Abstract
In the past ten years, the field of parasitology has witnessed an explosion of studies investigating gene regulation. In this review, we will describe recent advances largely stemming from the study of Toxoplasma gondii, a significant opportunistic pathogen and useful model for other apicomplexan protozoa. Surprising findings have emerged, including the discovery of a wealth of epigenetic machinery in these primitive eukaryotes, unusual histone variants, and a battery of plant-like transcription factors. We will elaborate on how these unusual features impact parasite physiology and potential therapeutics as we summarize some of the key discoveries from the last decade. We will close by proposing a few questions to address in the next ten years.
Keywords: chromatin, histone, transcription, differentiation, Apicomplexa, parasite
1. What’s so special about gene regulation in Toxoplasma gondii?
Toxoplasma (phylum Apicomplexa) is a medically important protozoan that can cause opportunistic disease in immunocompromised individuals. Toxoplasma can also cause spontaneous abortion or congenital birth defects in newborns if the mother contracts the parasite for the first time during pregnancy. Phylum Apicomplexa contains other dangerous parasites including Plasmodium (malaria), Cryptosporidium (cryptosporidiosis), and Babesia (babesiosis). Like most of these pathogenic protozoa, Toxoplasma undergoes a complex life cycle that involves multiple hosts and environments. Its life cycle is comprised of numerous developmental transitions, such as differentiation between rapidly growing tachyzoites and latent bradyzoite tissue cysts, the conversion into micro- and macrogametes within the gut of a feline definitive host, and the formation of sporozoites inside of an oocyst that remains infectious in the environment for up to a year. The ability to transform from one stage to another and adapt to changing environments demands precise regulation of gene expression [1]. Progression through the lytic cycle, which involves host cell invasion, asexual replication, and egress, also requires significant changes in the transcriptome.
Until recently, there was little reason to believe that the study of gene regulation in these primitive eukaryotes would be interesting - it was generally assumed that transcription in Toxoplasma would conform to the paradigm outlined for yeast or higher eukaryotes whereby a specific DNA-binding transcription factor (TF) binds cis-regulatory elements in the gene promoter to recruit the pre-initiation complex of general TFs. Apathy towards apicomplexan gene regulation began to evaporate in 1996 when Schmatz and colleagues made the notable discovery that a novel antiprotozoal agent called apicidin inhibited a parasite histone deacetylase (HDAC) [2]. While the importance of HDACs in gene regulation was only beginning to be realized (see below), the work with apicidin argued that enzymes involved in chromatin modification may be targeted to create effective new therapies against protozoal pathogens. Additionally, the failure of apicomplexan promoters to function in mammalian cells (and vise versa), foreshadowed a fundamental difference in how these primitive protozoa conduct gene expression [3].
Whole genome sequencing of various apicomplexans including Toxoplasma during the early 2000s brought additional surprises. Bioinformatic surveys of the sequenced genomes of Toxoplasma and Plasmodium revealed a peculiar absence of DNA-binding TFs that are well-conserved from yeast through humans. In contrast, a large repertoire of chromatin remodeling machinery is present in these primitive eukaryotes, especially Toxoplasma, supporting an idea that epigenetic events played a key role in gene regulation. It was also noted that many of these chromatin remodeling factors were quite divergent from eukaryotic counterparts, some of which is detailed below. Thus, some important questions began to emerge: what role does this vast chromatin remodeling machinery have in controlling gene expression? Are epigenetic factors important during developmental transitions? How are these complexes being recruited to target genes? Do parasite promoters have cis-regulatory elements that control stage-specific gene expression? How could apicomplexans regulate gene expression without the well-conserved TFs? Can the apicomplexan gene regulatory system be targeted pharmacologically? In this review, we will highlight work over the past decade that has begun to address these queries.
2. A brief history of epigenetics
Sometimes genes do not explain it all. Something beyond the gene is contributing to the phenotype – this is the literal meaning of “epi” and “genetics”. That “something” is now understood to be a number of phenomena, most of which impact the degree of gene expression. By varying levels of gene expression, the same genome can give rise to a wide variety of phenotypes. Methods that cells employ to alter gene expression levels include DNA methylation, nucleosomal remodeling, and covalent modification of histones. The latter has been the subject of intense investigation since 1996, when Brownell and Allis made the landmark observation that a Tetrahymena histone acetyltransferase (HAT) was homologous to a yeast transcriptional adaptor called GCN5 (general control nonderepressible-5) [4]. Histones are the primary constituent of chromatin and were first proposed to be an impediment to gene expression in 1950 [5]. In 1964, Allfrey presented evidence showing that acetylation of histones correlated with gene activation [6]. The discovery of additional histone modifications, such as methylation, phosphorylation, and sumoylation, as well as protein domains that can bind these modified residues, paved the way for “the histone code” hypothesis, which proposes that histone modifications (or marks) form a cellular code that is read by other proteins to produce specific downstream effects [7]. Three years after GCN5 was established to be a HAT that modulated gene expression, a GCN5 homologue was cloned and characterized in Toxoplasma gondii, ushering in a new frontier of research in Apicomplexa [8–9].
3. Identification of epigenetic machinery in Toxoplasma
Early forays into Toxoplasma epigenetics involved basic characterization of some of the major players. In addition to finding a GCN5 homologue in Toxoplasma, two more HATs were characterized belonging to the MYST family called TgMYST-A and -B [10]. Reports early in the decade also described the presence of TgSRCAP (Snf2-related CBP (CREB-binding protein) activator protein), one of at least 17 SWI/SNF ATP-dependent chromatin remodelers that non-covalently restructure nucleosomes [11]. The HAT counterparts, HDAC enzymes, as well as histone arginine methyltransferases (PRMTs), were first described in 2005 by Saksouk et al [12]. In 2007, Hakimi’s group reported significant duplication and divergence in the SET domain-containing proteins in Apicomplexa; SET domains are found in histone lysine methyltransferases (HKMTs) that either activate or repress gene expression depending upon the lysine modified [13]. Toxoplasma does not appear to have lysine methyltransferases of the DOT1 family, which lack the SET domain [14]. The latter half of the decade also witnessed the remarkable discovery that Toxoplasma possesses a second GCN5 HAT (distinguished TgGCN5-A and –B) [15]. Toxoplasma and its close relative Neospora are the only known invertebrates to have more than one GCN5 HAT. The TgGCN5s are also unusual for possessing lengthy, unconserved N-terminal extensions devoid of protein motifs. We have determined that at least one function of these N-terminal extensions is to localize the HAT to the parasite nucleus [16], but additional functions are likely. Table I is an up-to-date list of genes predicted or shown to be a part of the Toxoplasma epigenetic machinery. In brief, while the catalytic domain and domain architecture are generally conserved in the Toxoplasma homologues, much of the protein sequence outside the catalytic domain is unique and worthy of further investigation.
