Abstract
Sirtuins are ancient proteins widely distributed in all lifeforms of earth. These proteins are universally able to bind NAD+, and activate it to effect ADP-ribosylation of cellular nucleophiles. The most commonly observed sirtuin reaction, is the ADPribosylation of acetyllysine, which leads to NAD+-dependent deacetylation. Other types of ADP-ribosylation have also been observed, including protein ADP-ribosylation, NAD+ solvolysis and ADP-ribosyltransfer to 5,6-dimethylbenzimidazole, a reaction involved in eubacterial cobalamin biosynthesis. This review broadly surveys the chemistries and chemical mechanisms of these enzymes.
Sirtuins are a broadly conserved family of enzymes found in all phyla of life including archaea, eubacteria, yeast, plasmodia, metazoans, mammals and even viruses[1–3]. These are ancient proteins that have a common catalytic architecture, which allows these proteins to recognize universally, a metabolically central and abundant compound, NAD+. The most common reaction catalyzed by sirtuin enzymes is that of NAD+ dependent protein deacetylation, which consumes a mole equivalent of NAD+ per acetylgroup removed [1]. The reaction effects an acetyl group transfer to ADPR to form a novel compound call 2'-O-acetyl-ADPR (2'-AADPR, Scheme 1,[1]). Select sirtuins also catalyze other reactions, such as protein ADP-ribosyltransfer, NAD+ hydrolysis and many if not all seem to catalyze acetyllysine-dependent nicotinamide base-exchange into NAD+. Sirtuins have been implicated in organism adaptations to nutrient intake, and are regulators of aging in a variety of organisms, ranging from yeast, flies, worms and probably in mammals as well [4, 5].
Scheme 1.
Overall stoichiometry determined for the NAD+-dependent deacetylation reaction.
The detailed active site recognition of NAD+ has been delineated by X-ray structures of sirtuins co-complexed with this substrate [6, 7]. NAD+ complexed to a sirtuin active site is a chemically reactive form of NAD+ [6], capable of undergoing chemistry with nucleophiles such that ADP-ribosyl-transfer occurs with inversion of stereochemistry [8–12] (Scheme 2). Characterized general nucleophiles for NAD+ include acetyllysine substrates or thioacetyllysine inhibitors [11–13]. Close proximities of NAD+ and the acetyllysine group (or thioacetyl group) enforced by the active site structure leads to ADPribosylation of acetyllysine to form an imidate (or thioimidate, Scheme 1 top). The imidate is proposed to account for deacetylation as well as base exchange catalyzed by these enzymes [9, 12, 14, 15]. On several sirtuin enzymes, other nucleophiles have also been found to react directly with NAD+ such as methanol [10](Scheme 2 middle), and water [10], characteristic of an NAD+ glycohydrolase activity. NAD+ (and possibly NaMN) can also react with inversion with the nucleophile 5, 6-benzimidazole catalyzed by CobB, a Sir2 found in eubacteria (Scheme 2, bottom)[8, 16]. It has been suggested that some sirtuins catalyse a reaction where NAD+ reacts directly with protein nucleophilic amino acid side chains [16, 17]. Indeed, there are multiple examples in which sirtuins have been reported to catalyze ADPribosyltransfer to proteins [18–22].
Scheme 2.
Direct reaction of NAD+ with nucleophiles determined for distinct sirtuin enzymes. The top reaction depicts imidate and thioimidate formation. The second reaction depicts a direct solvolysis reaction characterized for a Plasmodium falciparum sirtuin enzyme. The bottom reaction represents an ADPribosyltransfer reaction determined for the CobB enzyme, a sirtuin from eubacteria.
The common mechanistic thread in all of these chemistries is that sirtuins activate NAD+ as a chemical partner to react with nucleophiles to effect different reaction outcomes. We continue to learn more about how sirtuin chemistry varies from species to species and from isoform to isoform. Nature has honed the chemistry platform provided by sirtuin enzymes to accomplish specificity, regulatory control, and linkage to metabolism, which in turn disposes these enzymes to regulate a variety of fundamental cellular processes. In this review, we examine the mechanisms of sirtuin chemistries with an emphasis on the relationships to known chemistries, the differences of chemistries discovered for different sirtuins, and we discuss the functional and chemical components of sirtuin reactions.
SIRTUIN DEACETYLATION REACTION
The most biologically relevant of the reactions that sirtuin enzymes catalyze is protein deacetylation [1, 3]. The reaction has been demonstrated for sirtuins isolated from a variety of phylogenetically distributed species, from archaea to human enzymes [2, 7, 9, 23]. The reaction stoichiometry in Scheme 1 is catalysed by sirtuins derived from yeast, archea, eubacteria and mammalian sirtuins [9, 24, 25]. Among other things, the reaction is unusual in that it generates 2'-AADPR [1, 9, 24]. The full characterization of 2'-AADPR established that sirtuins catalyze the synthesis of an ester from an amide, a thermodynamically challenging reaction paid for by NAD+ degradation [1]. This reaction outcome, the biological distribution of the reaction, the structural and sequence similarities of sirtuins provides evidence that sirtuins utilize a common mechanistic strategy to achieve deacetylation. The function of 2'-AADPR in cells, which spontaneously and non-enzymatically equilibrates with the 3'-AADPR isomer [9, 24], is currently poorly understood, but is the topic of an extended review in this journal issue.
The sirtuin deacetylation reaction, in addition to restoring the free amino group of lysine and producing AADPR, also generates nicotinamide as a product in all cases [9, 26, 27] indicating that the deacetylation reaction necessarily cleaves the nicotinamide bond to the anomeric carbon (C1') of ADPR (Scheme 2). It is also true that sirtuin enzymes are generally capable of reversible breakage and reformation of the N1-ribosyl bond [14, 26, 28]. The reversible cleavage of nicotinamide is observable by incubation of sirtuin reactions with [carbonyl- 14C] nicotinamide and by detection of newly formed 14C-NAD+ from initially unlabeled substrate NAD+. Interestingly, this base-exchange reaction requires the acetylated substrate to be present [26]. A detailed explanation of this mechanism will be presented in the following section. Finally, the overall sirtuin reaction requires stoichiometric consumption of water from solvent, as shown by 18O incorporation into the acetyl group of AADPR when reactions are performed in 18O water (Scheme 3, [9]).
Scheme 3.
Proposed reaction mechanism of sirtuin catalyzed NAD+-dependent deacetylation. The asterisk depicts radioactively labeled nicotinamide that can be used to monitor the base exchange reaction (reversal of imidate to NAD+). Bottom scheme shows spontaneous non-enzymatic equilibration of AADPR isomers that occurs in solution.
A detailed mechanism for the deacetylation reaction was proposed that we have since referred to as the ADPR peptidyl-imidate mechanism [1, 9] (Scheme 3). Several mechanistic clues led to this proposal, including the proper identification of 2'-AADPR. The regiochemistry of the acetyl group in this molecule implied that the acetyllysine was likely to react at the α-face of the NAD+ molecule. Isotope labeling experiments supported a reaction mechanism wherein ADPribosyltransfer occurs between acetyllysine oxygen with the C1' atom of the ADPribose moiety of NAD+ [1, 9]. An abundance of X-ray crystal structures demonstrate that the proposed direct reaction between acetylated lysine and NAD+ is readily accommodated at the active site, including a termolecular complex of acetylysine and NAD+ [6]. The proposed bond formation between the acetyl carbonyl oxygen and the ADPR was definitively established with synthesis of an 18O-isotopically labeled acetyllysine peptide substrate. This substrate was reacted with NAD+ catalyzed by a yeast HST2 and the 18O-isotope in the amide carbonyl oxygen was found to be transferred to the hydroxyl substituent at the C1'-position of the product AADPR, as predicted by the peptidylimidate mechanism [29].
