly, MS analyses of lipids have even been generalized as lipidomic analysis in literature. Although lipidomics has only emerged as a distinct field within the past few years [1, 2], numerous new discoveries and/or developments have already been made in the field and several special issues on lipidomics have been published including Prostaglandins & other Lipid Mediators (Vol. 77, 2005), Frontiers in Bioscience (Vol. 12, January, 2007), Methods in Enzymology (Vol. 432, 2007), European Journal of Lipid Science and Technology (Vol. 111(1), January 2009), and Journal of Chromatography B (Vol. 877 (26), 2009).
In general, ESI-MS analyses of lipids are classified into LC-MS and direct infusion (or shotgun lipidomics). Techniques such as selected reaction monitoring (SRM) or selected ion monitoring (SIM) or data-dependent analysis are usually associated with the HPLC-MS platforms while neutral loss and/or precursor-ion scans are the powerful tools applied for shotgun lipidomics. Multi-dimensional MS-based shotgun lipidomics (MDMS-SL) after intrasource separation represents one of the most powerful approaches in shotgun lipidomics [6, 9–11].
Multi-dimensional mass spectrometry-based shotgun lipidomics (MDMS-SL)
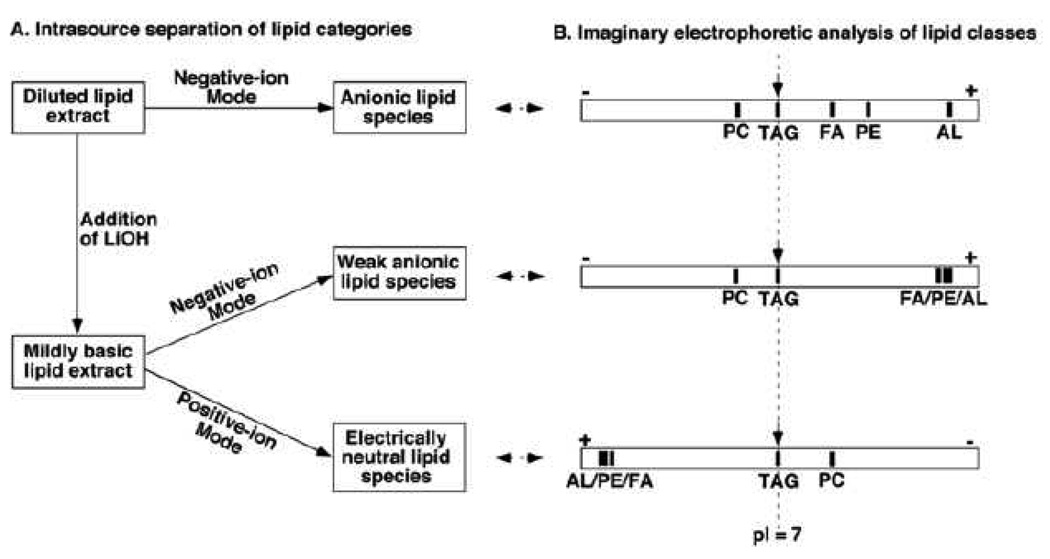
In MDMS-SL, the need for chromatography is largely replaced through exploiting the unique chemistries of different lipid classes including even extremely minor lipid classes. For example, differential hydrophobicity and differential sensitivity to base treatment are exploited during a multiplexed extraction approach [12]. Exploiting the differential electrical properties of lipid classes under multiplexed experimental conditions allows the ion source to selectively ionize a certain category of lipid classes at certain conditions. This is analogous to the electrophoretic separation of different compounds possessing different pI values as schematically illustrated (Figure 1).
Figure 1.
Schematic comparison of intrasource separation of lipid categories to the imaginary electrophoretic separation of lipid classes. Panel A schematically shows the selective ionization of different lipid categories under three different experimental conditions. Panel B schematically shows the imaginary chromatograms of lipid classes through electrophoretic analyses under corresponding experimental conditions. PC, TAG, FA, PE, and AL stand for phosphatidylcholine, triacylglycerol, free fatty acid, phosphatidylethanolamine, and anionic lipids, respectively. (Reprinted from ref. [93] with permission)
Following the selective ionization of a certain category of lipid classes, we could ramp the neutral loss of all potential fragments or monitor all potential fragment ions unit by unit (mass or mass to charge, respectively) in a mass range of interest to identify individual ion peaks. Each ramp should constitute a two-dimensional map of the molecular species in the determined mass range. The first dimension is the molecular ions (x-axis) in m/z values, while the second dimension is comprised of the mass corresponding to the neutrally lost fragments in mass values or the monitored fragment ions in m/z values (y-axis). The cross peaks of a given primary molecular ion in the first dimension with the second dimension represent the fragments of the given molecular ion. Analysis of these cross peaks (i.e. the individual fragments) thereby identifies the structure of the given molecular ion as well as its isobaric substituents [9]. This two-dimensional mass spectrometry (2D MS) is entirely analogues to two-dimensional NMR spectroscopy. The only difference between these mapping approaches is that the former uses units in the mass domain while the latter uses units in the frequency domain.
Since naturally occurring lipids are comprised of known building blocks, the ramping process described above can be readily simplified by monitoring only those building blocks that are characteristic of individual molecular species of a lipid class of interest (i.e. building blocks of the class of interest) by using tandem MS techniques of neutral loss and precursor ion scans. In this simplified 2D MS, the y-axis is the building blocks of a lipid class or a category of lipid classes. Individual lipid molecular species of a class of interest by 2D MS can be automatically identified [13].
Each 2D MS predictably varies with different infused solution conditions (e.g., lipid concentration, acidic/alkaline condition, and solvent polarity), ionization conditions (e.g., source temperature and spray voltage), and fragmentation conditions (e.g., collision gas pressure, collision energy, and collision gas). Each of these variables facilitates the construction of additional dimensions which can be built upon each 2D MS foundation, which collectively constitutes a new level of information directly obtainable from MS analysis of lipids, i.e., MDMS.
Identified lipid molecular species by MDMS can be quantified by using a two-step method [6, 11, 12, 14]. In this two-step methodology, the molecular species in the class of interest that are abundant and not overlapped with the species in any other classes are quantified by ratiometric comparison (see below) with the selected internal standard of the class by using a full MS scan. Then, some or all of these quantified molecular species plus the original internal standard are employed as standards to quantitate other low abundance and/or overlapped molecular species in the class by using one or more class-specific precursor-ion and/or neutral loss scans. Using this two-step methodology, the dynamic range of quantitation can be extended by at least two orders of magnitude as previously demonstrated [15] and can be easily achieved through automated bioinformatic analyses as described previously [13].
Given cu and ci are the contents of an individual species and the selected internal standard, respectively, while Iu and Ii are the peak intensities of the species and the selected internal standard, respectively, after 13C de-isotoping [6], ratiometric comparison with the selected internal standard of the class as follows:
| (1) |
can be derived from the linear correlation between the content (c) and the ion intensity (I) of a species. Within the linear dynamic range of an analytical method,
| (2) |
where a and b are the response factor-related parameter and background noise, respectively. When I >> b (e.g., S/N > 5),
| (3) |
The formula (1) is obtained when the response factors of different individual species of a lipid class are essentially identical. This holds true for polar lipid classes in the low concentration region after 13C de-isotoping, as validated with many studies by different laboratories [16–20]. However, the response factors of different non-polar lipid species to ESI-MS are quite different and have to be pre-determined for quantification [10].
The determined contents of those non-overlapping and abundant species by ratiometric comparison with the selected internal standard are the candidate standards for the second step of quantification. Note that the pre-selected exogenously added internal standard is always in the peak list of the candidate standards but not necessarily in the peak list of the final selected standards for the second step quantification [13]. It should be pointed out that ion intensities in the class-specific tandem MS scan(s) might depend on the profile of subclasses or subtypes of species [21]. Therefore, different standards from these subclasses or subtypes of species for the second step quantification are also considered [21].
Finally, the uniqueness of certain lipid classes can be exploited to identify and quantify their individual species. For example, the presence of two phosphate moieties in the cardiolipin chemical structure is unique, which is reflected as doubly-charged ions in mass spectra acquired under certain conditions [22]. By searching the [M – 2H + 1]2− isotopologues from the resulting doubly-charged ion pattern of cardiolipin species, cardiolipin molecular species can be readily recognized and quantified without interference from overlapping species from other classes within the mass range. The presence of a primary amine in phosphoethanolamine-containing species is also unique in the cellular lipidome and has been exploited to tag the phosphoethanolamine-containing lipid species using fluorenylmethoxylcarbonyl (Fmoc) chloride [23]. The facile loss of Fmoc from the tagged lipid molecular species allowed us to readily identify and quantify these species with unprecedented sensitivity.