Table I.
Predicted genes associated with epigenetic mechanisms in Toxoplasma gondii
| Predicted gene & in vitro substrate(s) | Accession # Release 5 / Model ID | GenBank | References | |
|---|---|---|---|---|
| Histone acetyltransferases | ||||
| GCN5-A | H3K18 | TGGT1_004130 / 52.m00008 | AF197953 | [8–9, 12, 15–16] |
| GCN5-B | H3K9, H3K14, H3K18 | TGME49_043440 / 49.m03346 | AY875982 | [15–16] |
| MYST-A | H4K5, H4K8, H4K12, H4K16 | TGGT1_122910 / 641.m01473 | AY578183 | [10] |
| MYST-B | TGME49_007080 / 23.m00146 | DQ104220 | [10, 31] | |
| Hat1, putative | TGME49_093380 / 83.m02144 | |||
| Elp3, putative | TGME49_105480 / 541.m01178 | |||
| TAF1/250 TFIID, putative | TGME49_076180 / 64.m00349 | |||
| Histone deacetylases | ||||
| HDAC1 | TGGT1_006030 / 74.m00425 | |||
| HDAC2 | TGME49_049620 / 50.m03318 | |||
| HDAC3 | H4K5, H4K8, H4K12 | TGME49_027290 / 42.m00014 | DQ004745 | [12, 33] |
| HDAC4 | TGME49_057790 / 55.m04722 | |||
| HDAC5 | TGME49_002230 / 20.m03703 | |||
| Sir2 | H3K56 | TGGT1_082510 / 42.m00081 | [25] | |
| Sir2-like | TGME49_067360 / 57.m01849 | |||
| Histone arginine methyltransferases | ||||
| PRMT1 | H4R3 | TGGT1_030400 / 38.m00018 | AY820756 | [12] |
| PRMT2 | TGGT1_073730 / 83.m01208 | |||
| PRMT3 | TGME49_052420 / 52.m01547 | |||
| PRMT4/CARM1 | H3R17 | TGME49_094270 / 83.m01256 | AY820755 | [12] |
| PRMT5 | H3R2 | TGME49_015560 / 33.m01376 | [25] | |
| CARM1-like (PRMT6/8) | TGME49_019520 / 38.m00018 | |||
| Histone lysine methyltransferases | ||||
| SET1 | H3K4 | TGME49_026810 / 42.m03532 | [25] | |
| SET2 | H3K36 | TGME49_057770 / 55.m04720 | [25] | |
| SET/SUV39 | TGME49_055970 / 55.m04640 | |||
| SET, putative | TGME49_088330 / 80.m02140 | |||
| SET, putative | TGME49_092170 / 80.m02357 | |||
| SET, putative | TGME49_094610 / 83.m01275 | |||
| SET, putative | TGME49_095610 / 86.m00376 | |||
| SET, putative | TGME49_001250 / 20.m03655 | |||
| SET, putative | TGME49_002490 / 20.m03722 | |||
| SET8 | H4K20 | TGME49_011730 / 27.m00875 | EF12195 | [13] |
| SET, putative | TGME49_016080 / 35.m00004 | |||
| SET, putative | TGME49_046910 / 50.m03139 | |||
| SET, putative | TGME49_076120 / 64.m00581 | |||
| SET, putative | TGME49_111660 / 583.m05442 | |||
| SET, putative | TGME49_119660 / 641.m01529 | |||
| SET, putative | TGME49_042870 / 49.m03319 | |||
| SET, putative | TGME49_062750 / 55.m05010 | |||
| SET13/KMTox | H4 and H2A | TGME49_081900 / 74.m00443 | [30] | |
| SET, putative | TGME49_018230 / 38.m02377 | |||
| SET, putative | TGME49_084160 / 76.m02672 | |||
| Histone demethylases | ||||
| JmjC domain, putative | TGGT1_055200 / 542.m00225 | |||
| JmjC domain, putative | TGME49_083890 / 76.m01561 | |||
| JmjC domain, putative | TGGT1_079840 / 42.m05839 | |||
| JMJD6 | H3R2 | TGGT1_008890 / 55.m04927 | [25] | |
| JmjC domain, putative | TGME49_012120 / 28.m00292 | |||
| JmjC domain, putative | TGME49_040840 / 49.m03238 | |||
| BHC110, putative | TGGT1_068080 / 63.m00151 | |||
| BHC110, putative | TGGT1_047400 / 49.m03290 | |||
| JARID1/LSD1 | H3K4 | TGME49_075420 / 63.m00151 | [25] | |
| LSD2, putative | TGME49_042420 / 49.m03290 | |||
| Histone Kinases | ||||
| Snf1, putative | TGME49_033900 / 44.m02822 | |||
| Snf1, putative | TGME49_091050 / 80.m02306 | |||
| Histone phosphatase | ||||
| PP1, putative | TGME49_110700 / 583.m05380 | |||
| Histone ADP-ribosylation | ||||
| PARP, putative | TGME49_070840 / 59.m03538 | |||
| PARG, related | TGME49_062760 / 55.m05011 | |||
| ATP-dependent remodeling enzymes | ||||
| SWI2/SNF2, Brahma-like, putative | TGME49_120300 / 641.m01573 | |||
| SWI2/SNF2, Brahma-like, putative | TGME49_078440 / 65.m01174 | |||
| SWI2/SNF2, ISWI-like (AT hook) , putative | TGME49_073870 / 59.m00063 | |||
| SWI2/SNF2, ISWI-like (SANT), putative | TGME49_121440 / 645.m00313 | |||
| SWI2/SNF2, Mi-2 (chromodomain) , putative | TGGT1_011870 / 55.m04750 | |||
| SWI2/SNF2, SRCAP/Ino80 | TGME49_080800 / 72.m00005 | AY061650 | [11–12, 51] | |
| SWI2/SNF2-containing protein, putative | TGME49_091090 / 80.m02309 | |||
| SWI2/SNF2-containing protein, putative | TGME49_077070 / 65.m01101 | |||
| SWI2/SNF2-containing protein, RAD5, putative | TGME49_118480 / 641.m01484 | |||
| SWI2/SNF2-containing PHD finger protein, putative | TGME49_036970 / 46.m01723 | |||
| SWI2/SNF2-containing protein, putative | TGME49_045720 / 50.m03086 | |||
| SWI2/SNF2-containing protein, RAD16, putative | TGME49_026440 / 42.m00128 | |||
| SWI2/SNF2-containing protein, putative | TGME49_029460 / 44.m02545 | |||
| SWI2/SNF2-containing protein, RAD54, putative | TGME49_032450 / 44.m02726 | |||
| SWI2/SNF2-containing protein, RAD26, putative | TGME49_063140 / 55.m05039 | |||
| SWI2/SNF2-containing protein, putative | TGME49_073780 / 59.m03701 | |||
| MORC | TGME49_105340 / 541.m01172 | |||
| Ubiquitination/Sumolyation | ||||
| Ubiquitin-conjugating enzyme E2, RAD6, putative | TGME49_016130 / 35.m0006 | |||
| Ubiquitin carboxyl-terminal hydrolase, Ubp8, putative | TGGT1_043540 / 55.m05059 | |||
| Ubc9 sumolyation conjugating enzyme, putative | TGME49_008780 / 25.m01826 | |||
| DNA Methyltransferases | ||||
| DNA methyltransferase, putative | TGME49_043610 / 49.m03360 | [18] | ||
| DNA methyltransferase 2, putative | TGME49_027660 / 42.m03580 | [18] | ||
Existing and predicted chromatin remodeling enzymes identified in Toxoplasma gondii using bioinformatic searches of the Toxoplasma database (ToxoDB - www.ToxoDB.org). For each gene, two accession numbers are listed: the first is the current gene assignment in ToxoDB 5.0 and the second is its previous designation. Strain ME49 gene assignments are listed unless the GT1 assignment provided a better prediction. GenBank accession are indicated where one exists, along with appropriate references. Following each gene name, the histone residue(s) modified by each enzyme is listed, if known.