As Scheme 3 shows, the first chemical step in the sirtuin deacetylation mechanism generates nicotinamide and a novel intermediate called a peptidylimidate intermediate (an intermediate we originally called an alkylamidate)[9]. Sauve et al. suggested that this intermediate has special properties, including the property that it could be formed reversibly, and thereby, could explain both the ability of sirtuins to catalyze base exchange, as well as the requirement for acetyllysine peptide to perform exchange chemistry [9]. In addition, the imidate can be envisioned to react as a reactive intermediate that is subject to intramolecular attack by the 2'-OH to eventually yield 2'-AADPR (Scheme 3). This downstream mechanism accounts for deacetylation, production of 2'-AADPR and can explain solvent labeling of 18O into the acetyl of AADPR (Scheme 3; [1, 9, 29]).
The proposed enzymatic mechanism in Scheme 3 is now supported by a variety of structural, kinetic, isotope labeling, computational and other studies. It has been exceptionally versatile for explaining a variety of features of sirtuin enzymes and is supported by a considerable body of experimental data. Some of these studies are reviewed here more specifically, and structural studies are extensively reviewed in this issue. Discussion on aspects of the mechanism that are less well understood will be included.
NAD+ BINDING GEOMETRY AND AFFINITIES
The reaction of NAD+ and a nucleophile is mediated by recognition of the NAD+ in an elongated trans-conformation, within a domain that has a Rossman-like fold [6, 7]. There are a variety of reports of NAD+ Km values for sirtuins. These measures establish that the enzyme affinity for NAD+ is not of high affinity and in the range of 100–550 µM for human enzymes, For example, several independent determinations of the Km of human SIRT1 have been published, ranging from 132 µM to 550 µM [30, 31]. The Sauve laboratory has determined a Km for NAD+ of 165 µM using a p53 derived peptide substrate (Sauve unpublished data). The Km value of SIRT2 was reported to be near 100 µM [32]. Corresponding values for yeast Sir2 can be estimated from reported data or taken from published values to be in the range 29–70 µM [26, 32, 33]. The likelihood that NAD+ levels in cells can regulate sirtuins is dependent on several factors, one of which is the Km for NAD+. Importantly, the value of Km (NAD+) for yeast Sir2 is quite low relative to the reported NAD+ levels in the yeast which are reportedly near or above mM concentration [33, 34], even when yeast are depleted of nicotinic acid [35]. These values suggest that NAD+ fluctuations alone in yeast would not be sufficient to regulate sirtuin function, although Belenky et al. found that at sub-mM (near 700 µM) NAD+ concentrations, the biological functions of yeast Sir2 appeared to be responsive to NAD+ increases [35]. These findings could imply other factors are important in the yeast that raise the apparent Km for NAD+. Among these factors could be allosteric effects, mediated by other proteins, which could be quite relevant since Sir2 is found complexed to other proteins in vivo. In addition, non-competitive inhibition effects on Km by nicotinamide can raise the apparent Km for NAD+[26]. Guarente has suggested that NADH levels in yeast might also increase apparent Km for NAD+ in vivo, raising it sufficiently so that alterations in NADH ratio or increases in NAD+ could provide Sir2 stimulatory effects [33]. In contradiction, the Denu laboratory, found Ki values for NADH to be in the 15 mM range, well above what would be relevant in vivo for this model of sirtuin regulation to be true [36]. A review on yeast regulation of Sir2 is presented in this issue.
Mammalian cell NAD+ concentrations are typically well below those found in yeast and are typically in the range from 200–600 µM (for recent measurements in mammalian cells and tissues see [37–39]). However, the mammalian Km values, for NAD+ for SIRT1 in particular, are high enough to suppose that direct NAD+ regulation of sirtuin biochemical activity in mammalian cells is likely and that the biochemical properties of at least some sirtuins in mammalian cells are consistent with roles as NAD+ sensors, suggesting they link directly to NAD+ fluctuations in vivo. Increasingly biological evidence supports this point of view [37, 38, 40, 41], although it is clear that limitations on accurately determining compartmentalized (organelle-specific) NAD+ concentrations still limits our ability to precisely link NAD+ concentration dynamics to regulation of sirtuins. The effect of NAD+ concentrations on sirtuin function in physiologic settings is beyond the scope of this review, but the idea of modulating NAD+ levels to influence sirtuin function is being pursued pharmacologically [42]. Nevertheless, the concept that NAD+ levels can regulate sirtuin activity provides one rationale for consuming metabolically expensive NAD+ as a price for deacetylation. Acetylation of proteins is metabolically expensive in its own right, and perhaps it is biologically logical to regulate removal of these groups by sensing centralized metabolic information in the cell. In time it appears that metabolic inputs transduced through sirtuin enzymatic activity provided a basis for profound adaptive responses to nutritional intake, via alteration of chromatin function and nuclear transcriptional programs which are highly sensitive to acetylation status.
The binding of NAD+ to sirtuins is mediated by a set of highly conserved catalytic residues. Some of the roles these residues play in mediating NAD+ recognition have been elucidated. For example, a universally conserved catalytic histidine has been identified that recognizes the 3'-OH group of NAD+. This residue is important for substrate binding, and for activation of the 2'-OH group for nucleophilic attack on the imidate to initiate downstream chemistry [7, 9, 29]. Furthermore, a conserved phenylalanine appears to be important for organizing and recognizing the nicotinamide riboside moiety into the active site [6, 12]. Both of these residues have also been argued to play important roles in catalysis. It has been proposed that the active site residues enforce a destabilization of the NAD+ on the active site which facilitates nicotinamide bond cleavage and downstream reaction of NAD+ with acetyllysine substrate or other nucleophiles [6, 12]. Currently the evidence that such a destabilization is important for catalytic activity of sirtuin enzymes is mostly from X-ray crystallographic data, and computational and other experimental evidence to support this model of NAD+ reactivity is currently not available. Ground state destabilization is one way to facilitate enzymatic catalysis, which leads indirectly to reducing the transition state barrier. The manner in which NAD+ is reacted in the first chemical step of deacetylation catalysis is now considered, with respect to views that have been presented on the nature of the transition state.
TRANSITION STATE AND MECHANISTIC PROPOSALS FOR ADPRIBOSYLATION REACTION WITH ACETYLLYSINE
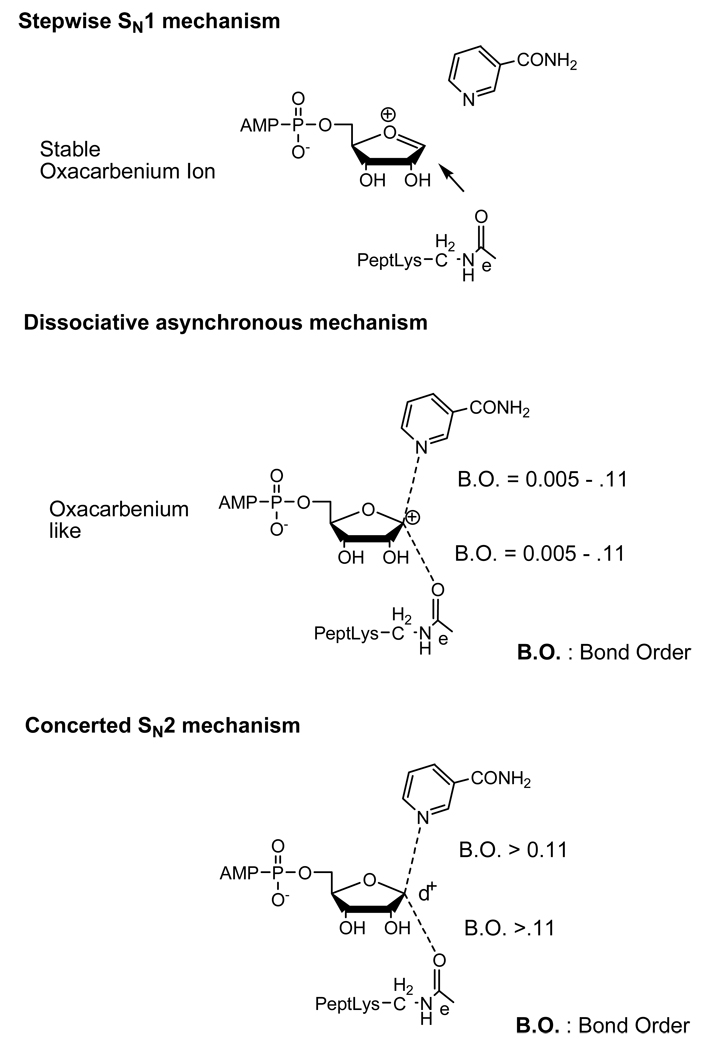
The proposed first chemical step catalyzed by sirtuins leading to acetyllysine deacetylation is ADP-ribosylation of acetyllysine, leading to formation of an ADPribosyl-peptidylimidate intermediate (Scheme 3). The formation of this species can be imagined to proceed via several distinct mechanistic processes, all of which can achieve the formation of the imidate. For simplification, we will present three mechanistic choices (Scheme 4) which have been proposed in the literature, although additional variations of these are possible. The first mechanism is a stepwise dissociative mechanism (formally SN1), in which full dissociation of the nicotinamide forms an enzyme-stabilized oxacarbenium ion, followed by collapse of the oxacarbenium ion by reaction with acetyllysine (Scheme 4, top). A second mechanism can be considered also, in which an oxacarbenium ion is formed at the transition state, but with extensive bond cleavage to the leaving group, and weak bond formation to the nucleophile (Scheme 4, middle). This dissociated, reaction mechanism is not stepwise, but since it lacks a discrete intermediate between the reactants and the imidate. The reaction is asynchronous, since the leaving group is largely cleaved prior to bond formation with the nucleophile. The third mechanism is characterized by associative effects of the nucleophile at the transition state, consistent with an SN2 mechanism (Scheme 4, bottom). This mechanism typically is characterized by a pentacoordinate transition state with significant bond orders to both nucleophile and leaving group. Data in support of each of these mechanisms has been produced, although, there is a lack of firm consensus on which of the mechanisms best explains sirtuin chemistry.