This MDMS-SL technology in combination with an automated sample injection device [15] is a powerful platform for global lipid analysis. The advances of this technology at least include, but are not limited to, the following:
MDMS-SL enables us to analyze samples in a limited amount of materials
By using this approach, we held the first study on the mass distribution and molecular species composition of triacylglycerols in ganglia with a sample size of less than 2 mg [24]. This study demonstrates five novel findings. First, unanticipated high levels of TAG were present in all examined ganglia from multiple species (e.g. mouse, rat and rabbit). Second, ganglial TAG mass content is location dependent. Third, the TAG mass levels in ganglia are species specific. Fourth, dorsal root ganglial TAG mass levels in streptozotocin-induced diabetic mice are dramatically depleted relative to those in untreated control mice. Fifth, mouse ganglial TAG mass levels decrease with age although molecular species composition is not changed. Collectively, these results indicate that TAG is an important component of ganglia and may potentially contribute to pathological alterations in peripheral neuronal function in diabetic neuropathy.
MDMS-SL provides an instant criterion to achieve representative sampling
One of the concerns in lipidomic analysis is how to determine the analyzed sample portion is a representative one of the entire sample of interest. For example, neurons are enriched in gray matter whereas oligodendrocytes are mainly present in white matter. One complication present in any study with brain samples, particularly those from large brains, e.g., human, is the varying degree of the cross presence of gray and white matter in the sampled tissues. Differences in the ratio of co-existing gray to white matter represent an unpredictable variable which may overshadow real differences between the samples from different states. Up to date, only MDMS-SL has addressed this concern and provides an instant criterion to direct a representative sampling during the analysis. It should be emphasized that representative sampling is critical for determination of the differences in lipidomes (particularly the neurolipidome) and other metabolomes as well as other measurements, including changes in gene expression and protein levels between different states. Representative sampling sets a solid foundation for identification of unambiguous alterations in lipids, metabolites, and proteins induced by disease, for investigation of the biochemical mechanisms underlying any disorders, and for discovery of biomarkers.
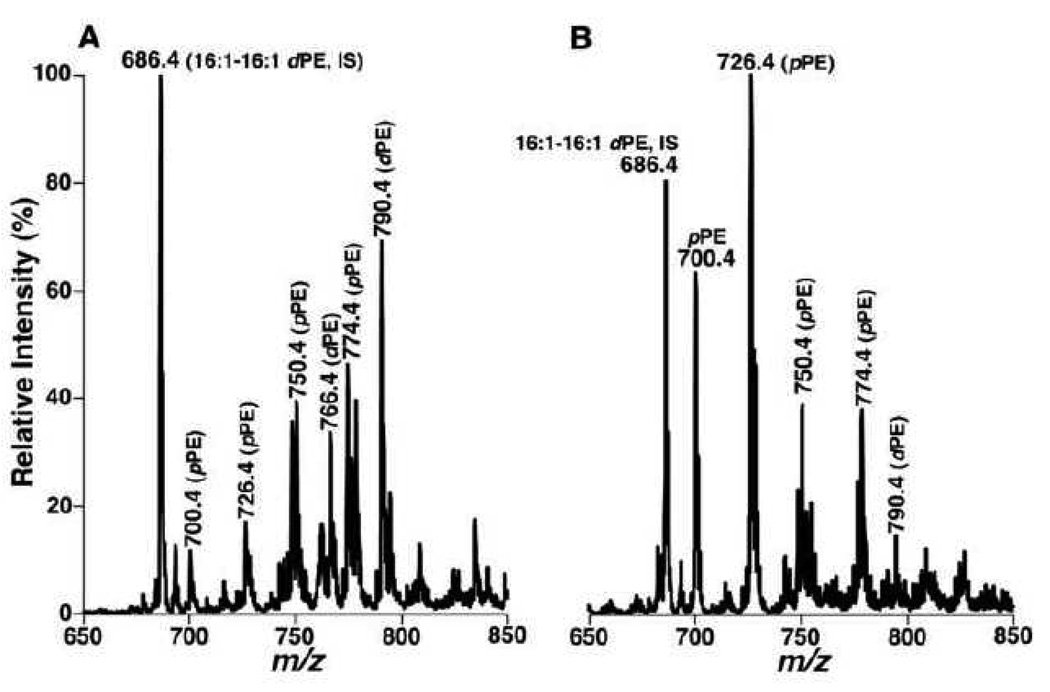
For example, MDMS-SL analysis of human brain samples demonstrates the presence of very distinct lipid profiles of ethanolamine glycerphospholipid (PE) molecular species in gray matter and white matter samples (Figure 2). Specifically, ESI-MS analysis of lipid extracts of cortex gray matter from post-mortem subjects has demonstrated multiple predominant deprotonated ion peaks corresponding to PE species (Figure 2A) in which over 80 mol% of PE species and 55–60% of plasmalogen PE (pPE) species contain polyunsaturated fatty acyl chains at the sn-2 position [25]. In contrast, ESI-MS analysis of lipid extracts of white matter from different brain regions has revealed the presence of one predominant peak at m/z 726.4 containing monounsaturated acyl chain (18:1–18:1 pPE) which represents over 85 mol% of the total pPE (Figure 2B). Accordingly, the distinct molecular species profiles of the PE classes between brain gray matter and white matter provide an important criterion to distinguish gray and white matters or determine the degree of cross contamination from co-existing gray and white matters. This degree of the cross-contamination can be accurately determined based upon the peak intensity ratios of ions at m/z 726.4 (18:1–18:1 pPE) and 790.4 (18:0–22:6 PE) following determination of a disease state with the entire profiles (Figure 2).
Figure 2.
Distinct profiles of ethanolamine glycerophospholipid molecular species in lipid extracts of cognitively normal human occipital gray and white matter. Brain samples were obtained from the brain bank of the Washington University ADRC Neuropathology/Tissue Resource Core and brain lipids were extracted by a modified procedure of Bligh-Dye [93]. Negative-ion ESI mass spectra of lipid extracts of occipital gray matter (Panel A) and white matter (Panel B) were acquired in the presence of a small amount of LiOH as previously described [6]. Individual molecular species corresponding to each ion peak were identified using multi-dimensional MS analysis as previously described [9]. Plasmenylethanolamine and phosphatidylethanolamine are abbreviated as “pPE” and “dPE”, respectively. “IS” denotes internal standard. (Han, unpublished data).
MDMS-SL determines a broad spectrum of lipid classes and individual molecular species
The primary goal of MDMS-SL is to allow one to screen in an unbiased fashion all molecular species of lipids present in a given biological sample. At its current stage, MDMS-SL enables us to automatically identify and quantitate the individual molecular species of most of the major and many of the minor lipid classes in cellular lipidomes, which collectively represent > 95% of the total lipid content and as many as 2,000 individual molecular species, directly from their chloroform extracts [13]. In combination with methods for multiplexed sample extraction and preparation, MDMS-SL allows us to routinely analyze approximately 30 lipid classes, including cardiolipins, choline glycerophospholipid (PC) including its all subclasses, PE including its all subclasses, phosphatidylinositol, phosphatidylglycerol, phosphatidylserine, phosphatidic acid, sphingomyelin, hexosylceramide, sulfatide, free fatty acid, triacylglycerol, lysoPC, lysoPE, lysophosphatidic acid, acylcarnitine, acyl-CoA, cholesterol, cholesterol esters, some oxysterols, ceramide (including dihydroceramide), sphingoid base-1-phosphate, sphingosine and sphinganine, psychosine, etc.
Lipid changes in Alzheimer’s disease
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and is the most common cause of dementia in the aging population. In the United States, its prevalence is ∼10% of those over 65 and is ∼50% in those over 85. Synapse loss, cholinergic system defects, and neuronal degeneration are some of the physiological sequelae of this devastating disease. The presence of neuritic plaques enriched with amyloid β (Aβ) peptides and neurofibrillary tangles containing hyperphosphorylated tau protein represent well-established AD pathological hallmarks [26, 27]. Many hypotheses addressing the causes responsible for AD have been investigated. These include Aβ cascade, tau deposition, oxidative stress, inflammation (i.e., arthritis-of-the brain hypothesis), metabolic disorder, and acetylcholine signaling defects, among others [28–38]. Although tremendous progress has been made toward understanding AD, particularly early-onset AD, the true biochemical mechanism(s) underlying the pathogenesis of the late-onset AD still remain unknown.