In addition to the modification of histones, higher eukaryotes modify genomic DNA itself as a means to regulate gene expression. 5-methylcytosine (5mC) is also present at repressed promoters in some unicellular eukaryotes, such as Entamoeba [17]. While Toxoplasma has two predicted cytosine-5 DNA methyltransferases (DNMT) homologues, 49.m03360 and 42.m03580, tachyzoites (and Cryptosporidium parvum) lack detectable cytosine methylation of DNA [18]. It remains possible that Toxoplasma may methylate DNA in another life cycle stage or that current techniques are not sensitive enough to detect it.
4. The histones: chromatin’s substrates
In addition to cataloguing and characterizing some of the histone modifiers [14], work has begun with respect to understanding the histone complement in Toxoplasma [19–20]. While histones H3 and H4 are highly conserved, apicomplexan parasites evidently lack the linker histone H1 and possesses unusual histone H2A and H2B variants [21–22]. Dalmasso et al. distinguished two separate lineages of H2B in Toxoplasma, one lineage consisting of TgH2Ba and TgH2Bb and the other being TgH2Bv1 [21]. While TgH2Ba and TgH2Bb are very similar, the latter was not detectable in tachyzoites or bradyzoites. Interestingly, TgH2Ba may be differentially regulated since its expression was shown to be higher in tachyzoites compared to bradyzoites. In contrast, TgH2Bv1 is highly expressed and not differentially regulated, leading the authors to suggest that TgH2Bv1 is the prominent H2B histone and TgH2Ba has a specialized role [21].
A study in 2009 identified TgH2A1 as the canonical H2A histone in the parasite and TgH2AX and TgH2AZ as variants [22]. Both TgH2A1 and TgH2AX have a C-terminal SQ motif and, like the H2A class of histones in general, are likely to be involved in the DNA damage response [23]. Interestingly, TgH2AZ was shown to dimerize with TgH2Bv1 while TgH2AX showed no interaction. Both TgH2AZ and TgH2Bv1 are associated with acetylated histones, consistent with their presence at active genes. Conversely, TgH2AX is present at repressed genes and its expression increases in bradyzoites, possibly as a consequence of the parasite preparing for quiescence. In addition, TgH2AX-containing nucleosomes do not seem to be located in close proximity to TgH2Bv1-containing nucleosomes. These pieces of evidence support the hypothesis that TgH2AZ and TgH2Bv1 are variants involved in the activation of transcription whereas TgH2AX and TgH2A1 may take part in chromatin remodeling during stress [22].
As mentioned above, a major epigenetic event is the post-translational modification (PTM) of histone proteins. Diagrams of the Toxoplasma histone sequences highlighting potential and proven PTMs have been published, as has a comprehensive map of PTMs as determined by mass spectrometry analysis of the highly similar histones in Plasmodium [20, 24–25]. In short, consistent with the large repertoire of histone modification enzymes in Table I, Toxoplasma histones have been shown to be acetylated, mono-, di-, or tri-methylated, phosphorylated, and sumolyated [12–13, 15, 22, 26–27].
5. ChIPing away at Toxoplasma histone code
A major advance in the study of epigenetic phenomena in protozoan parasites was the application of chromatin immunoprecipitation (ChIP) to the study of Toxoplasma. ChIP is a powerful technique to analyze protein-DNA interactions. The N-terminal tails of TgH3 and H4 are virtually identical to other species, so most of the commercial antibodies that recognize specific modified residues cross-react. This has allowed researchers to probe if certain histone modifications in the promoter of a gene correlate with the mRNA levels for that gene.
In a landmark 2005 study by Saksouk et al., ChIP was used in an apicomplexan parasite for the first time to show that histone acetylation and arginine methylation correlate with gene activation [12]. The utility of ChIP was greatly expanded in 2007 when Gissot and Kim used Toxoplasma microarray chips to identify the DNA sequences co-precipitated with select histone modifications [26]. Using this ChIP-chip method, Kim and colleagues were able to verify that acetylation of H3 at lysine 9 (H3K9), H4, and tri-methylation of H3K4 occur at promoters of actively expressed genes [26]. Similar methods were used to identify that tri-methylation of H3K9 and H4K20 is found at repressed genes localized at heterochromatic domains [13]. The identification of histone modifications may also facilitate mapping of active promoters and allows refinement of gene annotation. Expansion of ChIP-chip has been completed on a genome-wide scale and the results are currently available at the ToxoDB (www.toxoDB.org). It should be noted, however, that perfect correlations between acetylation levels and gene activation are not always observed [28]. For example, nucleosomes in the promoters of bradyzoite-specific genes have been found to be acetylated in low-passage tachyzoites from strains that differentiate more readily, arguing that histone activation marks may denote a gene “poised” for expression but not necessarily expressed yet [29].