Scheme 4.
Different transition states or intermediates proposed for ADP-ribosylation of acetyllysine discussed in the text. Bond angles as drawn between nicotinamide and C1' and carbonyl-oxygen and C1' are not strictly defined in these representations.
Sauve et al. first proposed the direct displacement reaction between NAD+ and acetyllysine and considered the reaction to proceed via a mechanism featuring a dissociated transition state (Scheme 4, middle) [9]. Acetyllysine is not a good nucleophile, and the authors proposed that electrophilicity of an oxacarbenium ion transition state likely accounted for the ability of sirtuins to ADP-ribosylate this unlikely sidechain nucleophile [9, 43]. An old example by Borch and co-workers demonstrated that O-alkylimidates could be generated from amides by use of very electrophilic alkylating agents, such as triethyloxonium tetrafluoroborate [44]. Interestingly Borch and co-workers determined that these imidates have a tendancy to decompose to esters and free amines when exposed to water, directly analogous to the chemistry proposed for sirtuins. The authors realized that NAD+ was not itself particularly electrophilic, but that enzyme catalyzed reactions of NAD+ involving ADP-ribosyltransfer commonly proceed through transition state oxacarbenium ions [45–48]. It is important to point out that in every case where there is a solved ADP-ribosyl transition state, the structures are highly dissociated, but still have very small bond orders between 0.001 to 0.11 to both nicotinamide leaving group and to nucleophile (consistent with Scheme 4, middle) [45–48]. These highly oxacarbenium-like transition states are characterized by extremely electrophilic sugars that would be envisioned to capture the acetyloxygen to form the envisioned imidate species [1, 9, 43].
The concept that the reaction might proceed differently than a conventional dissociated transition state was recently suggested by data obtained by Smith and Denu, in which it was suggested that an SN2-type reaction (Scheme 4, bottom) might better explain sirtuin chemistry in the initial ADP-ribosylation step [49]. To assess the nature of the transition state, Smith et al. found the rate of the ADP-ribosyltransfer reaction was strongly dependent on the nature of substitution at the α-position of the acetyllysine group, with a powerful decelerating effect associated with electron withdrawing groups such as fluorine [49]. Mono-fluoro, difluoro and trifluoro modifications of acetyl of acetyllysine reacted at relative rates 5.5 10−4, 6.9 10−6 and 1.6 10−6 versus that of the underivatized acetyllysine. This was interpreted to be an indication of “considerable nucleophilic participation” at the transition state. The authors argued that Sir2 deacetylase ribosyl-nicotinamide cleavage is consistent with an SN2 like mechanism where considerable bonding to the nucleophile occurs at the transition state (Scheme 4 bottom). The authors concluded that it was unlikely that a fully dissociated “stable” oxacarbenium ion (Scheme 4 top) as proposed by Marmorstein [50] and initially favored by Denu [14] would form at the transition state or prior to the ADP-ribosylation of the acetyllysine [49]. The authors argued that a step-wise mechanism involving a fully formed oxacarbenium ion (Scheme 4, top) would not be expected to provide rate differences for nicotinamide formation from NAD+ in the presence of different amide nucleophiles, as was observed. Interestingly, a fully dissociated yet enzyme-stabilized oxacarbenium ion has been favored by some authors in other instances of ADP-ribosylation, for instance, to explain the promiscuous active site chemistry of CD38 [51]. The existence of an enzyme stabilized oxacarbenium ion on sirtuin enzymes is still an open question. Interestingly, the catalytic properties of Plasmodium falciparum Sir2, which has promiscuous ADP-ribosyltransferase activity [10], could be explained by such an intermediate. Such an intermediate seems unlikely to be formed on yeast Hst2, the enzyme used in the Smith and Denu study [49]. The authors did not clearly suggest if the transition state was oxacarbenium like or non-oxacarbenium-like [49]. Thus the meaning of “considerable nucleophilic participation” was left quantitatively undefined in terms of transition state structure.
Hu et al. used computational methods to address this problem and obtain a more quantitatively defined geometry of the transition state [52]. These authors used QM/MM methods to determine that the Thermotoga maritime Sir2 transition state for imidate formation is highly dissociated, and highly oxacarbenium like, analogous to other solved ADP-ribosyltransfer transition states [45–48] with bond lengths of 2.60 angstrom to nicotinamide and 2.35 angstroms to the acetyllysine oxygen [52]. Their structure invokes weak nucleophilic participation at the transition state, to the extent of 0.14 bond order [52]. The Hu et al. result is largely consistent with our original suggestion that a highly dissociated oxacarbenium ion is likely formed at the transition state [1, 9, 43] and is in agreement with the result of Smith et al. that there is no fully dissociated oxacarbenium at the transition state [49]. In fact this computational result provides evidence that the sirtuin ADP-ribosyltransfer reaction is not stepwise (SN1, Scheme 4 top), but is a dissociative asynchronous mechanism (Scheme 4 middle) since the nicotinamide bond is largely cleaved before the acetyllysine bond is formed.
It is of interest to ask if the Smith and Denu rate data is more consistent with a highly-associated SN2 like transition state, or is consistent with the 0.14 ± 0.04 bond order to the nucleophile at the transition state as suggested by the Zhang study. A clear explanation of the Smith and Denu rate data remains to be provided, although the data at face value suggests a high degree of nucleophile associativeness, consistent with a concerted SN2 type mechanism [49]. Selectivity for the nucleophile can be understood to arise from the extent of nucleophilic involvement at the transition state in the bond forming reaction. Since the transition state marks the point of no return for the barrier crossing, it is intuitive that the more a bond is required to be formed to reach the transition state, the more selectivity there can be for making that bond. An interesting example at the minimal extreme is the selection of methanol versus water of 1:1 (taking into account mole fractions) for NAD+ solvolysis, suggesting a very weak bond to the nucleophile at the transition state [53]. Indeed Berti and Schramm found the nucleophile bond order at the transition state for this reaction was 0.005, consistent with extremely poor nucleophile selection in this reaction [54]. The Smith and Denu data set shows that 0the nicotinamide formation rate from NAD+ on the sirtuin HST2 displays a strong dependence on the nucleophile, seemingly inconsistent with a very weak bond to the nucleophile at the transition state.