To date, the only known major genetic risk factor for late-onset AD, including both familial and sporadic and accounting for over 95% of total cases, is the ε4 allele of apolipoprotein E (apoE4) [39, 40]. The mechanism(s) underlying the significance of the apoE4 allele for AD pathogenesis remain to be elucidated. Both in vitro and more recently in vivo data strongly suggest that the ability of apoE to modify Aβ deposition may underlie the importance of apoE4 as an AD risk factor [41].
Since apoE is a lipid transport protein [42, 43], apoE-mediated lipid alterations may also play an important role in pathogenesis of AD. To this end, investigators have determined lipid levels of AD brain samples by using different analytical techniques (e.g., chromatography and nuclear magnetic resonance spectroscopy) and found that there exist cell membrane defects in AD brain samples (e.g., [25, 44–47] and references cited). For example, up to 15% of PE was lost in cerebral gray matter [25]. Importantly, membrane defects correlate with the synapse loss and neurodegeneration in the cerebrum of AD patients. Nuclear magnetic resonance spectroscopic analyses have also revealed perturbations of neuronal membranes and alterations in high energy phosphate metabolism in AD brain samples [48]. However, these studies generally examined the samples from subjects at the advanced stages of AD. In addition, concerns related to sample representation as discussed above have not been addressed in these studies, from which the conclusion drawn might carry a certain degree of artifacts.
Carrying the hypothesis that alterations in apoE-mediated lipid trafficking and metabolism must play a role in AD pathogenesis, by using MDMS-SL, we [25, 44] have determined the altered lipid levels of pure gray and white matter from different brain regions of subjects with very mild AD (i.e., mild cognitive impairment (MCI) [49]). These clinical classifications were supported by neuropathological findings with a 93% diagnostic accuracy for AD [62, 63]. We discovered specific lipid changes in subjects with MCI relative to cognitively normal, age-matched controls (see [50, 51] for recent reviews). MDMS-SL analysis has uncovered three marked changes of lipid levels manifest in brain samples of subjects with MCI. These include the substantial loss of sulfatides [44], a significant increase and molecular species compositional change in ceramide [44], and a significant loss of plasmalogen content [25], which are outlined in next section. Furthermore, MDMS-SL plays an essential role in identification of causes responsible for these altered lipid levels at the MCI stage. Particularly, MDMS-SL facilitates the identification of apoE-mediated sulfatide metabolism which echoes our hypothesis that alterations in apoE-mediated lipid trafficking and metabolism must play a role in AD pathogenesis. Accordingly, the involvement of this metabolism pathway in sulfatide depletion in AD is also summarized in this mini review.
MDMS-SL has uncovered the altered lipid classes at the earliest clinically recognizable stage of AD (i.e., MCI)
Plasmalogen PE content is significantly reduced in subjects with MCI
Plasmalogen PE (pPE), in which a vinyl ether substituent at the sn-1 position of the glycerol backbone is present, is one of the subclasses of PE. Plasmalogen PE is a major component of neuronal cell membranes representing up to 90 mol% of PE, or ∼ 30 mol% of total phospholipids of these membranes [25, 52]. Specifically, pPE accounts for approximately 60 and 90 mol% of total PE content in human gray and white matter, respectively. It is interesting to note that pPE species in gray matter consist of predominantly species with polyunsaturated acyl chains (e.g., arachidonate and docosahexaenoate) whereas pPE species in white matter contain less unsaturated (almost exclusively monounsaturated) acyl constituents at sn-2 position (Figure 2).
MDMS-SL analysis has demonstrated a significant reduction (up to 15 mol% of total pPE) in the levels of pPE species in postmortem temporal cerebral and other examined gray matter regions from subjects with MCI in comparison to age-matched cognitively normal controls. This deficiency in gray matter worsens as AD progresses (∼ 30 mol% at the stage of very severe AD). Deficiencies in pPE (up to 50%) in mid-temporal cortex [53], frontal cortex [45] and other regions [48] from advanced AD subjects in comparison to controls have also been identified using conventional chromatographic methodologies and nuclear magnetic resonance spectroscopic techniques. Plasmalogen deficiency in cerebral gray matter may be directly related to neurodegeneration and synapse loss because a decrease in pPE content may induce membrane instability [54]. Previous studies have shown that cortical synapse loss is an early event in AD [55–57]. Since synaptic vesicles are comprised of over 60 mol% of pPE in total PE (unpublished data), we postulate that pPE deficiency might adversely affect synaptic structure and function in AD [50].
MDMS-SL analysis has also demonstrated more severe pPE deficiency (up to 40 mol% of total pPE) in white matter from all examined cerebral (temporal, frontal, and parietal) and cerebellar regions at the MCI stage [49]. The severity of this depletion does not worsen with AD dementia progression. This deficiency likely causes myelin sheath defects and axonal dysfunction, and thus may directly contribute to the dementia in AD patients with MCI, since white matter deterioration is a key event leading to cognitive impairment in aging primates [58].
The mass levels of pPE species in cerebral cortex and cerebellum of AD transgenic (Tg) mouse models (both PDAPP and APPsw (APP Swedish mutation K670N/M671L)) have also been examined by using MDMS-SL [25]. It has been found that a deficiency in pPE content is present (up to 10 mol% of total pPE at 18 months of age) in cerebral cortices, but is absent in cerebella from both animal models.
It is well known that pPE plays an important role in antioxidation in cellular membranes (e.g., [59–62]) since the products of enol ether oxidation do not propagate the oxidation of polyunsaturated fatty acids. On the other hand, oxidative stress may be a major risk factor for AD progression (see [32, 63] for reviews). Several factors that may contribute to oxidative stress in AD include age related decrements in energy availability, mitochondrial dysfunction, glutamatergic neurotransmission, and the accumulated Aβ toxicity. Indeed, treatment of cultured embryonic rat brain oligodendrocytes with Aβ causes a marked reduction of plasmalogen mass content [64]. Therefore, brain membrane pPE are apparently a target for reactive oxygen species that are generated under the various oxidative stress conditions in AD. The pPE deficiency in both cultured cells and animal AD-models strongly supports the oxidative stress hypothesis.
In addition to oxidative damage, other enzymatic or nonenzymatic factors may also play an important role in causing pPE deficiency (see [46, 65] for reviews). For example, hydrolysis of pPE species due to activation of phospholipase A2 activities may represent such an important factor. Very recently, it has been demonstrated that phospholipase A2 activities are activated in AD mouse models [66] or involved in neurodegenerative disorders [67]. Intriguingly, Farooqui and colleagues [68] have shown that brain intracellular phospholipase A2 selectively hydrolyzes plasmalogen molecular species and may participate in AD pathology. Their interesting studies have been extensively reviewed (e.g., [68]).
Sulfatide content is specifically depleted in patients with MCI
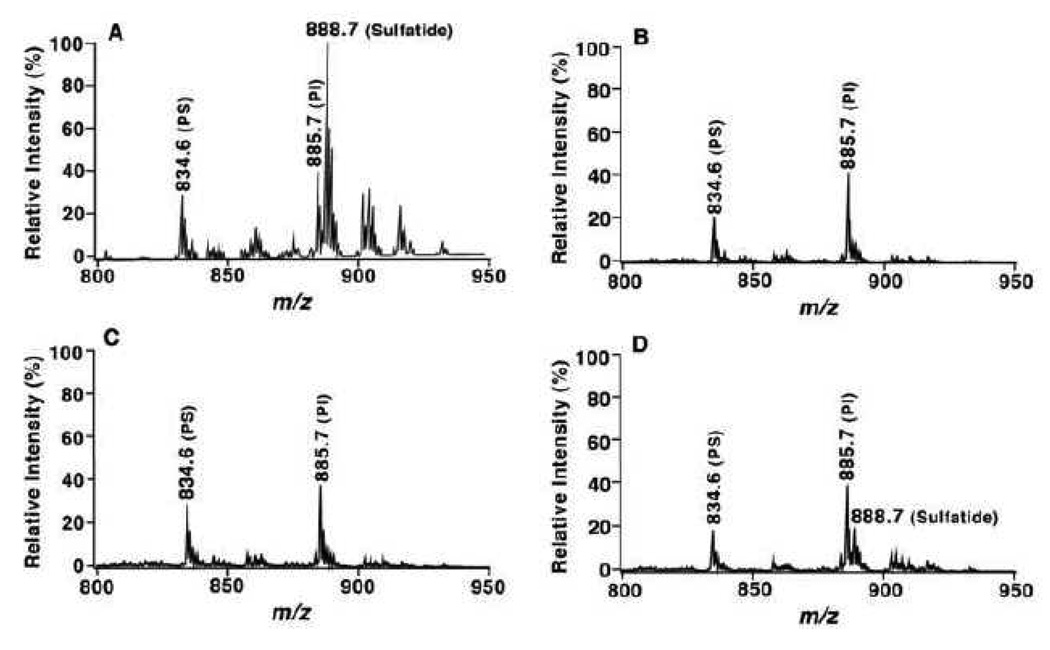
MDMS-SL analysis has demonstrated the substantial depletion (92 mol%) of sulfatide mass levels in gray matter of all examined regions from subjects at the clinical dementia rating (CDR) of 0.5 relative to controls without any molecular species preference. Figure 3 shows a few representative mass spectra (Panels B – D) acquired from lipid extracts of temporal cerebral gray matter from subjects with MCI. The spectra reflect the depleted content of sulfatides in comparison to that obtained from controls (Panel A). Identical depletions in sulfatide mass levels were also demonstrated in gray matter from other cerebral regions of subjects with MCI including frontal and parietal cortices [44]. The depletion in sulfatide levels in white matter of subjects with MCI relative to controls varies from 35 mol% in cerebellum to 58 mol% in temporal cortex. Intriguingly, the depletion of sulfatide levels does not significantly go further in either gray or white matter with the progression of AD severity.