The purification of recombinant forms of several histone modifiers for use in in vitro activity assays has also illuminated pieces of the parasite histone code. TgGCN5-A exhibits an unusual bias to acetylate only H3K18 in vitro while TgGCN5-B acts like other eukaryotic HATs, acetylating H3K9, H3K14, and H3K18 [12, 15]. The TgMYST-A HAT targets H4K5, H4K8, H4K12, and H4K16, consistent with MYST family members in other species [10]. Methylation of H3R17 has been shown to be mediated by TgCARM1 (coactivator-associated arginine methyltransferase 1), while methylation of H4R3 and H3R2 are mediated by TgPRMT1 and TgPRMT5, respectively [12, 25]. Interestingly, loci that contained methylated H3R17 also had enrichment of acetylated H3K18, perhaps hinting at a synergistic signature for gene activation that relies on cooperation between acetyl- and methyltransferase complexes [12]. H3K4, H3K9, H3K36 are methylated by TgSET1, TgSET3, and TgSET2, respectively [25]. TgSET8-related proteins can mono-, di-, and tri-methylate H4K20, which is of special note since human SET8 has only monomethylase activity [13]. KMTox/SET13 methylates lysines on histones H4 and H2A [30]. Finally, phosphorylation of serine 10 of TgH3 has been observed (H3S10), which peaks during mitosis simultaneously with monomethylation of H4K20 [13].
Likewise, enzymes serving to remove such modifications have been validated in Toxoplasma: TgHDAC3 removes acetyl groups from H4K5, H4K8, and H4K12, whereasTgSIR2 removes the acetyl group from H3K56 [25]. TgJMJD6 removes H3R2 methylation and TgJARID1/LSD1 removes H3K4 methylation [25].
ChIP has also allowed investigators to analyze non-histone proteins in association with DNA, including TFs and chromatin remodeling proteins. While it is technically challenging to perform ChIP using native antibody to histone modifying enzymes (likely due to their low abundance or tendency to form large multi-subunit complexes), success has been attained by expressing epitope tagged forms of the remodeler and ChIPing with antibody raised against the epitope tag. In this way, we have been able to deduce which histone modifying proteins may be controlling certain genes. For example, TgGCN5-A and TgHDAC3 work in concert to activate or repress stage-specific genes, respectively [12]. TgMYST-B has been found in association with promoters encoding ribosomal proteins (Sullivan, unpublished) as well as the ataxia telangiectasia mutated (ATM) kinase promoter [31]. Genome-wide analysis of the lysine methyltransferase KMTox using ChIP-chip revealed a large repertoire of genes involved in regulating antioxidant defenses and maintenance of cellular homeostasis [30]; in contrast, a similar analysis with TgSET8 showed that it was predominantly found at heterochromatin domains and intergenic regions, aligning with enrichments of methylated H4K20 and H3K9 [13].
6. Epigenetics playing vital roles in Toxoplasma biology
Ten years ago, it was still an open question as to whether epigenetics had a significant impact on parasite physiology, but research during the past decade has quickly revealed its widespread relevance. The first significant report regarding the importance of epigenetics on Toxoplasma physiology was first described in 2005, when researchers showed that histone-modifying complexes regulate gene expression pertinent to bradyzoite differentiation [12]. The report was the first to use ChIP to establish that bradyzoite-specific promoters are hypoacetylated in tachyzoites but become acetylated under differentiation conditions. The authors extended this analysis to show that TgGCN5-A was present at active promoters while TgHDAC3 was present at inactive promoters [12]. The same report also identified arginine methylation as a mark of gene activation in Toxoplasma, and found that pharmacological inhibition of TgCARM1 induces differentiation.
Additional studies have revealed epigenetics as a major contributor to stage conversion. Interestingly, the expression level of some histones appears to differ between life cycle stages; bradyzoites generated in vivo, which are arrested in G0, display increased TgH2AZ and TgH2AX [22]. The mRNA levels encoding the SWI2/SNF2 ATPase TgSRCAP increase during in vitro conversion to bradyzoites [11]. The use of Toxoplasma cDNA arrays to analyze parasites exposed to HDAC inhibitor apicidin showed that transcriptional activity during bradyzoite development may be regulated, at least in part, through gene-specific acetylation and deacetylation of histones [32]. Another HDAC inhibitor (FR235222), which selectively targets TgHDAC3 by virtue of a two amino acid insertion in the catalytic domain, was found to induce bradyzoite differentiation at sublethal concentrations [33]. New studies in our lab have implicated TgGCN5-A as a key player in stress-induced bradyzoite differentiation. While parasites lacking TgGCN5-A grow normally as tachyzoites [15], they do not recover from certain stresses as well as parental wild-type and fail to express stress-induced bradyzoite-marker genes (Sullivan, unpublished).
In contrast to TgGCN5-A, we have been unable to disrupt the loci encoding other HATs, including TgGCN5-B, TgMYST-A, and TgMYST-B. This may indicate that these HATs are essential for parasite viability, but that conclusion cannot be stated with certainty at present. Powerful new tools that facilitate knockout generation as well as the study of essential genes have recently become available for Toxoplasma that should be helpful in dissecting the role of these HATs. In the meantime, we have generated mutants that over-express each of these HATs. Parasites tolerate over-expression of TgGCN5-B [15], but do not tolerate additional TgMYST-A, which is lethal [10], or TgMYST-B, which produces a slow growth phenotype [31]. The detrimental effect on growth caused by over-expression of TgMYST-A or –B can be reversed if the HAT domain is mutated to be catalytically inactive, arguing that the parasite must maintain a strict balance on the acetylation activities of these HATs. Investigation of the mechanism behind the slow growth of parasites harboring additional TgMYST-B has lead to the discovery that TgMYST-B may be important for the parasite DNA damage response. These transgenic parasites are remarkably resistant to DNA damage induced by alkylating agents, exhibiting increased basal levels of ATM kinase and TgH2AX phosphorylation [31]. Pharmacological inhibitors of ATM kinase or HATs were able to reverse the slow growth phenotype seen in parasites over-expressing TgMYST-B.
TgSET8 may also have links to DNA repair as methylation of H4K20 is associated with maintaining genomic stability in other species, but this has not been validated in Toxoplasma to date. ChIP studies have linked TgSET8 and the repressive methylation marks on H3K9 and H4K20 to heterochromatin assembly [13]. Moreover, TgSET8 and monomethylation of H4K20 is cell cycle regulated, peaking during mitosis simultaneously with phosphorylation of H3S10. The authors also observed significant levels of monomethylated H4K20 in parasites undergoing cytokinesis, suggesting that epigenetic marks such as this could be transmitted to the daughter cell as a means of short-term memory. TgSET8 may also be active in the quiescent cyst as well, as suggested by high levels of monomethylated H4K20 in bradyzoites [13]. Intriguingly, a link has been reported between the mitotic period of the Toxoplasma cell cycle and development [34], and it is tempting to speculate that TgSET8 functions to repress tachyzoite gene expression, facilitating conversion and maintenance of bradyzoites.