However, there are some very important considerations and caveats for the modifications that Smith et al.[49] tested. These modifications may have caused the transition state position to shift to later and later character, requiring increased nucleophile participation to form an increasingly destabilized imidate. Weakening of the nucleophile basicity and application of Hammonds postulate would be the chemical explanation for this phenomenon. If so, the least basic of the amide groups (for example, the trifluoro-substituted) would require the most significant bonding at the transition state. Improvement of the imidate leaving group in the NAD+ reformation reaction (as predicted for the electron deficient amides) also increases the possibility that the rate of internal return to the Michaelis complex is enhanced for the least stable imidates, perhaps reducing the observed rates of nicotinamide formation. Finally, the geometric and electronic changes in substrates introduces the very real possibility that the modifications cause substrates to bind in altered geometries from those of the normal substrate, which could substantially reduce observed rates of reaction.
The Wolberger and Marmorstein laboratories have also produced X-ray crystallographic evidence for a highly dissociated transition state occurring on sirtuin enzymes [12, 50]. Since these are reviewed extensively elsewhere in this issue, we will not survey these data here, except in short. The Wolberger laboratory developed evidence for a dissociated transition state on the basis of structural analysis of distances between nucleophile and leaving group in several X-ray complexes, including a sirtuin-complexed dissociated NAD+ mimic [12]. They argued that the sirtuin ADP-ribosylation step is an example of “nucleophilic displacement by electrophilic migration” [12]. Electrophilic migration contends that the breakage of the leaving group bond largely precedes the formation of the nucleophile bond, and is caused by the migration of the sugar electrophile between leaving group and nucleophile positions that are largely fixed through the reaction coordinate [55]. This mechanism is found for a number of N-ribosyltransferase enzymes and explains asynchronous mechanisms for substitution reactions in this family of enzymes [55, 56].
The debate on transition state structure of the ADP-ribosylation of acetyllysine catalyzed by sirtuins is still unresolved, partly because there is no clear intellectual reconciliation of the variety of different data sets that have emerged on this question. In addition, the possibility that distinct sirtuin isoforms might stabilize variants of the theoretically available transition states, or even proceed through different mechanisms, remains experimentally untested. Although it is clearly of interest to explain the biochemical and structural basis for the ability of sirtuins to catalyze ADP-ribosyltransfer, investigations to address this issue arguably require a more thorough elucidation of sirtuin transition states. An expected solution is in the experimental determination of the transition states by kinetic isotope effects with interpretations supported by computational analyses. Such methods have the potential to probe the transition state structure directly, without perturbation of substrate steric or electronic structure. The Schramm laboratory in particular has shown that this approach, combined with computational methods, can provide remarkably well determined structural determinations of transition states, including several ADP-ribosyltransfer transition states [45–48]. In several cases, using the transition state structures as blueprints, inhibitors of N-ribosyltransferase enzymes have been designed and synthesized and bind their targets with nM-fM affinity [57, 58]. It is expected that similar studies will provide a more time resolved and mechanistically defined view of the ADP-ribosyltransfer step mediated by specific sirtuin enzymes, and can lead logically to studies which probe the enzymatic features that direct and accelerate sirtuin ADP-ribosyltransfer chemistry.
IMIDATE AND CHEMISTRIES FROM IMIDATE
Whatever the transition state of reaction between acetyllysine and NAD+ it is now widely agreed that the ADPR-peptidyl imidate (originally called an 1'-O-alkylamidate [1, 9] is the species that is formed by the reaction of the two main reaction participants after the first chemical step (Scheme 3). Nevertheless direct evidence for the existence of this catalytic species is still unavailable. Supportive but indirect evidence that this intermediate is formed on the enzyme has come from a number of observations. For example, the amide is required for base exchange [26], and 1-O-glycosyl imidates are now commonly used in coupling of sugars to nucleophiles in organic synthesis [59, 60], demonstrating the intrinsic reactivity of this type of functionality at the anomeric center. The imidate formation is relatively insensitive to removal of an active site histidine from the enzyme, which coordinates to the 3' and 2' OH in different crystal structures and does not significantly decelerate sirtuin base-exchange [7, 29]. This suggests that the enzyme is not typically reliant on acid-base catalysis to form or reverse the proposed imidate. Consistently, the Denu laboratory has shown that β-2'-deoxy-2'-fluororibo NAD+ can be accepted as a base exchange substrate with the HST2 enzyme [14].
The most direct evidence in support of the imidate is a crystal structure of TmSir2 with the adduct formed between NAD+ and a thioacetylated peptide on the enzyme [12]. The crystal structure reveals a 1'-thio-imidate formed on the enzyme (Scheme 2 top, where S is bonded to C1'). This analogue of the reactive imidate is surprisingly unreactive, and causes potent inhibition of sirtuin enzymes [13, 61]. Moreover, in several sirtuins sensitive to inhibition by thioacetylpeptides, base exchange cannot be supported by a thioacetylpeptide (Sauve unpublished results), arguing that the thioimidate is formed irreversibly from NAD+. The competency of the thioimidate to eventually complete deacetylation chemistry has been demonstrated by Denu and coworkers, who showed that the thioacetylgroups on sirtuin enzymes eventually yield a compound consistent with the structure 1'-thio-2'-AADPR [61]. Collectively these results provide compelling evidence that an imidate species formed on sirtuin enzymes is responsible for the sirtuin deacetylation mechanism.
The imidate is proposed to have two fundamental reaction pathways. The first is collapse of the 2'-OH group onto the imidate by nucleophilic attack (Scheme 3, Step 2), and the other is reversal to NAD+ via β-face attack of nicotinamide on the anomeric carbon (Scheme 3, Step 1 reversed). Evidence in favor of the 2'-OH being involved in the decomposition of the imidate as a means to obtain deacetylation has come from mutation of a universally conserved catalytic histidine that was first observed at the 3'-OH position of complexed NAD+ and AF1Sir2 [7]. The mutation His to Ala does not appear to significantly alter base-exchange, but significantly slows deacetylation catalysis [7, 29]. This histidine is posited to play an important role as an activating base that deprotonates the 2'-OH group during Step 2, Scheme 3 downstream of imidate formation, leading to imidate decomposition [9, 12, 29]. In addition, β-2'-deoxy-2'-fluororibo-NAD+ which lacks a 2'-OH group, reacts to form an apparent imidate on HST2, but only releases acetate when treated with strong alkaline quench, consistent with a requirement of the 2'-OH to complete deacetylation chemistry under multiple turnover conditions of the enzyme [14].
The other important chemistry of the imidate is nucleophilic attack at the β-face. In the presence of nicotinamide, nicotinamide is proposed to reverse the imidate back to NAD+ (Scheme 3 reverse Step 1). This reversal from the imidate has been demonstrated to be the mechanism whereby nicotinamide can inhibit deacetylation chemistry [14, 15]. Consistent with the idea that the imidate has a reactivity to nucleophilic substitution at the anomeric carbon is the observation that histidine mutants which cannot achieve deacetylation and have low nicotinamide in media, instead react by hydrolysis at the β-face [29]. This was shown for the Hst2 enzyme, wherein addition of methanol caused the imidate to form β-1'-O-methyl-ADPR with selection of methanol over water of 26 : 1 [29]. The Sauve laboratory has also identified a wildtype enzyme from Plasmodium falciparum which is a slow deacetylase, but can perform base-exchange very efficiently [10]. In the absence of nicotinamide, but in the presence of NAD+ and acetylated peptide the enzyme makes ADPR more rapidly than it deacetylates peptides [10]. As such, it is the first wildtype sirtuin to have a demonstrated NAD+ glycohydrolase function [10]. When methanol is present the enzyme forms β-1'-O-methyl-ADPR consistent with reaction of the imidate with methanol at C1[10]. Interestingly, when nicotinamide is added to reaction mixtures β-1'-O-methyl-ADPR formation is inhibited, consistent with nicotinamide and methanol competing for the same anomeric carbon in the same active site pocket (Scheme 5) [10].
Scheme 5.
Methanolysis of imidate observed for a mutant and wildtype sirtuin. Sensitivity to nicotinamide inhibition is depicted by competitive attack of nicotinamide in base exchange.
