Abstract
Tumor cells have a high tolerance for acidic and hypoxic microenvironments, also producing abundant lactic acid through accelerated glycolysis in the presence or absence of O2. While the accumulation of lactate is thought to be a major contributor to the reduction of pH-circumscribing aggressive tumors, it is not known if other endogenous metabolic products contribute this acidity. Furthermore, anaerobic metabolism in cancer cells bears similarity to homo-fermentative lactic acid bacteria, however very little is known about an alternative pathway that may drive adenosine triphosphate (ATP) production independent of glycolysis. In this study, we quantify over 40 end-products (amines, acids, alcohols, aldehydes, or ketones) produced by malignant neuroblastoma under accelerated glycolysis (+glucose (GLU) supply 1–10 mM) ± mitochondrial toxin; 1-methyl-4-phenyl-pyridinium (MPP+) to abate aerobic respiration to delineate differences between anaerobic vs. aerobic cell required metabolic pathways. The data show that an acceleration of anaerobic glycolysis prompts an expected reduction in extracellular pH (pHex) from neutral to 6.7±0.006. Diverse metabolic acids associated with this drop in acidity were quantified by ionic exchange liquid chromatography (LC), showing concomitant rise in lactate (Ctrls 7.5±0.5 mM; +GLU 12.35±1.3 mM; +GLU + MPP 18.1±1.8 mM), acetate (Ctrl 0.84±0.13 mM: +GLU 1.3±0.15 mM; +GLU + MPP 2.7±0.4 mM), fumarate, and a-ketoglutarate (<10μM) while a range of other metabolic organic acids remained undetected. Amino acids quantified by o-phthalaldehyde precolumn derivatization/electrochemical detection–LC show accumulation of L-alanine (1.6±.052 mM), L-glutamate (285±9.7μM), L-asparagine (202±2.1μM), and L-aspartate (84.2±4.9μM) produced during routine metabolism, while other amino acids remain undetected. In contrast, the data show no evidence for accumulation of acetaldehyde, aldehydes, or ketones (Purpald/2,4-dinitrophenylhydrazine—Brady’s reagent), acetoin (Voges–Proskauer test), or alcohols (NAD+-linked alcohol dehydrogenase). In conclusion, these results provide preliminary evidence to suggest the existence of an active pyruvate–alanine transaminase or phosphotransacetylase/acetyl-CoA synthetase pathway to be involved with anaerobic energy metabolism of cancer cells.
Keywords: Cancer, Anaerobic, Glycolysis, Acetate, Alanine
Introduction
In 1930, Otto Warburg described the etiology of cancer to involve an inherent abnormality of glucose metabolism, one that is divergent from the normal oxidative metabolic processes of eukaryotic cells. Subsequently, the classical “Warburg effect” describes a high glycolytic activity that occurs in cancer cells, even in the presence of oxygen (O2), accounting for disproportionately high lactic acid production by tumor tissue. Since then, studies continue to report that tumor cells display superior anaerobic capability, where faulty mitochondrial respiration parallels acceleration of glycolysis, overexpression of hypoxia-responsive glucose transporters (GLUT1 and GLUT3), excessive production of lactic acid, and a drop in extracellular pH (Koukourakis et al. 2007; De Milito and Fais 2005; Airley and Mobasheri 2007). It is now understood that these processes occur in response to the lack of O2 at the core of a growing tumor, where a rise in inducible cytosolic hypoxia inducible factor (HIF)-1α hetero-dimerizes with HIF-1β translocating to the nucleus where it activates gene expression of glycolytic enzymes (i.e., pyruvate kinase M, hexokinase-2, lactic acid dehydrogenase (LDH)-5), proteins involved with angiogenesis, tumor proliferation, and metastasis (i.e., vascular endothelial growth factor, matrix metalloproteinase 2, cathepsin D, and keratin; Ziello et al. 2007; Kim et al. 2007; Koukourakis et al. 2006) and downregulates genes that modify expression of proteins involved with the mitochondrial tricarboxylic acid cycle (TCA) i.e., pyruvate dehydrogenase (PDH)-1. All of these events assist with survival under low-O2 environment by promoting anaerobic metabolism.
Anaerobic metabolism in tumor tissue renders excessive lactate which leads to reduction of pH in the tumor microenvironment which amplifies the fermentative processes by potentiating mitochondrial pyruvate dehydrogenase inhibition (Bongaerts et al. 2006) and preventing von Hippel–Lindau tumor suppressor protein from degrading HIF-1α (Mekhail et al. 2004). Aggressive malignancies are often marked by heightened concentrations of lactic acid, LDH protein expression (Walenta et al. 2004), acidity, hypoxia (De Milito and Fais 2005; Herrmann and Herrmann 2007), and a growing resistance to acidity in part due to HIF-1-mediated expression of carbonic anhydrase (Pastorekova et al. 2008). These adaptive changes in response to an acidic and hypoxic environment may also be responsible for the greater incidence of anaerobic yeast and bacterial infections occurring in cancer patients (Rafailidis et al. 2008).