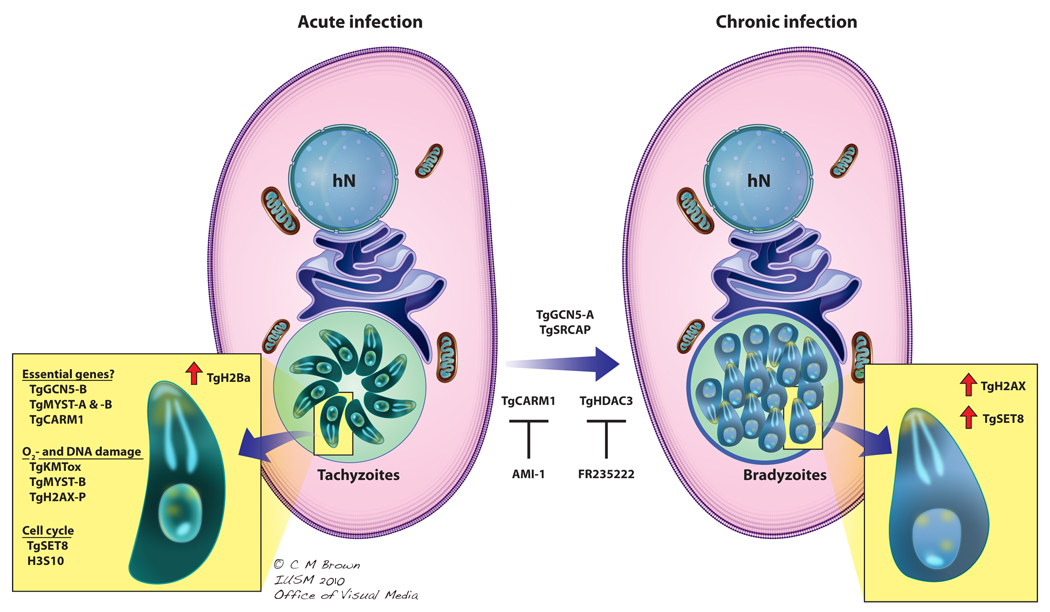
Another SET-domain protein contributes to the parasite’s oxidative stress response. Sautel et al. showed that KMTox (formerly TgSET13) is a lysine methyltransferase that displays enhanced interaction with 2-cys peroxiredoxin-1 (TgPrx1) under oxidative conditions [30]. Parasites over-expressing KMTox were more resistant to hydrogen peroxide, and ChIP-chip analysis linked KMTox to antioxidant defense genes, chromatin and transcription genes, heat-shock proteins/chaperone genes, and genes involved in translation and carbohydrate metabolism [30]. A summary of the potential roles of epigenetics in Toxoplasma biology is shown in Figure 1.
Figure 1. Diverse roles for epigenetics in Toxoplasma biology.
Schematic diagram shows two host cells (hN = host cell nucleus) infected with either a parasite vacuole containing proliferative tachyzoites (acute infection, left) or a cyst containing slow-growing bradyzoites (chronic infection, right). The inserts depict a single Toxoplasma parasite in either the tachyzoite stage (left) or the bradyzoite stage (right) and list the known epigenetic characteristics associated with each life-cycle stage. In tachyzoites, TgGCN5-B, TgMYST-A and –B, and TgCARM1 are histone modifying enzymes that may be required for propagation, based on the inability to disrupt the genetic loci. KMTox and TgMYST-B have been linked to the parasite’s response to reactive oxygen species (O2Ȓ) and DNA damage, respectively. Additionally, the histone variant TgH2AX is phosphorylated in response to DNA damage in tachyzoites. TgSET8 has been implicated in Toxoplasma cell cycle regulation, as has the phosphorylation of histone H3 on Serine-10, which is delivered by an uncharacterized kinase. TgH2Ba appears to be a variant histone exclusive to tachyzoites. TgGCN5-A and TgSRCAP have been implicated in the differentiation of tachyzoites into bradyzoites. The inhibition of TgCARM1 or TgHDAC3 by AMI-1 or FR235222, respectively, has been shown to induce cyst formation. The expression of the histone variant TgH2AX increases during bradyzoite differentiation. Presently, TgSET8 is the only chromatin remodeler identified to be associated in bradyzoite biology.
7. The transcription factors im‘planted’ in the Toxoplasma genome
As in other eukaryotes, the histone modifying enzymes in Toxoplasma lack DNA-binding domains; these enzymes complexes appear to be recruited by DNA-bound TFs. As mentioned earlier, apicomplexan genomes are devoid of conventional eukaryotic TFs, creating a quandary as to how the epigenetic modifiers are recruited to target DNA. A potential solution to this enigma appeared in 2005 when Aravind and colleagues discovered in silico evidence for a lineage-specific expansion of proteins containing motifs similar to AP2 (Apetala2)-integrase DNA-binding domains [35]. AP2 proteins are critical for developmental transitions and the stress response in plants [36]. In other words, Apicomplexa appears to use plant-like TFs rather than those that are prevalent in metazoans. “Guilt-by-association” evidence was already appearing in the literature that same year, suggesting that these apicomplexan AP2s (ApiAP2s) were indeed linked to transcription and gene control. For example, the purification of an TgHDAC3 complex revealed an associated ApiAP2 (TgCRC350, TGME49_072710) [12]. Additionally, a protein interaction network for Plasmodium falciparum showed ApiAP2 proteins (PF10_0075 and MAL8P1.153) in association with PfGCN5; incidentally, PfGCN5 is the most interconnected protein in the parasite and these interactions occurred at the unusual N-terminal extension [37]. In collaboration with Dr. Hakimi, we have also observed AP2 proteins co-purifying with TgGCN5-B (unpublished results). These observations parallel what has been observed in plants: CBF1, an Arabidopsis AP2 factor, associates with the GCN5 complex [38]. Together, the data argues that ApiAP2s represent at least one set of TFs that interact with epigenetic machinery to regulate parasite gene expression.
A demonstration that ApiAP2 proteins bind DNA was shown in 2008 by virtue of a protein binding microarray (PBM). De Silva et al. used such PBMs to elucidate DNA-binding motifs for select Plasmodium AP2 factors, and found that the motifs were present upstream of coordinately regulated genes [39]. Functional validation was subsequently established by Yuda and colleagues in a study showing that a Plasmodium AP2 factor (PF11_0442, AP2-O) directly activates ookinete stage-specific genes by binding to a six-base sequence, TAGCTA [40]. Interestingly, at least 40 predicted sequences with a canonical AP2 domain have been identified in Toxoplasma compared to 27 in Plasmodium and 17 in Cryptosporidium [41]. Apart from the AP2 domain, the predicted protein sequences are very diverse in size and composition. Probing the role of ApiAP2s in gene regulation will undoubtedly be an area of intense investigation in the near future.