PARTITIONING OF THE INTERMEDIATE (NATURE OF NICOTINAMIDE INHIBITION)
Of interest to the understanding of the mechanism of sirtuins, and for understanding their biological function is the appreciation that a variety of sirtuins in different organisms are sensitive to physiologic nicotinamide concentrations [62, 63]. Nicotinamide is the first product formed in sirtuin reaction chemistry, as has been demonstrated by rapid-mix rapid-quench studies [32] and from observations that nicotinamide product formation is reversible [26]. In depth studies of the nicotinamide base-exchange process and the nicotinamide inhibition of deacetylation have determined them to be intimately connected [14, 15]. For example, independent laboratories have shown that nicotinamide Ki and base exchange Km values are similar [14, 15]. Inhibition depletes the imidate intermediate (Scheme 3) and leads to return of the imidate to the Michaelis complex, which explains the correspondence of the two kinetic parameters, since they both measure this effect using inhibition and base exchange respectively as readouts. This type of inhibition mechanism is expected to produce non-competitive inhibition [64]. Sirtuins are non-competitively inhibited [26, 63], and Ki values are in the 200 µM range or below for a number of sirtuins. Influential work by the Sinclair laboratory has shown that nicotinamide is a potent disruptor of yeast silencing and can prevent lifespan extension as measured by replication of mother yeast cells [62, 63]. There is sufficient biological and biochemical data to suggest that nicotinamide is an endogenous negative regulator of sirtuin activity in yeast, flies and even in mammalian cells.
Several structural studies have been performed looking at how nicotinamide reacts on sirtuin enzymes, and they are reviewed elsewhere in this issue [65]. We look now at the biochemical and kinetic studies that have been performed to look more in depth at how inhibition of sirtuins results from the reactivity of nicotinamide with the imidate complex (Scheme 3). A condition of the ability of sirtuins to sense nicotinamide concentrations within a cell is that nicotinamide has to have the opportunity to dissociate and reassociate prior to the intermediate on the enzyme progressing forward (Scheme 3). This condition is required for base-exchange to be observed and base exchange has been observed for a variety of sirtuin enzymes [7, 14, 15, 26]. This intermediate stability speaks to the existence of an alpha-ADPR covalent complex, capable of reversal, and is consistent with the proposed imidate [9, 14, 15]. Interestingly, other stabilized ADPR intermediates have been identified on enzymes like CD38, where nicotinamide also causes base exchange and can inhibit enzyme activity, such as NAD+ glycohydrolase activity [64]. The CD38 intermediate is stabilized by an active site Glu residue [66] which has been claimed by different groups to stabilize covalent [66] or non-covalent [51] forms of ADPR on the enzyme. Interestingly covalent intermediates have been formed on CD38, using ara-F-NMN[66] and 2'-deoxy-nicotinamide riboside [67]. Interestingly, these two compounds, which covalently inhibit CD38 by ribosylating the catalytic residue, are less reactive and more reactive respectively, compared with their 2'-OH substituted counterparts [68]. The enzyme-bound ribosyl intermediates formed from these compounds are able to support base-exchange chemistries occurring on CD38 [66, 67]. The covalent complex formed from ara-F-NMN has been identified by MS [66] and has been solved by X-ray crystallography [51]. In spite of the demonstration of covalent intermediates that can support chemical mechanisms observed with NAD+ on CD38, the identity of the ADP-ribosyl intermediate on these enzymes remains undetermined and controversial. Although a report of a crystallized non-covalent ribosyl cation on CD38 has appeared [51], the thorough demonstration that this species is a cation is not available. Sauve et al. reported KIE data for reactions catalyzed by CD38 suggesting the equilibrium isotope effect (versus NAD+) for an intermediate with a 2'-OH group is not consistent with a cation [66].
Studies indicate that in general, the nicotinamide bond cleavage step is usually faster than the downstream chemistry of the imidate intermediate (Figure 1A) [14, 15, 32]. Indeed not only is the nicotinamide bond cleavage step not rate limiting, but in a variety of cases it has been shown that the nicotinamide base-exchange rate proceeds faster than the deacetylation reaction rate [14, 15]. Both the Schramm and Denu laboratories have agreed that the efficiency of nicotinamide inhibition typically depends on the ability of nicotinamide to react faster (k–1: Figure 1) than the attack of the 2 hydroxyl on the imidate (k2 Figure 1) [14, 15] and it has been argued that k2 can often be rate limiting for the enzyme [15], an idea that appears not to be universal since HST2 is limited by product release [29] but this situation has been demonstrated for the Plasmodium falciparum Sir2 enzyme [10]. If k2 is usually limiting or close to limiting for deacetylation rate, then by rule of thumb, nicotinamide inhibition is typically efficient when nicotinamide base exchange rate greatly exceeds the deacetylation rate. Exceptions to this pattern have proven interesting for testing the model. For example, on the enzyme Af2Sir2 the base exchange process proceeded slower than the deacetylation rate [15]. Inhibition of deacetylation on this enzyme caused by nicotinamide does not exceed 50%, even when nicotinamide saturates base exchange [15]. This type of inhibition is called hyperbolic or non-linear inhibition. With reference to reaction coordinate Figure 1B, this is best explained by having a slower rate of return for the imidate than for forward chemistry (k−1 < k2), given that both base exchange and deacetylation share the imidate forming step k1. This causes nicotinamide to be slower in reacting with the intermediate, relative to forward chemistry and results in hyperbolic inhibition. In this case, the inhibition can be viewed as a purely kinetic phenomenon determined by the partitioning of the imidate between the two reaction pathways (Figure 1B).
Figure 1.
Depictions of fate of imidate complex partitioned between base exchange and deacetylation pathway. The formation of imidate is governed by the rate constant k1, its reversal to the Michaelis complex is k−1 and k2 is the rate of attack of the 2'-hydroxyl of the imidate. The two reaction coordinates are for enzymes that are sensitive to nicotinamide inhibition of deacetylation, and the bottom is proposed for the enzyme Af2Sir2, which is relatively insensitive to nicotinamide inhibition of deacetylation. The significance of these reaction coordinates is discussed in the text.
We have suggested that for most sirtuins, in which nicotinamide functions as a potent inhibitor, that both barriers of the nicotinamide formation and reversal are lower than the hill that controls the rate of the deacetylation reaction downstream of the intermediate (Figure 1A [15]). We have contended that under these conditions, the intermediate must also be destabilized with respect to the Michaelis complex (Figure 1A)[10, 15]. Since generally sirtuins have very fast rate constants k1 and k−1 barriers relative to k2, pseudoequilibration of the imidate and Michaelis complex would be predicted to occur when base-exchange is at maximum rate. If the energies of imidate and Michaelis complex were similar, the imidate would only be depleted 50% at maximal exchange and less so if the imidate was more stable, causing hyperbolic inhibition. What phenomenon can account for the imidate being relatively unstable to NAD+? This is not completely understood, but one likelihood is that the imidate remains protonated during catalysis, and since an amide leaving group has negligible basicity even when compared to nicotinamide, the amide remains the superior leaving group and correspondingly the imidate is thermodynamically less stable. Interestingly, base exchange rates for sirtuins do not vary much across extended pH ranges (Sauve unpublished data) suggesting that the imidate pKa on the sirtuin enzyme is well above 8, since deprotonation of it would likely stabilize the intermediate to base exchange. Interestingly, protonation of the imidate also activates this imidate for attack by the 2'-OH on the imidate. Imidates are normally easily deprotonated by amines, such as triethylamine, suggesting that the active site kinetically and/or thermodynamically stabilizes this proton to deprotonation even at pHs as high as 9. In fact the Wolberger laboratory has identified a residue which is likely important for interacting with the imidate proton, a Val 160 in the TMSir2 structure in which a backbone carbonyl is within hydrogen-bonding distance to the N of the imidate in the structurally determined thioimidate [12].