Figure 3.
Loss of sulfatide molecular species in lipid extracts of temporal gray matter from subjects with mild cognitive impairment relative to control. Brain samples were obtained from the brain bank of the Washington University ADRC Neuropathology/Tissue Resource Core and lipids were extracted by a modified method of Bligh-Dyer [93]. The mass region showing sulfatide molecular ions is displayed after normalization to an internal standard which is not included in the mass region. Each spectrum was from a negative-ion ESI mass spectrum of the lipid extract from temporal gray matter of cognitively normal control (Panel A), or of subjects with MCI (Panels B to D). PI denotes phosphatidylinositol and PS stands for phosphatidylserine. (Han, unpublished data)
Sulfatides are a class of myelin-specialized glycosphingolipids, i.e., sulfated galactosylceramides in the CNS. Molecular species of sulfatides differ either in sphingoid backbone or fatty acyl chains linked to the amine of sphingoid bases. Sulfatides are involved in various biological processes such as cell growth, protein trafficking, signal transduction, cell-cell recognition, neuronal plasticity, and cell morphogenesis (see [69, 70] for reviews). Sulfatide accumulation, however, is associated with human diseases such as metachromatic leukodystrophy [71, 72]. A mouse model deficient in sulfatide shows multiple abnormalities including abnormal axonal function, dysmyelinosis, and loss of axonal conduction velocity [73]. Thus, substantial sulfatide depletion in the very early stage of AD development may play an important role in AD pathogenesis and may be linked with early events in the pathological process of AD including neurodegeneration, synapse loss, and synaptic dysfunction.
Sulfatide content in severe AD subjects has been analyzed previously by using conventional methodologies, such as high performance thin layer chromatography. However, conflicting results were observed. One group reported significantly higher sulfatide mass levels in AD subjects than in normal controls [74], but another found 38% lower sulfatide content in late-onset AD cases compared to controls [56]. These differences probably resulted from cross contamination between gray and white matter during sampling which is inevitable when conventional methodologies were used, in which several grams of tissue are commonly required. Recently, it has been determined that the levels of sulfatides in membrane rafts isolated from AD patients were not significantly different from those in control samples [75, 76]. This finding may reflect a unique lipid profile in isolated brain membrane rafts.
To determine the specificity of altered sulfatide levels in AD among other neurodegenerative diseases, we have determined the levels of sulfatides in postmortem brain samples from subjects with non-AD related Parkinson’s disease (PD), dementia with Lewy bodies (DLB) [64] as well as multiple sclerosis and frontotemporal dementia (Han, unpublished data). In contrast to AD cases, the sulfatide contents of all examined brain regions in both gray matter and white matter from PD subjects were dramatically elevated compared to cognitively normal controls. The levels of sulfatides in both gray matter and white matter in other examined neuronal disorders were comparable to those observed in controls. These studies suggest that sulfatide deficiency in subjects with MCI is specific among examined neurodegenerative diseases.
The levels of sulfatides in brain samples of AD animal models which transgenically express amyloid precursor protein (APP) mutants have also been recently examined [77]. MDMS-SL analysis has demonstrated that the sulfatide levels in brain tissues are reduced beginning at approximately 6 months of age in PDAPP mouse brain cortex and at 9 months of age in APPsw Tg mouse cortex relative to their respective non-APP Tg littermates. This reduction increases in both APP Tg mice as they aged. The reduced levels of sulfatides in cerebellum are much lower relative to those in cortex.
Ceramide levels are significantly higher in subjects with MCI in comparison to controls
Ceramides are the core constituents of most sphingolipids. Cellular ceramides are either de novo synthesized from fatty acyl-CoA and sphingosine or produced from the enzymatic hydrolysis of sphingolipids. In most cell lines, the major enzymatic activities of sphingolipid hydrolysis are sphingomyelinases (see [78, 79] for recent reviews). However, in the nervous system, the production of ceramides from the hydrolysis of other classes of sphingolipids (e.g., galactosylceramides, sulfatides, gangliosides, etc.) may be significant due to their abundant presence in neuronal cells. Ceramide has proven to be a powerful second messenger that regulates diverse cellular processes and activates a number of enzymes involved in stress signaling cascades including both protein kinases and protein phosphatases (see [80] for reviews). Accumulation of ceramides causes diverse biological consequences including up-regulation of cytokines, generation of reactive oxygen species, interruption of the mitochondrial respiratory chain, and apoptosis [80].
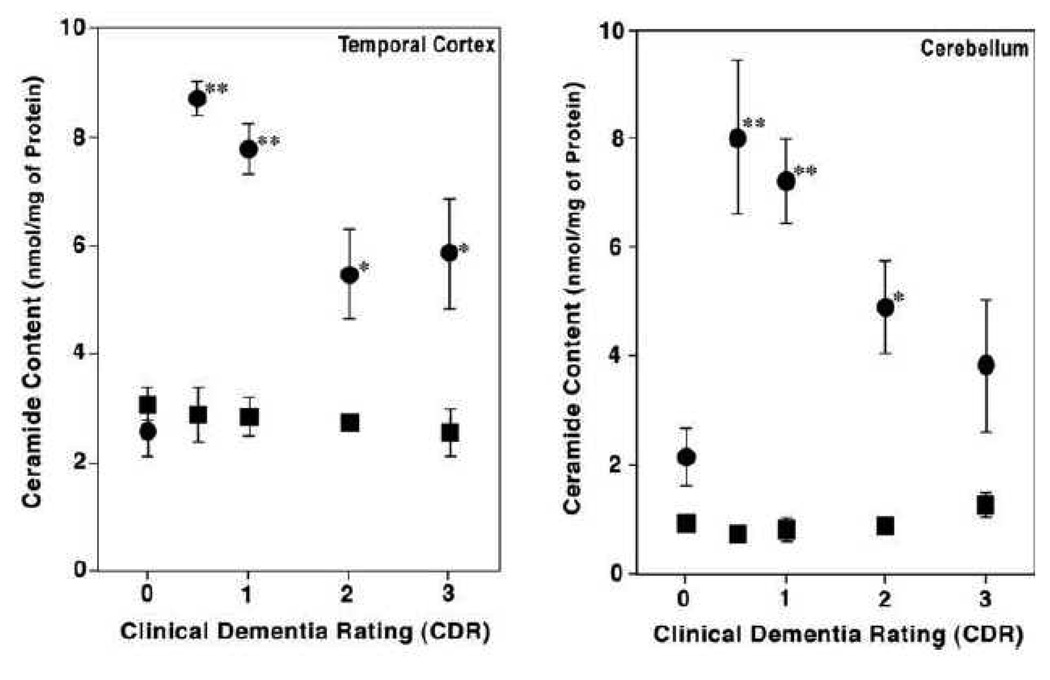
MDMS-SL analysis has uncovered a dramatic elevation of the ceramide level in white matter of postmortem brain samples from subjects with MCI relative to that from aged-matched control individuals [44]. The mass levels of ceramides in white matter from AD patients are over 3 folds higher than those in controls in all examined brain regions including cerebellum (Figure 4). Intriguingly, the levels of ceramides in white matter do not increase, but decrease as dementia becomes more severe than the MCI stage (Figure 4). In contrast to the elevated levels of ceramides in white matter, the content of ceramide in gray matter from all examined brain regions is nearly unchanged at all stages of AD and is comparable to that of controls. Recently, Cutler and colleagues [75] have also found that ceramide mass content is significantly higher in cerebral tissues from subjects with advanced AD in comparison with that in controls.