Another method investigators have been using to study transcription in Toxoplasma involves the mapping of cis-regulatory elements in gene promoters. Early studies by Soldati and Boothroyd in 1995 identified a series of “TGAGACG” repeats in the promoter of the SAG1 gene, which encodes the major surface antigen in tachyzoites [42]. This element, which functions both as enhancer and selector of transcription initiation, has since been reported in numerous other Toxoplasma genes [43]. With the arrival of sequenced genomes, it was revealed that coordinately expressed genes are scattered across the chromosomes, which indicates that they are likely to be transcriptionally regulated by trans-acting factors that bind to cis-regulatory elements in gene promoters [1]. However, well-described cis-regulatory elements in other eukaryotes appear to be lacking in Toxoplasma; this is not surprising in light of the discovery that apicomplexans employ plant-like ApiAP2s as TFs. A few in silico studies were published that took advantage of the genomic data and bioinformatic search algorithms to identify over-represented sequences in the upstream regions of gene sets encoding functionally related proteins. Using this approach with ribosomal protein genes, Schaap and colleagues were able to identify two putative promoter motifs designated TRP1 and TRP2 (Table II) [44]. Strikingly, the TRP2 motif (5’YGCATGCR) matches the PF14_0633 ApiAP2-binding sequence elucidated by PBM analysis (5 TGCATGCA) [39], creating the possibility that the Toxoplasma homologue (TGME49_110950/583.m05398) could be operating as a potential TF at ribosomal protein genes. Kissinger and colleagues were able to identify and validate additional promoter elements for several different sets of genes believed to be coordinately regulated, including microneme genes, ribosomal protein genes, and genes involved in nucleotide metabolism and glycolysis (Table II) [45].
Table II.
Cis-regulatory elements identified in Toxoplasma gondii
| Motif sequence | Gene | Reference |
|---|---|---|
| [A/T]GAGACG | TUB1, SAG1, GRA1, GRA2, GRA5, GRA6, DHFR-TS, HSP70 | [42, 46, 54–55] |
| TCAGTTT | NTP1, NTP3, SAG1 | [56] |
| [A/G][C/T]TTGTTTT[T/C]T | NTPaseII, G6PD, GAPDH, PDI-like, cAMP-dependent protein kinase, calmodulin, IMCI, MLCI, SAG1–3, MIC4–6, 11, GRA2–7, ROP9, ribosomal P protein. |
[16] |
| AGGGG or CCCCT (STRE) |
ENO1, ENO2, HSP70 | [46–47] |
| nTTCn, nGAAn (HSE) |
ENO1, HSP70 | [46–47] |
| TCGGCTTATATTCGG [T/C]GCATGC[G/A] |
Ribosomal protein genes (TRP-1) Ribosomal protein genes (TRP-2) |
[44–45] |
| TACTGG | BAG1 | [29] |
| TGTGTG CAGC |
B-NTPase | [29] |
| GCTGCCTC TGCAGTGT |
HK, G6PI, PFK, ALD, TPI, GAPDH, PGK, PGM, ENO, Pyk | [45] |
| GCAAAGGA TTTTCGC |
AK, CTPS, DCDA, DHFR-TS, GMPS, RDPR, UPRT, AT | [29, 45] |
| GCGTCGCA CATGCAGT |
MIC1–11, M2AP | [45] |
The list of cis-regulatory elements found to date in Toxoplasma. Presumed genes are indicated as follows ALD, aldolase; AK, adenosine kinase; AT, adenosine transporter; BAG1, bradyzoite antigen gene 1; B-NTPase, bradyzoite-specific nucleoside-triphosphatase; CTPS, cytidine synthase; DCDA, deoxycytidine deaminase; DHFR-TS, dihydrofolate reducatase-thymidine synthase; ENO, enolase; GAPDH, glyceraldehydye-3-phosphate dehydrogenase; GMPS, guanidine monophosphate synthase; GRA, dense granule protein; G6PI, glucose-6-phosphate-isomerase; HK, hexokinase; HSE, heat shock element; HSP70, heat shock protein 70; IMC1, Inner membrane complex 1; M2AP, microneme 2-associating protein; MIC1–12, microneme 1–12; NTP, nucleoside triophosphate hydrolase; PDI, protein disulfide isomerase; PFK, phosphofructokinase; PGK, phosphoglycerate kinase; PGM, phosphoglucomutase; PyK, pyruvate kinase; RDPR, ribonucleotide diphosphate reductase; ROP9, rhoptry protein 9; SAG, surface antigen; STRE, Stress-related elements; TPI, triose-phosphate-isomerase; TUB1, α-tubulin 1; TRP, Toxoplasma ribosomal protein; UPRT, uracil phosphoribosyl transferase.
Several functional studies have provided convincing evidence of cis-regulatory factors. A pH-regulated region of the hsp70 gene locus was identified that had similarities to heat shock elements described in other eukaryotes [46]. A 2005 study by Kibe et al examined the regulation of ENO1 and ENO2, genes that are specifically expressed in bradyzoites and tachyzoites, respectively. The authors described a sequence in the ENO1 promoter that resembled a yeast stress response element (STRE) and bound nuclear protein in a stress-dependent manner [47]. A 2008 study by White and colleagues mapped minimal cis-regulatory elements to 6–8 bp in bradyzoite (Bz) promoters (BAG1 and a novel Bz-NTPase) that were capable of converting a constitutive promoter to one that is induced by bradyzoite conditions [29]. Collectively, these studies argue that Toxoplasma promoters contain regulatory information vital to gene expression, including the transcriptional changes necessary for stage conversion. A summary of described cis-regulatory elements identified in Toxoplasma is shown in Table II.
Many of the factors necessary for general transcription [41] including homologues for RNA polymerases are found in Apicomplexa genomes. In short, while many of the general transcription factors (GTFs) comprising the basal transcriptional complex are less conserved in Apicomplexa, most of them are present [43]. One unusual feature of note, however, is the apparent lack of TATA boxes in Toxoplasma gene promoters, yet the presence of a TATA-binding protein (TBP) homologue (TGME49_058680/55.m04779). There are examples of TBPs binding TATA-less promoters in other eukaryotes [48], and this should prove an interesting area for further study in Toxoplasma.
8. Questions for the next decade and beyond
In summary, the data generated over the past ten years of epigenetic research in Toxoplasma have unveiled a tightly regulated program of gene regulation that relies on chromatin remodeling and the use of at least one novel class of TFs. The parasites use a large repertoire of chromatin modifiers to modulate gene expression that is critical to many different aspects of their life cycle and pathogenesis. To close, we would like to point out a few areas that deserve attention in the upcoming decade.