In the interesting case of Af2Sir2, where nicotinamide is a poor inhibitor of this enzyme and fails to inhibit deacetylation fully we have hypothesized that the intermediate is stabilized relative to the Michaelis complex in this case (See discussion above and Figure 1B). We do not have evidence that this stabilization is due to deprotonation of the imidate, since deacetylation remains active. We suppose instead that the intermediate is likely stabilized with respect to the Michaelis complex by some other means, which causes nicotinamide attack to be poorly competitive with 2'-OH attack on the imidate (Figure 1B). What accounts for this stabilization is also not well understood. One possibility has been suggested by structural work done by the Wolberger laboratory, who showed that a Phe residue interacts with the β-face of the ribose. This residue is highly conserved in sirtuins and can regulate the imidate reaction with nicotinamide [12]. Mutation of this residue was found to profoundly accelerate the base-exchange rate of the TmSir2 enzyme, suggesting that this residue can shield the intermediate kinetically, and consequently produce stabilization [12]. It is also possible that the Phe forms a pi-cation interaction with the imidate which also contributes to stabilization, given that the sugar is calculated to retain approximately 0.5 positive charge in the imidate complex [52]. Evidence that this Phe residue is mobile and adopts different geometries in different types of sirtuin complexes solved crystallographically fully supports the idea that this residue can modulate stability of imidate complexes [12]. It has also been suggested that this Phe residue can prevent indiscriminate reactivity of the imidate with solvent, although increased hydrolysis relative to deacetylation in a Phe mutant has not been demonstrated [12].
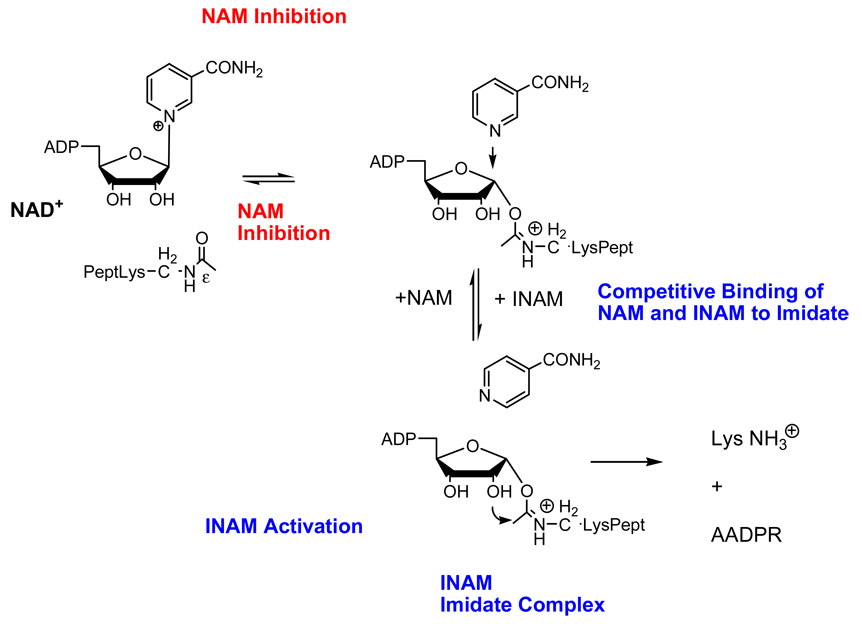
NICOTINAMIDE DEREPRESSION
The realization that nicotinamide regulates sirtuins in vivo, and that it negatively impacts sirtuin deacetylation activity through the base-exchange reaction led to the idea that competitive inhibition with nicotinamide in the base-exchange reaction could lead to relief or derepression of sirtuin inhibition [1, 15, 28, 42]. A requirement for this would be a small molecule that would only bind to the imidate complex and only compete for the nicotinamide binding pocket and would not itself react with the imidate, or otherwise inhibit deacetylation. The author identified a small molecule that had the required property, in the form of a nicotinamide isostere, called isonicotinamide [28]. Isonicotinamide does not inhibit deacetylation, it does not compete with NAD+ or acetyllysine binding to yeast Sir2, and it does not react with the imidate, but it can competitively inhibit nicotinamide binding to this sirtuin (Scheme 6). As such it inhibits the base exchange reaction selectively and thereby activates sirtuin activity that is inhibited by nicotinamide (Scheme 6) [28]. The authors showed that isonicotinamide increases the apparent Km value for nicotinamide base exchange, and also increases apparent Ki values of nicotinamide inhibition of yeast Sir2 [28]. This causes nicotinamide to be weakened as an inhibitor, a phenomenon we have called nicotinamide derepression [42]. The compound isonicotinamide is a weak binder to yeast Sir2 and only derepresses nicotinamide inhibition at millimolar concentrations. However, this compound is inexpensive, is relatively non-toxic to yeast and to mammalian cells and can be dissolved in nearly molar concentrations in water. It also readily penetrates cells and is very stable.
Scheme 6.
Mechanism of activation of sirtuin deacetylation reaction by isonicotinamide (INAM). INAM is able to compete with nicotinamide (NAM) to occupy position above b-face of imidate which pushes deacetylation reaction forward, and avoids NAM induced chemical reversal of the imidate, which is inhibitory to the deacetylation reaction.
In a biological test designed to test if nicotinamide is a relevant negative regulator in yeast cells, yeast were treated with isonicotinamide to determine if this compound could cause Sir2 activation. These yeast experienced significant enhancements in gene silencing and could even correct for deletion of PNC1, which confers a silencing defect because of highly increased endogenous nicotinamide, measured to be 10 fold over wildtype [42]. There is no published data on the effects of isonicotinamide on human sirtuins, although isonicotinamide acts on mammalian cells in a manner consistent with activation of SIRT1, and can duplicate some of the biological effects of resveratrol, albeit at much higher concentrations [69–72].
POSSIBLE MECHANISMS OF ADPRIBOSYL TRANSFER
In the final section we consider a set of observations that have been made for sirtuin enzymes, that they are able to catalyze stable ADP-ribosylation of proteins. Interestingly an ADP-ribosyltransferase activity of yeast Sir2 was determined prior to the discovery that NAD+ could also stimulate histone deacetylation [16, 17]. Numerous reports have appeared since that time reporting sirtuin dependent ADP-ribosylation, although there is little agreement on its possible mechanism [10, 18–22, 73, 74] nor are there definitive characterizations of the reaction products themselves. For example, the identities of residues that have been subject to this type of ADP-ribosylation have not been determined, thus complicating the assessments of the nature of this activity. Rather, most of the data obtained has been by observation of radiolabeled proteins when sirtuins and proteins are co-incubated with 32P NAD.
We will not provide an extensive review of the reported claims of ADP-ribosylation of sirtuins, but merely focus on several examples: SIRT6, Trypanosoma. brucei Sir2, SIRT4 and Plasmodium falciparum Sir2. We will review each of these examples, taking a critical view of what has been learned and what remains undetermined about protein ADP-ribosylation by sirtuins.
SIRT6
The Guarente laboratory authored a very interesting report on Sirt6, which was reported to auto ADPribosylate itself [21], and interestingly enough in this same report was presented evidence that SIRT1 could catalyze ADP-ribosylation of histones [21]. Later workers have shown that SIRT6 also has an NAD+ dependent histone deacetylase activity [75–78]. ADP-ribosylation activities characterized by Liszt et al. were deduced by protein labeling with 32P NAD+, wherein it could be shown that label was transferred covalently to the respective proteins [21]. Skepticism about the ADP-ribosylation activity attributed to sirtuins has focused on the likelihood of ADP-ribosylation being a consequence of non-enzymatic reaction of ADP-ribose, which had been shown to react with lysine histones at pH 7.5 and increasingly at higher pH values [79, 80]. This was in fact recently, re-demonstrated for histones by the Denu laboratory [18].
The observed SIRT6 auto-ADP-ribosylation was very weak, and did not exceed 20 % of the available SIRT6 in solution after single hour incubations of protein and [32P]NAD+[21]. Interestingly the activity is stimulated by the presence of core histones [21]. The auto-ADPribosylation required enzymatic activity, as demonstrated using catalytically inactive mutants, and of some note the reaction appeared to be intramolecular [21]. This was determined by co-incubation of catalytically active and inactive SIRT6 together, in which it was demonstrated that the label could not be transferred to the catalytically inactive SIRT6. This argues that the active enzyme does not transfer the label to another protein, but rather ADP-ribosylates itself with NAD+[21]. The amino acid residue that is so modified is unknown. The effect of this ADPribosylation on catalytic function of SIRT6 has not been determined, although the ubiquitous presence of NAD+ in cells and the ability of SIRT6 to bind NAD+ in the absence of other substrates suggests the possibility that this modification could occur in vivo, and could be of regulatory significance to SIRT6 itself. This would be especially true if the ADP-ribosylation occurred at or near the active site of the enzyme, which would be likely for an intramolecular mechanism.