Figure 4.
Ceramide contents in post mortem brain tissue samples from cognitively normal individuals (CDR = 0) and AD patients. The total content of ceramide in chloroform extracts of white matter (Black circles) and gray matter (Black squares) from temporal cortex (Panel A) and cerebellum (Panel B) was determined using negative-ion ESI/MS as described previously [44]. * p < 0.001 and ** p < 0.0001 compared with controls. (Modified from ref. [44] with permission from International Society for Neurochemistry, Copyright 2002)
The substantial elevation of ceramide content in AD might result from the degradation of sulfatide with a loss of a galactosylsulfate, the increases in de novo biosynthesis, or the increased hydrolysis of sphingomyelin and/or cerebroside. The dramatic loss of sulfatide, but not sphingomyelin and cerebroside, in subjects with MCI might account for the substantial increase in the mass content of ceramides. The molecular species profile of ceramides in samples from subjects with MCI more closely matched the molecular species profile of sulfatides than the profiles of sphingomyelin and cerebroside molecular species [44]. Therefore, ceramide elevation in AD might be mainly due to sulfatide degradation mediated by an unknown factor. Elucidation of such mechanism(s) will likely provide essential insights into the mechanism(s) underlying AD pathogenesis.
Cutler and colleagues [75] and Lee and colleagues [81] have demonstrated that treatment of cultured cells (either primary neurons or primary oligodendrocytes, respectively) with Aβ activated neutral sphingomyelinase and increased sphingomyelin hydrolysis which led to the significant accumulation of ceramide content. Intriguingly, Lee and colleagues [81] demonstrated that the ceramide accumulation induced by Aβ treatment was maximized after Aβ treatment for approximately 12 h. This phenomenon is analogous to the pattern of ceramide content at different severe stages of AD where ceramide content has peaked at the MCI stage [44]. This observation supports the hypothesis that Aβ accumulation is likely one of the factors that leads to the dramatic accumulation of ceramide content at this stage of AD.
The altered levels of sulfatides and ceramides in AD suggest the existence of a pre-MCI stage of AD development
AD pathology in gray matter has been well characterized with respect to its being diagnostic for AD. The lack of alterations in ceramide content in gray matter of AD subjects does not appear to be consistent with gray matter AD pathology. However, we have found an intriguing phenomenon from MDMS-SL analysis that a sharp loss of sulfatide and a maximal content of ceramide are present at the MCI stage of AD. At more severe stages of AD dementia, minimal additional losses of sulfatide content occur relative to that at the MCI stage and a decreased rate of ceramide accumulation is manifest in comparison to that at the MCI stage. These observations suggest the existence of a pre-MCI stage of AD. In this pre-MCI stage, sulfatides would be dramatically decreasing in both gray and white matter compared to those in normal controls while in a higher content compared to those in the MCI stage, and ceramide content in gray matter would be peaking among all other stages. In parallel, the ceramide content in white matter at this pre-MCI stage should also be higher than that at the MCI stage. Further studies to determine or define a pre-MCI stage are necessary to provide greater understanding of the mechanism(s) underlying AD pathogenesis and potentially even earlier diagnosis than MCI.
This speculation is consistent with autopsy studies which show that approximately 30% of cognitively normal subjects who died in their mid 70’s have a marked AD pathology (i.e., the presence of large amounts of plaques and tangles), but do not yet have the marked neuronal cell loss that is present in subjects who died with MCI [82, 83]. Therefore, this finding strongly supports the notion that extensive AD pathology (i.e., plaques and tangles) likely develops over a 10–20 year period prior to any cognitive impairment or neuronal cell death [82] and indicates that alterations in sulfatide and ceramide levels are among the earliest events of AD pathogenesis.
MDMS-SL facilitates identification of apoE-mediated sulfatide trafficking and metabolism which is likely involved in sulfatide depletion in subjects with MCI
During our studies to elucidate the mechanism(s) underlying the sulfatide deficiency in subjects with MCI, we have uncovered in both the CNS and the PNS that apoE modulates sulfatide metabolism [14, 84]. MDMS-SL analysis in combination with immunoprecipitation has demonstrated that sulfatides are specifically associated with apoE-containing lipoprotein particles [84]. This finding indicates that sulfatides can be transported by apoE-associated lipoproteins to brain interstitial fluid and cerebrospinal fluid (CSF). This finding also suggests that apoE can modulate sulfatide content in the CNS and the PNS through endocytotic recycling of apoE-associated lipoprotein particles containing sulfatides through the low density lipoprotein (LDL) receptor or LDL receptor family members such as LDL receptor-related proteins (LRP). MDMS-SL analysis of lipid extracts of nervous tissues from either the CNS or the PNS in apoE knockout mice showed accumulation of sulfatides relative to their wild-type littermates [14, 84]. These studies provide strong evidence that sulfatide metabolism is tightly associated with apoE particles in both the CNS and the PNS. Moreover, by using MDMS-SL, we have also determined the sulfatide levels in brain samples of human apoE Tg mice and found that the modulation of sulfatide content in the CNS is apoE-isoform dependent. Specifically, the levels of sulfatides in the nervous system of apoE4 Tg mice are significantly lower than those of apoE3 transgenic mice in which sulfatide content is comparable with that of wild-type littermates [84]. This observation has been validated by analysis of sulfatide content in human CSF from cognitively normal subjects with different apoE genotypes by using MDMS-SL. We have found that the levels of sulfatides in human CSF from cognitively normal subjects with one or two alleles of apoE4 are significantly higher than those with homozygous apoE3 [84]. This finding could explain why the sulfatide content in brain tissues of apoE4 Tg mice is lower than that of apoE3 Tg mice, i.e., apoE4-containing lipoprotein particles can carry more sulfatide content to the metabolic pathway than apoE3-containing lipoproteins.
All these findings not only support the role of apoE in the regulation of sulfatide metabolism in the nervous system, but also suggest that apoE may be involved in the sulfatide loss in AD patients even with MCI. To date, the reason for the marked sulfatide deficiency in the CNS of Alzheimer’s patients with MCI is still unresolved, although it may reflect axonal damage/degeneration or abnormal metabolism of apoE-associated lipoproteins or both. The determination of CSF sulfatide levels alone or in combination with other biomarkers may be useful for early diagnosis of AD as well as in assessing response to any therapeutic treatment.