1. With respect to characterizing the chromatin remodeling machinery, we have only explored the tip of the iceberg (Table I). We cannot put together a complete picture of how Toxoplasma coordinates gene expression until all of the pieces are turned over and studied. On a related note, new technologies such as ChIP-seq (coupling immunoprecipitated DNA to deep sequencing) should be adapted to increase the throughput and sensitivity in resolving where chromatin remodelers and their corresponding modifications occur in the genome under varying conditions. Mapping of all the post-translational modifications on Toxoplasma histones by mass spectrometry should also be performed in the near future. In addition to the histone modifications highlighted in this review, it would be of interest to study other epigenetic events such as the influence of subnuclear positioning on gene expression, which has been reported in Plasmodium [49], or the role of recently discovered noncoding RNAs. Small RNAs (rdsRNAs and satRNAs) may contribute to heterochromatin at repeat and satellite DNAs, and a Toxoplasma argonaute has been found in association with t he TgCRC complex, including TgHDAC3 and a TgAP2 factor (Ali Hakimi, personal communication).
2. How is the chromatin remodeling machinery mobilized? What cellular signals occur that communicate when a specific area of chromatin needs to be remodeled for activation or repression? In other species, master regulator transcription factors such as GCN4 are preferentially translated during times of stress, bind cis-regulatory elements and recruit chromatin remodelers. In Apicomplexa, elements of translation control have been identified [50], but it is not known if TFs like ApiAP2s are regulated in this manner.
3. How chromatin remodeling complexes are recruited to their target DNA sequence remains unclear. A tantalizing model is that ApiAP2s recognize cis-regulatory elements in gene promoters and recruit chromatin remodeling machinery upon DNA binding. Indeed, some ApiAP2s and histone modifying enzymes have been co-purified or identified as interacting proteins through yeast two-hybrid [12, 37]. There may be additional TFs in Toxoplasma performing this role as well: in Plasmodium, for instance, PfMyb is a probable TF that associates with PfGCN5 [37]. A number of zinc finger proteins are also evident in apicomplexan genomes that may have DNA-binding activity. Finally, TgLZTR was isolated from a yeast two-hybrid screen using a portion of TgSRCAP as bait [51]. TgLZTR is a predicted Leucine Zipper-like Transcriptional Regulator (TGME49_098600/125.m00070), but it has not been demonstrated to be a functional TF at present.
4. Could epigenetics be exploited in novel approaches to chemotherapy? New treatments to combat Toxoplasma and other apicomplexan pathogens are urgently needed and epigenetic-based gene regulation may be rich with potential drug targets. The potent anti-proliferative activity of many HDAC inhibitors in vitro argues that components of histone acetylation are viable drug targets, but there are no reports to date of these compounds being tested against in vivo Toxoplasma infection [2, 33, 52]. Despite the intense research into HATs over the past decade, surprisingly few HAT inhibitors are available. Cui et al have shown that anacardic acid and curcumin have anti-proliferative activity against Plasmodium and can inhibit PfGCN5 in vitro, but these compounds are believed to have many off-target effects [53]. It would be of great interest to test novel inhibitors of chromatin machinery as they become available, or initiate small molecule inhibitor screens to find them.
In conclusion, the past decade of epigenetic research in Toxoplasma has illuminated a novel path for exploration in this primitive protozoan. The mile marks passed so far have revealed that Toxoplasma utilizes several means of epigenetic modulation along with a set of unique transcription factors to regulate its gene expression. Answers to some of the questions posed above will teach us a great deal about parasite physiology and will likely provide new avenues for therapeutics.
Acknowledgements
Research in the Sullivan laboratory is supported through grants from the National Institute of Allergy and Infectious Disease (NIAID) at the National Institutes of Health (AI077502 and AI0834031). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors thank Drs. Ali Hakimi and Michael White for critically reading the manuscript and providing helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. The transcriptome of Toxoplasma gondii. BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darkin-Rattray SJ, Gurnett AM, Myers RW, Dulski PM, Crumley TM, Allocco JJ, et al. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc Natl Acad Sci U S A. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horrocks P, Dechering K, Lanzer M. Control of gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1998;95:171–181. doi: 10.1016/s0166-6851(98)00110-8. [DOI] [PubMed] [Google Scholar]
- 4.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Stedman E. Cell specificity of histones. Nature. 1950;166:780–781. doi: 10.1038/166780a0. [DOI] [PubMed] [Google Scholar]
- 6.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan WJ, Jr, Smith CK., 2nd Cloning and characterization of a novel histone acetyltransferase homologue from the protozoan parasite Toxoplasma gondii reveals a distinct GCN5 family member. Gene. 2000;242:193–200. doi: 10.1016/s0378-1119(99)00526-0. [DOI] [PubMed] [Google Scholar]
- 9.Hettmann C, Soldati D. Cloning and analysis of a Toxoplasma gondii histone acetyltransferase: a novel chromatin remodelling factor in Apicomplexan parasites. Nucleic Acids Res. 1999;27:4344–4352. doi: 10.1093/nar/27.22.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith AT, Tucker-Samaras SD, Fairlamb AH, Sullivan WJ., Jr MYST Family Histone Acetyltransferases in the Protozoan Parasite Toxoplasma gondii. Eukaryot Cell. 2005;4:2057–2065. doi: 10.1128/EC.4.12.2057-2065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan WJ, Jr, Monroy MA, Bohne W, Nallani KC, Chrivia J, Yaciuk P, et al. Molecular cloning and characterization of an SRCAP chromatin remodeling homologue in Toxoplasma gondii. Parasitol Res. 2003;90:1–8. doi: 10.1007/s00436-002-0814-1. [DOI] [PubMed] [Google Scholar]
- 12.Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, et al. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol Cell Biol. 2007;27:5711–5724. doi: 10.1128/MCB.00482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan WJ, Jr, Hakimi MA. Histone mediated gene activation in Toxoplasma gondii. Mol Biochem Parasitol. 2006;148:109–116. doi: 10.1016/j.molbiopara.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Bhatti MM, Livingston M, Mullapudi N, Sullivan WJ., Jr Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii. Eukaryot Cell. 2006;5:62–76. doi: 10.1128/EC.5.1.62-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatti MM, Sullivan WJ., Jr Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J Biol Chem. 2005;280:5902–5908. doi: 10.1074/jbc.M410656200. [DOI] [PubMed] [Google Scholar]