With an understanding of the importance of the hypoxic–acidic nature by which tumor cells thrive, novel therapies to fight cancer may involve targeting proteins that impair anaerobic energy metabolism within the tumor. However, there is a need for research which defines clearly how malignant tumor cells produce ATP from glucose under anaerobic conditions and to determine if there are other fermentation pathways involved independent of the Embden–Meyerhof pathway (EMP) pathway. Tumor metabolism bears an overt resemblance to primitive anaerobic microbes which produce energy from carbon substrates under both anaerobic and aerobic conditions, showing preference for slightly acidic pH to optimize survival. The term fermentation often applies to these microbes describing the oxidation (loss of electrons) of an energy substrate (i.e., sugar or amino acids) to an electron acceptor such as NAD+, where oxidation–reduction reactions drive the synthesis of ATP by substrate-level phosphorylation (SLP) in the absence of O2. In these microbes, a mitochondrion is not required to produce energy and as a result a diverse range of metabolic products accumulate including organic acids, alcohols, aldehydes, and gasses as by-products of SLP.
This study aims to gain greater understanding of the anaerobic nature of tumor cell energy metabolism by analyzing the presence of a diverse range of fermentative products and acids routinely produced during glucose metabolism contiguous to the drop in pH, rise in lactic acid, and forced anaerobic conditions by blocking mitochondrial function in malignant tumor cells in vitro.
Methods and materials
Neuro-2A (N-2A) brain neuroblastoma cells were purchased from American Type Culture Collection (Manassas, VA, USA); 2′,7′-bis(2-carboxyethyl)-5 (6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM) was obtained from In Vitrogen (Eugene, OR, USA); Dulbecco’s modified Eagle medium (DMEM), L-glutamine, fetal bovine serum (FBS) heat-inactivated, phosphate buffered saline (PBS), Hank’s balanced salt solution (HBSS), penicillin/streptomycin, MPP+, and all other materials were purchased from Sigma Chemical (St. Louis, MO, USA).
Cell culture
N2-A neuroblastoma cells are commonly used for in vitro studies and display distinct malignant phenotype with parallel adeptness for use in evaluation of chemotherapy drugs to treat cancer (Biedler et al. 1978; Finklestein et al. 1975). N-2A cells were grown in DMEM with phenol red, 10% FBS, 4 mML-glutamine, 20μM sodium pyruvate, and penicillin/streptomycin (100 U per 0.1 mg/ml). The cells were maintained at 37°C in 5% CO2/atmosphere. Every 2–5 days, the medium was replaced and the cells were subcultured. The experimental plating media consisted of DMEM minus phenol red, 1.8% FBS, penicillin/streptomycin (100 U per 0.1 mg/ml), 2 mM sodium pyruvate, and 3 mML-glutamine. For experiments, cells were plated in 96-well plates at a density of approximately 0.5×106 cells per milliliter. A stock solution for each experimental compound was prepared in HBSS+5 mM (N-[2-hydroxyethylpiperazine]-N′-[2-ethanesulfonic acid]) (HEPES) and adjusted to a pH of 7.4.
Cell metabolism
Cell metabolism/viability was assessed using resazurin (AlamarBlue) indicator dye (Mazzio and Soliman 2004). A working solution of resazurin was prepared in sterile PBS minus phenol red (0.5 mg/ml), added (15% v/v) to the samples, and returned to the incubator for 6–8 h. Reduction of the dye by viable cells was quantitatively analyzed using a microplate fluorometer, Model 7620, version 5.02 (Cambridge Technologies Inc, Watertown, Mass) with settings established at [550/580], [excitation/emission].
Glucose quantification
Glucose was assessed using an enzymatic assay containing glucose oxidase (20 U/ml) and a chromogenic solution containing 1 mM vanillic acid, 500μM of 4-aminoantipyrine and 4 U/ml purpurogallin of horse-radish peroxidase—type II (Mazzio and Soliman 2004). Both solutions were prepared in distilled water containing 10 mM HEPES, adjusted to a pH of 5.1. The combined reagent was added to each sample and returned to the incubator for 5 min at 37°C. Glucose was quantified on a UV microplate spectrophotometer, Model 7600 version 5.02, Cambridge Technologies Inc. (Watertown, MA, USA) with wavelength settings at 490 nm.