Trypanosoma brucei Sir2
This enzyme was cloned from a trypanosome that causes sleeping sickness in humans. This Sir2 is a telomere associated protein that was initially reported to have both deacetylase and ADP-ribosylation activities [22]. Reactions containing 33P NAD+ were used to show sirtuin-catalyzed ADP-ribosylation of BSA and histones at high pH = 8.8 [22]. Both deacetylase and ADP-ribosylation activities were confirmed independently by another laboratory [18]. The Denu laboratory investigation of this enzyme focused on the relative strengths of the two activities, and determined that the deacetylase activity was a much more robust activity than the ADP-ribosylation activity [18]. Addition of acetylated peptide was able to stimulate the observed ADP-ribosylation [18]. However, a priori if the mechanism of ADP-ribosylation is catalytic generation of AADPR or ADPR, followed by a non-enzymatic pathway of protein ADP-ribosylation from these compounds, then the stimulation by acetylpeptide is predicted. The question then is what is the likelihood that the enzyme itself directly mediates some or all of the observed ADP-ribosylation. It was concluded that part of the observed ADP-ribosylation was likely to be non-enzymatic, and that a major part was due to reaction of lysine residues that can be ADP-ribosylated by an imidate-dependent ADPribosylation mechanism, wherein the imidate transfers ADPR with β-stereochemistry in the final product [18]. Most importantly, the overall ADP-ribosylation activity was found to proceed 5 orders of magnitude slower than the deacetylase reaction [18]. With such a meager activity in evidence Kowieski et al advocated skepticism [18] about the potential biological relevance of reported protein ADP-ribosyltransfers of sirtuins, a view also advocated by another laboratory [73]. Nevertheless, the caveat still applies that the actual ADPribosylation target was not identified, and that a still unidentified substrate could provide a higher catalytic rate. Also possible is that the modification is supposed to be slow, and performs a currently unidentified regulatory function.
SIRT4
The most potentially interesting of the ADP-ribosylations identified to date is attributed to the enzyme SIRT4. SIRT4 ADP-ribosylation is implicated in an important biological process, that of regulating the activity of glutamate dehydrogenase (GDH)[20]. Glutamate dehydrogenase was known to be ADP-ribosylated in vivo, although the actual responsible ADPribosyltransferase was unidentified [81]. ADPribosylation was found to inhibit this enzyme which interconverts glutamate and α-keto-glutarate. The latter metabolite is a part of the TCA cycle. Increased activity of GDH can increase mitochondrial activity by increasing the dominant upstream feeder of oxidative phosphorylation, the TCA cycle. Thus, ADPribosylation of GDH is predicted to downregulate ATP synthesis in mitochondria, and to weaken ATP production from fuel sources such as glutamine and glutamate.
SIRT4 was found to interact directly with GDH in cells using immunoprecipitation, and it was also demonstrated that SIRT4 and GDH colocalize in tissues in mice and within cells, to mitochondria [20]. SIRT4 ADPribosylation of proteins was demonstrated by 32P blots and this ADP-ribosylation was found to inhibit the activity of the enzyme [20] consistent with the effects of ADP-ribosylation identified previously [81]. The specific residue on GDH that is ADP-ribosylated has not been determined, and the catalytic mechanism for SIRT4 ADPribosylation also remains undetermined. However, SIRT4 knockout animals have increases in amino acid-induced insulin release, consistent with the concept that GDH activity is upregulated, which is predicted to increase ATP synthesis in the pancreatic β-cell [20]. ATP synthesis is an important metabolic process in nutrient sensing by these cells, and is directly coupled to the insulin releasing pathway.
SIRT4 has proven difficult to study in vitro, and a deacetylation activity for this human sirtuin isoform is thus far unreported. This suggests that this sirtuin could indeed be an ADP-ribosyltransferase. It is of interest to consider that in the PARP family, many of the PARP isoforms are in fact not polymerase enzymes but are protein mono-ADPribosytransferases, providing an example where common scaffolds that catalyze ADP-ribosyltransfer can achieve divergent chemical functions. One laboratory concluded that it is unlikely that SIRT4 has an ADP-ribosyltransferase activity after finding no activity. However, with use of modified NAD+s, and no activity for SIRT4 determined, it is difficult to determine if the SIRT4 used in these studies was catalytically functional or compatible with the modified substrates [73].
Plasmodium falciparum Sir2
The parasite Plasmodium falciparum is the pathogen most responsible for human malarial infections. It encodes two Sir2s implicated in regulating silencing in a large set of functionally redundant genes called var that encode isoforms of a virulence factor [82–84]. This virulence factor is expressed on the surface of infected erythrocytes. Persistence of malaria in the host can occur by a mechanism wherein var gene expression is switched so that the epitopic identity of the var product is changed, allowing the malarial parasite to avoid the host immunity. Thus, while one gene is typically expressed, all other var genes are kept silent. Deletion of Sir2 leads to dysregulated silencing for a large set of var genes [82–84]. It has been suggested that the activity most responsible for doing this is deacetylation of core histones within the parasite nucleus at sites of chromatin that encode these genes [82–84]. In fact independent laboratories have confirmed histone deacetylase activity for this enzyme [10, 19].
In addition to deacetylase activity, the enzyme appears to be able to ADP-ribosylate histones as shown by labeling of histones when incubated with 32P NAD and recombinant pfSir2 at pH 8 [19]. This activity was found to be mostly inhibited by 10 mM nicotinamide but not completely [19], implying that the bulk of the ADP-ribosylation mechanism requires formation of the imidate, but another imidate independent mechanism could also be contributing. Given the known ability of histones to be ADPribosylated by ADPR, or AADPR [18, 85] it is quite possible that the ADPribosylation process observed is non-enzyme mediated ADPribosyltransfer of ADPR and AADPR, as we have previously discussed.
Interestingly, PfSir2 has an acetyllysine peptide dependent and an acetyllysine peptide independent NAD+ glycohydrolase activity, both of which exceed the deacetylase activity of the enzyme [10]. These activities could be important to explaining the ADPribosylation of histones observed for this enzyme [19]. Our work found that the acetyllysine dependent hydrolysis activity was much greater than the acetyllysine independent hydrolysis activity [10]. The acetyllysine dependent activity proceeds through hydrolysis of the imidate, which only slowly proceeds forward in the deacetylation reaction [10]. The imidate hydrolysis could be inhibited by nicotinamide, which leaves a residual hydrolytic activity uninhibited which proceeds through the peptide independent route [10]. A scheme that accounts for all of these activities is shown in Scheme 7. The high production of ADPR from the imidate by this enzyme could explain the observed histone ADP-ribosylation and the observed majority but incomplete sensitivity to nicotinamide [19]. However, the alternative still exists that some of the observed ADPribosylation is directly enzyme catalyzed [10, 19]. This is a topic of current investigation in our laboratory.
Scheme 7.
General mechanistic scheme that accounts for ADPribosyltransfers with inversion, retention and deacetylation as observed for Plasmodium falciparum Sir2 enzyme. In addition, nicotinamide reaction from imidate causes inhibitions of imidatesolvolysis and deacetylation. Nicotinamide does not inhibit the direct ADP-ribosyltransfer reaction shown by top arrow.
SUMMARY
We have reviewed here the varieties of chemistries that sirtuins catalyse and possible mechanisms by which these reactions proceed. Investigations into this class of enzymes continues to expand as new enzymatic activities are discovered and new biology develops. Important questions remain unresolved. This includes the mechanisms of sirtuin activation, especially by small molecules such as resveratrol. The mechanisms of ADPribosyltransfer are still in debate. In addition, mechanistic studies to further clarify the inner workings of these complex enzymes are ongoing in several laboratories to resolve questions about transition states and what regulates the chemical reactivities of imidate complexes. Importantly, elucidating the biochemical mechanisms that link these enzymes to NAD+ metabolism is highly relevant to understanding how these enzymes function as transducers of metabolic information in cells. Undoubtedly, this research will impact ongoing medicinal chemistry efforts and drug development that target this very interesting enzyme class.