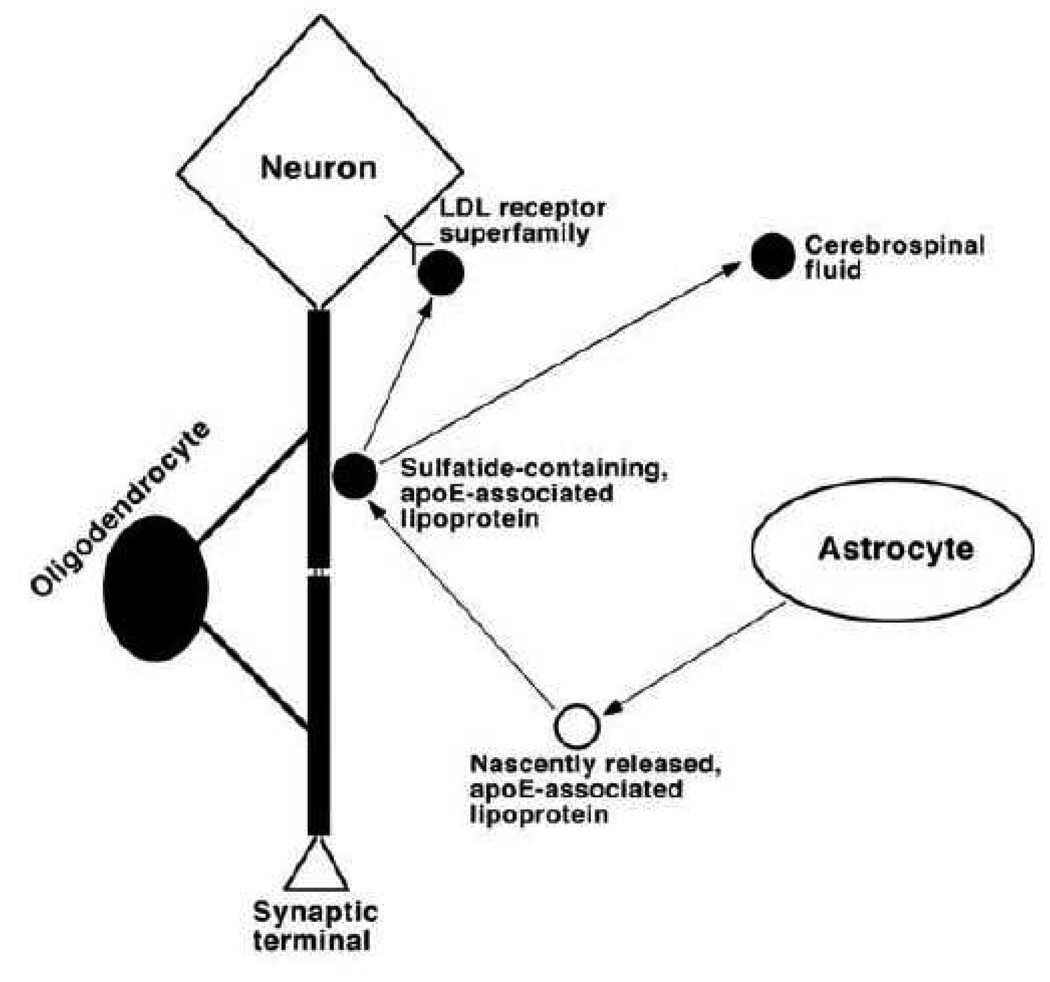
Furthermore, the results from lipidomic studies by using MDMS-SL also lead us to establish a working model for understanding sulfatide metabolism and elucidating potential biochemical mechanism(s) underlying sulfatide depletion in subjects with MCI (Figure 5). In this model, apoE-associated lipoproteins are released from astrocytes and acquire sulfatides from the myelin sheath for which the mechanism is unknown, but likely through a “kiss-and-run” mechanism. Then the sulfatide-containing apoE-associated lipoproteins can be metabolized through an endocytotic pathway by neuronal cells possessing LDL receptors or its family members such as LRP. Alternatively, these sulfatide-containing apoE-associated lipoproteins can be transported to destinations in the peripheral system through the CSF.
Figure 5.
A proposed working model for the metabolism of apolipoprotein E-associated lipoproteins, which mediate sulfatide homeostasis. In the model, apoE-associated lipoproteins are released from astrocytes, acquire sulfatide from myelin sheath, and then either metabolized through endocytotic pathways via LDL receptors or its family members such as the LDL receptor-related protein or transported to cerebrospinal fluid. Therefore, any factors that modulate apoE-associated lipoprotein metabolism can alter sulfatide levels in the nervous system. (Reprinted from ref. [77] with permission)
This model is supported by all of our currently available experimental data. For example, sulfatide is present in CSF [85]; apoE4-associated lipoprotein articles contain a significantly higher sulfatide content in CSF than their apoE3-associated counterparts [84], thereby accounting for the lower sulfatide levels in apoE4 Tg mouse brain tissue relative to that in apoE3 mice [84]; and apoE deficiency leads to the accumulation of sulfatide in the brain [84] and in the ganglia [14]. This model not only suggests that apoE is a risk factor for AD, but that it is also involved in the abnormal sulfatide metabolism of AD pathogenesis, thereby accounting for the sulfatide loss in AD patients. Moreover, other reported AD risk factors such as the LDL receptor, LRP [86, 87], and heparan sulfate proteoglycans (which have been proposed to be associated with lipoprotein metabolism [88]) are linked together with this model.
Therefore, it could be speculated that any factors that alter this apoE-mediated sulfatide metabolic pathway would change the levels of sulfatide in the CNS. It has previously been documented that the expression levels of apoE, LDL receptor and LRP are higher in AD patients than those in controls [89–92]. It can be anticipated based upon our working model that increased expression of these apolipoproteins and receptors can lead to accelerated sulfatide metabolism, thereby resulting in sulfatide depletion in AD patients. It can also be speculated that this accelerated sulfatide depletion would occur at the relatively earlier stages in apoE4 carriers than other apoE isoform carriers since apoE4 lipoproteins carry more sulfatide content than their apoE3 counterparts in the CNS [84].
It should also be emphasized that our working model does not exclude the possibility that factors which affect apoE-mediated sulfatide metabolism may also alter the intra-cellular metabolic pathways of sulfatide in particular and of the sphingolipidome in general. As well known, the metabolism of lipid classes and individual lipid molecular species in a cellular lipidome is interwoven. The factors that influence apoE-mediated sulfatide metabolism could directly or indirectly affect the entire sphingolipidome, thereby causing dramatic changes in ceramide levels present in AD [44].
Conclusions
In this mini review, we have discussed the powerful MDMS-SL technology and its application in discovery of the altered lipid content in AD including plasmalogen PE, sulfatides, and ceramides. MDMS-SL analysis-assisted discoveries related to the mechanisms responsible for these altered lipids from the studies of cell or animal models are also discussed. A recently proposed working model which concludes currently available experimental data involving sulfatide metabolism is also outlined. This model is useful for understanding the potential biochemical mechanism(s) responsible for sulfatide depletion in subjects with MCI. Collectively, MDMS-SL technology not only allows us to uncover altered lipids at a disease state, but also facilitates the identification of biochemical mechanisms underlying the alterations.
Acknowledgement
This work was supported by National Institute on Aging Grants R01 AG23168 and R01 AG31675. The author is grateful to Dr. Kui Yang for her comments and Ms Hua Cheng for her technical assistance.
Abbreviation Used
- AD
Alzheimer’s disease
- AL
anionic lipids
- apoE
apolipoprotein E
- APP
amyloid precursor protein
- APPsw
APP Swedish mutation K670N/M671L
- CDR
clinical dementia rating
- CNS
central nervous system
- CSF
cerebrospinal fluid
- ESI
electrospray ionization
- FA
fatty acyl or fatty acid
- m:n
acyl chain containing m carbons and n double bonds
- LDL
low density lipoprotein
- LRP
LDL receptor-related proteins
- MCI
mild cognitive impairment
- MDMS-SL
multi-dimensional mass spectrometry-based shotgun lipidomics
- MS
mass spectrometry
- PC
choline glycerophosphospholipid(s)
- PD
Parkinson’s disease
- PDAPP
human APP V717F mutation
- PE
ethanolamine glycerophosphospholipid(s)
- PNS
peripheral nervous system
- pPE
alkenyl-acyl PE (i.e., plasmalogen PE)
- TAG
triacylglycerol(s)
- Tg
transgenic
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Lagarde M, Geloen A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim. Biophys. Acta. 2003;1634:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths WJ. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. Rev. 2003;22:81–152. doi: 10.1002/mas.10046. [DOI] [PubMed] [Google Scholar]
- 5.Byrdwell WC. APCI-MS lipid analysis. Oily Press Lipid Library. 2003;16:171–253. [Google Scholar]
- 6.Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 7.Hsu F-F, Turk J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of complex lipids: structural characterization and mechanism of fragmentation. In: Byrdwell WC, editor. Modern Methods for Lipid Analysis by Liquid Chromatography/Mass Spectrometry and Related Techniques. Champaign, Ill: AOCS Press; 2005. pp. 61–178. [Google Scholar]
- 8.Schiller J, Suss R, Fuchs B, Muller M, Zschornig O, Arnold K. MALDI-TOF MS in lipidomics. Front. Biosci. 2007;12:2568–2579. doi: 10.2741/2255. [DOI] [PubMed] [Google Scholar]
- 9.Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev. Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 11.Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal. Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Cheng H, Yang K, Gross RW, Han X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal. Biochem. 2007;371:135–145. doi: 10.1016/j.ab.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multi-dimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, Jiang X, Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: A shotgun lipidomics study. J. Neurochem. 2007;101:57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Yang K, Gross RW. Microfluidics-based electrospray ionization enhances intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: Development of an automated high throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Gubitosi-Klug RA, Collins BJ, Gross RW. Alterations in individual molecular species of human platelet phospholipids during thrombin stimulation: electrospray ionization mass spectrometry-facilitated identification of the boundary conditions for the magnitude and selectivity of thrombin-induced platelet phospholipid hydrolysis. Biochemistry. 1996;35:5822–5832. doi: 10.1021/bi952927v. [DOI] [PubMed] [Google Scholar]