Determination of H+ concentration
Extracellular pH(ex) was determined using phenol red indicator dye and dual determination using a HANNA handheld portable pH checker (Sigma Chemical, MO, USA). Briefly, a phenol red stock solution (0.3 mg/ml in distilled H20) was added (15% v/v) to the cell supernatants after 24 h. The change in pH was assessed by measuring the drop in OD against a scanning range of 350–650 nm using a Spectra 190 max UV detector (Molecular devices, Sunnydale, CA, USA). Results were corroborated with a handheld portable pH device.
Intracellular cytosolic H+ ion concentration was assessed with BCECF-AM (Ozkan and Mutharasan 2002). A stock solution of BCECF-AM was prepared in absolute ethanol and diluted with HBSS (1:9) to a concentration of 48.8μg/ml. After 24 h, cells were loaded with BCECF-AM (6μM; 13% v/v) for 60 min. The supernatant was discarded; the cells were washed with PBS. Samples were analyzed using an Olympus IX-70 inverted microscope with a mercury burner [490/530] [excitation/emissions]; images were captured by an MD35 Electronic Eyepiece (Zhejiang Jincheng Sci and Tech Co., Ltd., China), and photographic data were obtained with C-imaging systems PCI-Simple software (Compix Inc. Cranberry Township, PA, USA).
Amino acid quantification
Amino acids were detected by reverse-phase high-performance liquid chromatography (HPLC) with precolumn derivatization (Montero et al. 2001). A pilot study was preformed to validate there was no variation in amino acids detected on fresh samples verses those stored at −80°C or lysed with slow thaw to room temperature. A modification to this method was made to remove Tris from the derivatization solution which was not required. After experimentation, samples were stored at −80°C and subsequently lysed by slow thaw at room temperature prior to evaluation. Briefly, a derivation stock solution was prepared consisting of 0.027 g o-phthalaldehyde (OPA) in 1 ml of methanol, 5μl of β-mercaptoethanol (βMe) which was diluted to 10 ml with 0.1 M tetraborate buffer (pH9.3). Prior to derivatization, a 25% (v/v) solution of OPA/βMe stock was prepared in 0.1 M tetraborate buffer, and the samples were derivatized for 3 min prior to injection. Briefly, 6μl of sample was dissolved in 15μl of distilled deionized water to which 15μl of OPE/βMe was added. After derivatization, samples were brought to 700μl with mobile phase. The mobile phase consisted of 191 ml of water—prefiltered through C18 guard column, 50 ml methanol, 8.75 ml of acetonitrile, and 3.58 g of anhydrous sodium phosphate monobasic. The mobile-phase pH was initially equilibrated to 3.5 with phosphoric acid and reestablished at pH6.6 using sodium hydroxide 5 N. Samples were run on an HPLC Waters system (isocratic) using a Waters 510 pump, HR-80 C18 column, 3-μM particle size (4.6×80 mm), and an ESA electrochemical detector columchem II detector with settings at: Guard 650 mV CH1: E 150 mV R: 100μA 2 s CH1 output 1 V, 0% CH2 E 550 mV R 5μA, filter 5 s Output 1 V 0% voltage. The flow rate was 1.4 ml/min and injection volume 20μl.
Organic acid quantification
Cell lysates were prepared by addition of H2SO4 (final concentration 0.01 N), centrifuged at 6,500 rpm for 5 min, and filtered (0.45μm). Briefly, organic acids were quantified using an Interaction Ion-300 anion exchange column on a Waters 2487/Millennium 32 version 3.20 system controller with 2690 Separations Module and UV detector (210 nm; Silva et al. 2002). The mobile phase consisted of 0.01 N H2SO4; the flow rate was set at 0.4 ml/min, and column temperature was sustained at 40°C.
Aldehyde quantification
Acetaldehyde concentration was assessed using the Purpald reagent (Sigma Aldrich Technical Bulletin AL-145). Briefly, 20 mg of Purpald was dissolved in 2 ml of 1 N NAOH. A standard curve for acetaldehyde (2μM–2 mM) was prepared in PBS in 96-well plates, and the method was deemed sensitive and reliable. Quantification of acetaldehyde was assessed by OD at 550 nm using a Spectra Max 190 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). Purpald reagent was found to be reactive with experimental media containing FBS; therefore, 24-h cell culture studies were preformed in PBS supplemented with glucose (0–10 mM)–FBS, both in the presence or absence of MPP+. The data indicate an absence of acetaldehyde accumulation in the media during cellular metabolism at 24 h; however, the method also establishes the absence of other aldehydes which would be detected by this dye including 5-hydroxypentanal, crotonaldehyde, benzaldehyde, 2-nitrobenzaldehyde, cinnamaldehyde, acetamidobenzaldehyde, and 2-chloro-6-nitrobenzaldehyde (Sigma Aldrich Technical Bulletin AL-145).