ACKNOWLEDGEMENTS
The author acknowledges support from NIH grant R01 DK 074366, NY State Spinal Cord Injury Board C023832 and Ellison Medical Foundation New Scholar Award in Aging (2007–2011). AAS is a Scientific Advisory Board Member for Sirtris Pharmaceuticals (A GlaxoSmithKline Company) and has financial interests related to sirtuins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 2.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 4.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 5.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 6.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 8.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 9.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 10.French JB, Cen Y, Sauve AA. Plasmodium falciparum Sir2 is an NAD+- dependent deacetylase and an acetyllysine-dependent and acetyllysineindependent NAD+ glycohydrolase. Biochemistry. 2008;47:10227–10239. doi: 10.1021/bi800767t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith BC, Denu JM. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J Biol Chem. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 12.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatkins DG, Monnot AD, Zheng W. Nepsilon-thioacetyl-lysine: a multi-facet functional probe for enzymatic protein lysine Nepsilon-deacetylation. Bioorg Med Chem Lett. 2006;16:3651–3656. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- 14.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 15.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 16.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 17.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 18.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 19.Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 21.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. Embo J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb Symp Quant Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 24.Jackson MD, Denu JM. Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 25.Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 26.Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 27.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 29.Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 32.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 33.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin SJ, Bench G. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J Sep Sci. 2008;31:3202–3211. doi: 10.1002/jssc.200800238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 37.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 38.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;352:282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 41.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 42.Sauve AA. Pharmaceutical strategies for activating sirtuins. Curr Pharm Des. 2009;15:45–56. doi: 10.2174/138161209787185797. [DOI] [PubMed] [Google Scholar]
- 43.Sauve AA, Schramm VL. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr Med Chem. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- 44.Borch RF. A New Method for Reduction of Secondary and Tertiary Amides. Tetrahedron Letters. 1968:61–64. [Google Scholar]
- 45.Scheuring J, Berti PJ, Schramm VL. Transition-state structure for the ADP-ribosylation of recombinant Gialpha1 subunits by pertussis toxin. Biochemistry. 1998;37:2748–2758. doi: 10.1021/bi972594x. [DOI] [PubMed] [Google Scholar]
- 46.Berti PJ, Blanke SR, Schramm VL. Transition State Structure for the Hydrolysis of NAD Catalyzed by Diphtheria Toxin. J Am Chem Soc. 1997;119:12079–12088. doi: 10.1021/ja971317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheuring J, Schramm VL. Pertussis toxin: transition state analysis for ADP-ribosylation of G-protein peptide alphai3C20. Biochemistry. 1997;36:8215–8223. doi: 10.1021/bi970379a. [DOI] [PubMed] [Google Scholar]
- 48.Scheuring J, Schramm VL. Kinetic isotope effect characterization of the transition state for oxidized nicotinamide adenine dinucleotide hydrolysis by pertussis toxin. Biochemistry. 1997;36:4526–4534. doi: 10.1021/bi962841h. [DOI] [PubMed] [Google Scholar]
- 49.Smith BC, Denu JM. Sir2 deacetylases exhibit nucleophilic participation of acetyl-lysine in NAD+ cleavage. J Am Chem Soc. 2007;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao K, Harshaw R, Chai X, Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc Natl Acad Sci U S A. 2004;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Q, Kriksunov IA, Jiang H, Graeff R, Lin H, Lee HC, Hao Q. Covalent and noncovalent intermediates of an NAD utilizing enzyme, human CD38. Chem Biol. 2008;15:1068–1078. doi: 10.1016/j.chembiol.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu P, Wang S, Zhang Y. Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamics simulations. J Am Chem Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson RW, Marschner TM, Oppenheimer NJ. Pyridine-Nucleotide Chemistry - a New Mechanism for the Hydroxide-Catalyzed Hydrolysis of the Nicotinamide Glycosyl Bond. Journal of the American Chemical Society. 1988;110:2257–2263. [Google Scholar]
- 54.Berti PJ, Schramm VL. Transition state structure of the solvolytic hydrolysis of NAD(+) Journal of the American Chemical Society. 1997;119:12069–12078. doi: 10.1021/ja971317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 56.Schramm VL. Enzymatic transition state theory and transition state analogue design. J Biol Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 57.Evans GB, Furneaux RH, Kelly PM, Schramm VL, Tyler PC. Transition state analogue inhibitors of N-ribosyltransferases: new drugs by targeting nucleoside processing enzymes. Nucleic Acids Symp Ser (Oxf) 2007:63–64. doi: 10.1093/nass/nrm032. [DOI] [PubMed] [Google Scholar]
- 58.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5'-methylthioadenosine/Sadenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt RR, Kinzy W. Anomeric-Oxygen Activation for Glycoside Synthesis - the Trichloroacetimidate Method. Advances in Carbohydrate Chemistry and Biochemistry. 1994;Vol 50:21–123. doi: 10.1016/s0065-2318(08)60150-x. 50. [DOI] [PubMed] [Google Scholar]
- 60.Zhu XM, Schmidt RR. New Principles for Glycoside-Bond Formation. Angewandte Chemie-International Edition. 2009;48:1900–1934. doi: 10.1002/anie.200802036. [DOI] [PubMed] [Google Scholar]
- 61.Smith BC, Denu JM. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry. 2007;46:14478–14486. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 62.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 64.Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37:13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- 65.Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: What we know and what we don't. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.09.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sauve AA, Deng HT, Angeletti RH, Schramm VL. A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. Journal of the American Chemical Society. 2000;122:7855–7859. [Google Scholar]
- 67.Sauve AA, Schramm VL. Mechanism-based inhibitors of CD38: a mammalian cyclic ADP-ribose synthetase. Biochemistry. 2002;41:8455–8463. doi: 10.1021/bi0258795. [DOI] [PubMed] [Google Scholar]
- 68.Handlon AL, Xu C, Mullersteffner HM, Schuber F, Oppenheimer NJ. 2'-Ribose Substituent Effects on the Chemical and Enzymatic-Hydrolysis of Nad(+) Journal of the American Chemical Society. 1994;116:12087–12088. [Google Scholar]
- 69.Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, Wu SY, Samols D, Hakimi P, Chiang CM, Hanson RW. Activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating hepatic nuclear factor 4alpha. J Biol Chem. 2009;284:27042–27053. doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Backesjo CM, Haldosen LA, Lindgren U. Resveratrol inhibits proliferation and promotes apoptosis of osteosarcoma cells. Eur J Pharmacol. 2009;609:13–18. doi: 10.1016/j.ejphar.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. Cells Tissues Organs. 2009;189:93–97. doi: 10.1159/000151744. [DOI] [PubMed] [Google Scholar]
- 72.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 73.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogs and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 74.Hawse WF, Wolberger C. Structure-based mechanism of ADP ribosylation by sirtuins. J Biol Chem. 2009;284:33654–33661. doi: 10.1074/jbc.M109.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacobson EL, Cervantes-Laurean D, Jacobson MK. ADP-ribose in glycation and glycoxidation reactions. Adv Exp Med Biol. 1997;419:371–379. doi: 10.1007/978-1-4419-8632-0_49. [DOI] [PubMed] [Google Scholar]
- 80.Cervantes-Laurean D, Minter DE, Jacobson EL, Jacobson MK. Protein glycation by ADP-ribose: studies of model conjugates. Biochemistry. 1993;32:1528–1534. doi: 10.1021/bi00057a017. [DOI] [PubMed] [Google Scholar]
- 81.Herrero-Yraola A, Bakhit SM, Franke P, Weise C, Schweiger M, Jorcke D, Ziegler M. Regulation of glutamate dehydrogenase by reversible ADPribosylation in mitochondria. Embo J. 2001;20:2404–2412. doi: 10.1093/emboj/20.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 83.Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LM, Scherf A, Ivens A, Speed TP, Beeson JG, Cowman AF. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 85.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J Biol Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]