- 18.Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 2006;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 20.Kim HY, Wang TC, Ma YC. Liquid chromatography/mass spectrometry of phospholipids using electrospray ionization. Anal. Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 21.Yang K, Zhao Z, Gross RW, Han X. Systematic analysis of choline-containing phospholipids using multi-dimensional mass spectrometry-based shotgun lipidomics. J. Chromatogr. B. 2009;877:2924–2936. doi: 10.1016/j.jchromb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J. Lipid Res. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J. Lipid Res. 2005;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H, Guan S, Han X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: Implications for the role of altered triacylglycerols in diabetic neuropathy. J. Neurochem. 2006;97:1288–1300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X, Holtzman DM, McKeel DW., Jr Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 26.Yankner BA. New clues to Alzheimer's disease: unraveling the roles of amyloid and tau. Nat. Med. 1996;2:850–852. doi: 10.1038/nm0896-850. [DOI] [PubMed] [Google Scholar]
- 27.Morishima-Kawashima M, Ihara Y. Alzheimer's disease: beta-Amyloid protein and tau. J. Neurosci. Res. 2002;70:392–401. doi: 10.1002/jnr.10355. [DOI] [PubMed] [Google Scholar]
- 28.Selkoe DJ. Alzheimer disease: mechanistic understanding predicts novel therapies. Ann. Intern. Med. 2004;140:627–638. doi: 10.7326/0003-4819-140-8-200404200-00047. [DOI] [PubMed] [Google Scholar]
- 29.Armstrong RA, Winsper SJ, Blair JA. Aluminium and Alzheimer's disease: review of possible pathogenic mechanisms. Dementia. 1996;7:1–9. doi: 10.1159/000106845. [DOI] [PubMed] [Google Scholar]
- 30.Cummings JL, Cole G. Alzheimer disease. Jama. 2002;287:2335–2338. doi: 10.1001/jama.287.18.2335. [DOI] [PubMed] [Google Scholar]
- 31.McGeer EG, McGeer PL. Inflammatory processes in Alzheimer's disease. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2003;27:741–749. doi: 10.1016/S0278-5846(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 32.Albers DS, Beal MF. Mitochondrial dysfunction and oxidative stress in aging and neurodegenerative disease. J. Neural Transm. Suppl. 2000;59:133–154. doi: 10.1007/978-3-7091-6781-6_16. [DOI] [PubMed] [Google Scholar]
- 33.Mattson MP. Will caloric restriction and folate protect against AD and PD? Neurology. 2003;60:690–695. doi: 10.1212/01.wnl.0000042785.02850.11. [DOI] [PubMed] [Google Scholar]
- 34.Tabet N. Acetylcholinesterase inhibitors for Alzheimer's disease: anti-inflammatories in acetylcholine clothing! Age Ageing. 2006;35:336–338. doi: 10.1093/ageing/afl027. [DOI] [PubMed] [Google Scholar]
- 35.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat. Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 36.Onyango IG, Khan SM. Oxidative stress, mitochondrial dysfunction and stress signaling in Alzheimer's disease. Curr. Alz. Res. 2006;3:339–349. doi: 10.2174/156720506778249489. [DOI] [PubMed] [Google Scholar]
- 37.Sivaprakasam K. Towards a unifying hypothesis of Alzheimer's disease: cholinergic system linked to plaques, tangles and neuroinflammation. Curr. Med. Chem. 2006;13:2179–2188. doi: 10.2174/092986706777935203. [DOI] [PubMed] [Google Scholar]
- 38.Hauptmann S, Keil U, Scherping I, Bonert A, Eckert A, Mueller WE. Mitochondrial dysfunction in sporadic and genetic Alzheimer's disease. Exp. Gerontol. 2006;41:668–673. doi: 10.1016/j.exger.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer's disease. Annu. Rev. Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- 40.Cedazo-Minguez A, Cowburn RF. Apolipoprotein E: a major piece in the Alzheimer's disease puzzle. J. Cell. Mol. Med. 2001;5:254–266. doi: 10.1111/j.1582-4934.2001.tb00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtzman DM. In vivo effects of ApoE and clusterin on amyloid-beta metabolism and neuropathology. J. Mol. Neurosci. 2004;23:247–254. doi: 10.1385/JMN:23:3:247. [DOI] [PubMed] [Google Scholar]
- 42.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 43.Han X. The role of apolipoprotein E in lipid metabolism in the central nervous system. Cell. Mol. Life Sci. 2004;61:1896–1906. doi: 10.1007/s00018-004-4009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X, Holtzman DM, McKeel DW, Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: potential role in disease pathogenesis. J. Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- 45.Guan Z, Wang Y, Cairns NJ, Lantos PL, Dallner G, Sindelar PJ. Decrease and structural modifications of phosphatidylethanolamine plasmalogen in the brain with Alzheimer disease. J. Neuropathol. Exp. Neurol. 1999;58:740–747. doi: 10.1097/00005072-199907000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Klein J. Membrane breakdown in acute and chronic neurodegeneration: Focus on choline-containing phospholipids. J. Neural Transm. 2000;107:1027–1063. doi: 10.1007/s007020070051. [DOI] [PubMed] [Google Scholar]
- 47.Mulder C, Wahlund LO, Teerlink T, Blomberg M, Veerhuis R, van Kamp GJ, Scheltens P, Scheffer PG. Decreased lysophosphatidylcholine/phosphatidylcholine ratio in cerebrospinal fluid in Alzheimer's disease. J. Neural Transm. 2003;110:949–955. doi: 10.1007/s00702-003-0007-9. [DOI] [PubMed] [Google Scholar]
- 48.Pettegrew JW, Panchalingam K, Hamilton RL, McClure RJ. Brain membrane phospholipid alterations in Alzheimer's disease. Neurochem. Res. 2001;26:771–782. doi: 10.1023/a:1011603916962. [DOI] [PubMed] [Google Scholar]
- 49.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 50.Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: Implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr. Alz. Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- 51.Han X. Potential mechanisms contributing to sulfatide depletion at the earliest clinically recognizable stages of Alzheimer's disease: a tale of shotgun lipidomics. J. Neurochem. 2007;103(s1):171–179. doi: 10.1111/j.1471-4159.2007.04708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horrocks LA, Sharma M. Plasmalogens and O-alkyl glycerophospholipids. In: Hawthorne JN, Ansell GB, editors. Phospholipids. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1982. pp. 51–93. [Google Scholar]
- 53.Ginsberg L, Rafique S, Xuereb JH, Rapoport SI, Gershfeld NL. Disease and anatomic specificity of ethanolamine plasmalogen deficiency in Alzheimer's disease brain. Brain Res. 1995;698:223–226. doi: 10.1016/0006-8993(95)00931-f. [DOI] [PubMed] [Google Scholar]
- 54.Ginsberg L, Xuereb JH, Gershfeld NL. Membrane instability, plasmalogen content, and Alzheimer's disease. J. Neurochem. 1998;70:2533–2538. doi: 10.1046/j.1471-4159.1998.70062533.x. [DOI] [PubMed] [Google Scholar]
- 55.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 56.Svennerholm L, Gottfries CG. Membrane lipids, selectively diminished in Alzheimer brains, suggest synapse loss as a primary event in early-onset form (type I) and demyelination in late-onset form (type II) J. Neurochem. 1994;62:1039–1047. doi: 10.1046/j.1471-4159.1994.62031039.x. [DOI] [PubMed] [Google Scholar]
- 57.DeKosky ST, Scheff SW, Styren SD. Structural correlates of cognition in dementia: quantification and assessment of synapse change. Neurodegeneration. 1996;5:417–421. doi: 10.1006/neur.1996.0056. [DOI] [PubMed] [Google Scholar]
- 58.Peters A, Rosene DL, Moss MB, Kemper TL, Abraham CR, Tigges J, Albert MS. Neurobiological bases of age-related cognitive decline in the rhesus monkey. J. Neuropathol. Exp. Neurol. 1996;55:861–874. doi: 10.1097/00005072-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Zoeller RA, Morand OH, Raetz CR. A possible role for plasmalogens in protecting animal cells against photosensitized killing. J. Biol. Chem. 1988;263:11590–11596. [PubMed] [Google Scholar]