- 17.Fisher O, Siman-Tov R, Ankri S. Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica. Nucleic Acids Res. 2004;32:287–297. doi: 10.1093/nar/gkh161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gissot M, Choi SW, Thompson RF, Greally JM, Kim K. Toxoplasma gondii and Cryptosporidium parvum lack detectable DNA cytosine methylation. Eukaryot Cell. 2008;7:537–540. doi: 10.1128/EC.00448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan WJ., Jr Histone H3 and H3.3 variants in the protozoan pathogens Plasmodium falciparum and Toxoplasma gondii. DNA Seq. 2003;14:227–231. doi: 10.1080/1042517031000089496. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan WJ, Jr, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cell Microbiol. 2006;8:1850–1861. doi: 10.1111/j.1462-5822.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- 21.Dalmasso MC, Echeverria PC, Zappia MP, Hellman U, Dubremetz JF, Angel SO. Toxoplasma gondii has two lineages of histones 2b (H2B) with different expression profiles. Mol Biochem Parasitol. 2006;148:103–107. doi: 10.1016/j.molbiopara.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Dalmasso MC, Onyango DO, Naguleswaran A, Sullivan WJ, Jr, Angel SO. Toxoplasma H2A variants reveal novel insights into nucleosome composition and functions for this histone family. J Mol Biol. 2009;392:33–47. doi: 10.1016/j.jmb.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escargueil AE, Soares DG, Salvador M, Larsen AK, Henriques JA. What histone code for DNA repair? Mutat Res. 2008;658:259–270. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J Proteome Res. 2009;8:3439–3450. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- 25.Bougdour A, Braun L, Cannella D, Hakimi MA. Chromatin Modifications: Implications in the Regulation of Gene Expression in Toxoplasma gondii. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2010.01446.x. In press. [DOI] [PubMed] [Google Scholar]
- 26.Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. Epigenomic Modifications Predict Active Promoters and Gene Structure in Toxoplasma gondii. PLoS Pathog. 2007;3:e77. doi: 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun L, Cannella D, Pinheiro AM, Kieffer S, Belrhali H, Garin J, et al. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int J Parasitol. 2009;39:81–90. doi: 10.1016/j.ijpara.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Behnke MS, Radke JB, Smith AT, Sullivan WJ, Jr, White MW. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol. 2008;68:1502–1518. doi: 10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sautel CF, Ortet P, Saksouk N, Kieffer S, Garin J, Bastien O, et al. The histone methylase KMTox interacts with the redox-sensor peroxiredoxin-1 and targets genes involved in Toxoplasma gondii antioxidant defences. Mol Microbiol. 2009;71:212–226. doi: 10.1111/j.1365-2958.2008.06519.x. [DOI] [PubMed] [Google Scholar]
- 31.Vonlaufen N, Naguleswaran A, Coppens I, Sullivan WJ., Jr MYST-family lysine acetyltransferase facilitates ataxia telangiectasia mutated (ATM) kinase-mediated DNA damage response in Toxoplasma gondii. J Biol Chem. 2010 doi: 10.1074/jbc.M109.066134. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle JP, Saeij JP, Cleary MD, Boothroyd JC. Analysis of gene expression during development: lessons from the Apicomplexa. Microbes Infect. 2006;8:1623–1630. doi: 10.1016/j.micinf.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Bougdour A, Maubon D, Baldacci P, Ortet P, Bastien O, Bouillon A, et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med. 2009;206:953–966. doi: 10.1084/jem.20082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radke JR, Guerini MN, Jerome M, White MW. A change in the premitotic period of the cell cycle is associated with bradyzoite differentiation in Toxoplasma gondii. Mol Biochem Parasitol. 2003;131:119–127. doi: 10.1016/s0166-6851(03)00198-1. [DOI] [PubMed] [Google Scholar]
- 35.Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 38.Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci U S A. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, et al. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol Microbiol. 2009;71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
- 41.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Soldati D, Boothroyd JC. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meissner M, Soldati D. The transcription machinery and the molecular toolbox to control gene expression in Toxoplasma gondii and other protozoan parasites. Microbes Infect. 2005;7:1376–1384. doi: 10.1016/j.micinf.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Van Poppel NF, Welagen J, Vermeulen AN, Schaap D. The complete set of Toxoplasma gondii ribosomal protein genes contains two conserved promoter elements. Parasitology. 2006;133:19–31. doi: 10.1017/S0031182006009954. [DOI] [PubMed] [Google Scholar]
- 45.Mullapudi N, Joseph SJ, Kissinger JC. Identification and functional characterization of cis-regulatory elements in the apicomplexan parasite Toxoplasma gondii. Genome Biol. 2009;10:R34. doi: 10.1186/gb-2009-10-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma YF, Zhang Y, Kim K, Weiss LM. Identification and characterisation of a regulatory region in the Toxoplasma gondii hsp70 genomic locus. Int J Parasitol. 2004;34:333–346. doi: 10.1016/j.ijpara.2003.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kibe MK, Coppin A, Dendouga N, Oria G, Meurice E, Mortuaire M, et al. Transcriptional regulation of two stage-specifically expressed genes in the protozoan parasite Toxoplasma gondii. Nucleic Acids Res. 2005;33:1722–1736. doi: 10.1093/nar/gki314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 49.Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci U S A. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, et al. Translation Regulation by Eukaryotic Initiation Factor-2 Kinases in the Development of Latent Cysts in Toxoplasma gondii. J Biol Chem. 2008;283:16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nallani KC, Sullivan WJ., Jr Identification of proteins interacting with Toxoplasma SRCAP by yeast two-hybrid screening. Parasitol Res. 2005;95:236–242. doi: 10.1007/s00436-004-1291-5. [DOI] [PubMed] [Google Scholar]
- 52.Strobl JS, Cassell M, Mitchell SM, Reilly CM, Lindsay DS. Scriptaid and suberoylanilide hydroxamic acid are histone deacetylase inhibitors with potent anti-Toxoplasma gondii activity in vitro. J Parasitol. 2007;93:694–700. doi: 10.1645/GE-1043R.1. [DOI] [PubMed] [Google Scholar]
- 53.Cui L, Miao J, Furuya T, Fan Q, Li X, Rathod PK, et al. Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot Cell. 2008;7:1200–1210. doi: 10.1128/EC.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercier C, Lefebvre-Van Hende S, Garber GE, Lecordier L, Capron A, Cesbron-Delauw MF. Common cis-acting elements critical for the expression of several genes of Toxoplasma gondii. Mol Microbiol. 1996;21:421–428. doi: 10.1046/j.1365-2958.1996.6501361.x. [DOI] [PubMed] [Google Scholar]
- 55.Matrajt M, Platt CD, Sagar AD, Lindsay A, Moulton C, Roos DS. Transcript initiation, polyadenylation, and functional promoter mapping for the dihydrofolate reductase-thymidylate synthase gene of Toxoplasma gondii. Mol Biochem Parasitol. 2004;137:229–238. doi: 10.1016/j.molbiopara.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Nakaar V, Bermudes D, Peck KR, Joiner KA. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol Biochem Parasitol. 1998;92:229–239. doi: 10.1016/s0166-6851(97)00220-x. [DOI] [PubMed] [Google Scholar]