- 60.Scherrer LA, Gross RW. Subcellular distribution, molecular dynamics and catabolism of plasmalogens in myocardium. Mol. Cell. Biochem. 1989;88:97–105. doi: 10.1007/BF00223430. [DOI] [PubMed] [Google Scholar]
- 61.Brosche T, Platt D. The biological significance of plasmalogens in defense against oxidative damage. Exp Gerontol. 1998;33:363–369. doi: 10.1016/s0531-5565(98)00014-x. [DOI] [PubMed] [Google Scholar]
- 62.Sindelar PJ, Guan Z, Dallner G, Ernster L. The protective role of plasmalogens in iron-induced lipid peroxidation. Free Radic. Biol. Med. 1999;26:318–324. doi: 10.1016/s0891-5849(98)00221-4. [DOI] [PubMed] [Google Scholar]
- 63.Bassett CN, Montine TJ. Lipoproteins and lipid peroxidation in Alzheimer's disease. J. Nutr. Health Aging. 2003;7:24–29. [PubMed] [Google Scholar]
- 64.Cheng H, Xu J, McKeel DW, Jr, Han X. Specificity and potential mechanism of sulfatide deficiency in Alzheimer's disease: An electrospray ionization mass spectrometric study. Cell. Mol. Biol. 2003;49:809–818. [PubMed] [Google Scholar]
- 65.Farooqui AA, Horrocks LA, Farooqui T. Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem. Phys. Lipids. 2000;106:1–29. doi: 10.1016/s0009-3084(00)00128-6. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Mejia RO, Newman JW, Toh S, Yu GQ, Zhou Y, Halabisky B, Cisse M, Scearce-Levie K, Cheng IH, Gan L, Palop JJ, Bonventre JV, Mucke L. Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer's disease. Nat Neurosci. 2008;11:1311–1318. doi: 10.1038/nn.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mancuso DJ, Kotzbauer PT, Wozniak DF, Sims HF, Jenkins CM, Guan S, Han X, Yang K, Sun G, Malik I, Conyers S, Green KG, Schmidt RE, Gross RW. Genetic ablation of calcium-independent phospholipase A2{gamma} leads to alterations in hippocampal cardiolipin content and molecular species distribution, mitochondrial degeneration, autophagy and cognitive dysfunction. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farooqui AA, Horrocks LA. Plasmalogen-selective phospholipase A2 and its involvement in Alzheimer's disease. Biochem. Soc. Trans. 1998;26:243–246. doi: 10.1042/bst0260243. [DOI] [PubMed] [Google Scholar]
- 69.Vos JP, Lopes-Cardozo M, Gadella BM. Metabolic and functional aspects of sulfogalactolipids. Biochim. Biophys. Acta. 1994;1211:125–149. doi: 10.1016/0005-2760(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 70.Marcus J, Popko B. Galactolipids are molecular determinants of myelin development and axo-glial organization. Biochim. Biophys. Acta. 2002;1573:406–413. doi: 10.1016/s0304-4165(02)00410-5. [DOI] [PubMed] [Google Scholar]
- 71.von Figura K, Gieselmann V, Jaeken J. Metachromatic leukodystrophy: Lysosomal disorders. In: Sachdev HS, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited diseases. McGraw-Hill, New York: 2001. pp. 3695–3724. [Google Scholar]
- 72.Eckhardt M, Hedayati KK, Pitsch J, Lullmann-Rauch R, Beck H, Fewou SN, Gieselmann V. Sulfatide storage in neurons causes hyperexcitability and axonal degeneration in a mouse model of metachromatic leukodystrophy. J. Neurosci. 2007;27:9009–9021. doi: 10.1523/JNEUROSCI.2329-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcus J, Honigbaum S, Shroff S, Honke K, Rosenbluth J, Dupree JL. Sulfatide is essential for the maintenance of CNS myelin and axon structure. Glia. 2006;53:372–381. doi: 10.1002/glia.20292. [DOI] [PubMed] [Google Scholar]
- 74.Majocha RE, Jungalwala FB, Rodenrys A, Marotta CA. Monoclonal antibody to embryonic CNS antigen A2B5 provides evidence for the involvement of membrane components at sites of Alzheimer degeneration and detects sulfatides as well as gangliosides. J. Neurochem. 1989;53:953–961. doi: 10.1111/j.1471-4159.1989.tb11798.x. [DOI] [PubMed] [Google Scholar]
- 75.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morishima-Kawashima M, Han X, Tanimura Y, Hamanaka H, Kobayashi M, Sakurai T, Yokoyama M, Wada K, Nukina N, Fujita SC, Ihara Y. Effects of human apolipoprotein E isoforms on the amyloid beta-protein concentration and lipid composition in brain low-density membrane domains. J. Neurochem. 2007;101:949–958. doi: 10.1111/j.1471-4159.2006.04400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng H, Zhou Y, Holtzman DM, Han X. Apolipoprotein E mediates sulfatide depletion in amyloid precursor protein transgenic animal models of Alzheimer’s disease. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2008.07.020. doi:10.1016/j.neurobiolaging.2008.1007.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goni FM, Alonso A. Sphingomyelinases: enzymology and membrane activity. FEBS Lett. 2002;531:38–46. doi: 10.1016/s0014-5793(02)03482-8. [DOI] [PubMed] [Google Scholar]
- 79.Gulbins E, Kolesnick R. Acid sphingomyelinase-derived ceramide signaling in apoptosis. Subcell Biochem. 2002;36:229–244. doi: 10.1007/0-306-47931-1_12. [DOI] [PubMed] [Google Scholar]
- 80.Ruvolo PP, Johnson CR, Jarvis WD. Ceramide-driven stress signals in cancer and aging. Adv. Cell Aging Gerontol. 2003;12:47–69. [Google Scholar]
- 81.Lee J-T, Xu J, Lee J-M, Ku G, Han X, Yang D-I, Chen S, Hsu CY. Amyloid-b peptide induces oligodendrocyte death by activating the neutral sphingomyelinase-ceramide pathway. J. Cell Biol. 2004;164:123–131. doi: 10.1083/jcb.200307017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price JL, Morris JC. Tangles and plaques in nondemented aging and "preclinical" Alzheimer's disease. Ann. Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 83.Morris JC, Storandt M, Miller JP, McKeel DW, Price JL, Rubin EH, Berg L. Mild cognitive impairment represents early-stage Alzheimer disease. Arch. Neurol. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- 84.Han X, Cheng H, Fryer JD, Fagan AM, Holtzman DM. Novel role for apolipoprotein E in the central nervous system: Modulation of sulfatide content. J. Biol. Chem. 2003;278:8043–8051. doi: 10.1074/jbc.M212340200. [DOI] [PubMed] [Google Scholar]
- 85.Han X, Fagan AM, Cheng H, Morris JC, Xiong C, Holtzman DM. Cerebrospinal fluid sulfatide is decreased in subjects with incipient dementia.[see comment][erratum appears in Ann Neurol. 2003 Nov;54(5):693] Ann. Neurol. 2003;54:115–119. doi: 10.1002/ana.10618. [DOI] [PubMed] [Google Scholar]
- 86.Van Uden E, Kang DE, Koo EH, Masliah E. LDL receptor-related protein (LRP) in Alzheimer's disease: towards a unified theory of pathogenesis. Microsc. Res. Tech. 2000;50:268–272. doi: 10.1002/1097-0029(20000815)50:4<268::AID-JEMT3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 87.Abdulkarim Y, Hameed Z. Is the LDL receptor involved in cortical amyloid protein clearance? Neurochem. Res. 2006;31:839–847. doi: 10.1007/s11064-006-9084-0. [DOI] [PubMed] [Google Scholar]
- 88.Mulder M, Terwel D. Possible link between lipid metabolism and cerebral amyloid angiopathy in Alzheimer's disease: A role for high-density lipoproteins? Haemostasis. 1998;28:174–194. doi: 10.1159/000022429. [DOI] [PubMed] [Google Scholar]
- 89.LaFerla FM, Troncoso JC, Strickland DK, Kawas CH, Jay G. Neuronal cell death in Alzheimer’s disease correlates with apoE uptake and intracellular Abeta stabilization. J. Clin. Invest. 1997;100:310–320. doi: 10.1172/JCI119536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laws SM, Hone E, Taddei K, Harper C, Dean B, McClean C, Masters C, Lautenschlager N, Gandy SE, Martins RN. Variation at the APOE −491 promoter locus is associated with altered brain levels of apolipoprotein E. Mol Psychiatry. 2002;7:886–890. doi: 10.1038/sj.mp.4001097. [DOI] [PubMed] [Google Scholar]
- 91.Ulery PG, Strickland DK. LRP in Alzheimer’s disease: friend or foe? J. Clin. Invest. 2000;106:1077–1079. doi: 10.1172/JCI11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christie WW, Han X. Fourth ed. Bridgwater, England: The Oily Press; 2010. Lipid Analysis: Isolation, separation, identification and lipidomics analysis. [Google Scholar]