Aldehyde and ketone quantification
Dual confirmation for quantitative assessment of both aldehydes and ketones was performed using the Brady’s reagent which yields an orange precipitate (dinitrophenylhydrazone) as a positive indicative marker. Briefly, 0.3 g of 2,4-dinitrophenylhydrazine (DNPH) was dissolved in 1.5 ml of concentrated sulfuric acid. The DNPH was added slowly to a solution containing 7 ml of 95% ethanol and 2 ml of distilled water and homogenized. One hundred sixty microliters of glutaraldehyde standard or samples were placed in 2 ml of 95% ethanol solution to which 1.5 ml of Brady’s reagent was added. The samples were vortexed, allowed to settle for 20 min at room temperature, and examined for precipitate.
Acetoin quantification
Quantitative analysis of acetoin was performed using the Voges–Proskauer (VP) test. Briefly, samples and acetoin standards were diluted with 6% Barritt Reagent A: 5% α-naphthol (1-naphthol) dissolved in absolute ethanol and 2% Barritt Reagent B: 40% potassium hydroxide/distilled water (Fluka Analytical-Sigma Chemical, St. Louis, MO, USA). An acetoin standard curve was prepared and along with samples was added to the VP reagent and incubated at room temperature for 25 min. Quantification of acetylmethylcarbinol was assessed by monitoring OD at 550 using a Spectra Max 190 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
Alcohol and β-hydroxybutyrate quantification
Ethanol concentration was determined using alcohol dehydrogenase (EC 1.1.1.1) derived from Saccharo-myces cerevisiae (Sigma, Tech. Bulletin A3263). Briefly, 3.5 mM (final concentration) of β-nicotinamide adenine dinucleotide solution (β-NAD) dissolved in PBS was added to the wells, and a prereading documenting OD at 350nM was acquired. A standard curve containing ethanol was prepared according to the experimental design in solvent of DMEM media—phenol red (LOD was established at three times noise=0.002% ethanol). To each sample, either 0.75 U/ml of ADH (final concentration) in 10 mM solution of sodium phosphate/water (+Enzyme) or an equal volume of 10 mM sodium phosphate without the enzyme (−E) was added. Quantification of NADH was assessed by OD at 350 using a Spectra Max 190 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). Alcohol content of samples was also reconfirmed with a QED® A150 test kit using corresponding standards for butanol, isopropanol, and ethanol (CLIAwaived, San Diego, CA). And β-hydroxybutyric acid was determined using Precision Xtra Blood β-Ketone Strips (Abbot Laboratory, Alameda, CA, USA).
Data analysis
Statistical analysis was performed using GraphPad Prism (version 3.0; GraphPad Software Inc. San Diego, CA, USA) with significance of difference between the groups assessed using a one-way ANOVA, followed by Tukey post hoc means comparison test or Student’s t test.
Results
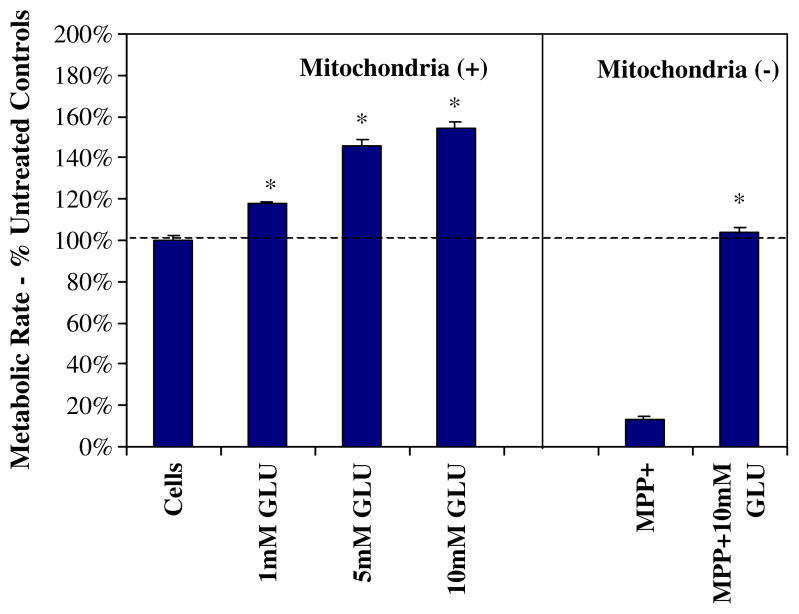
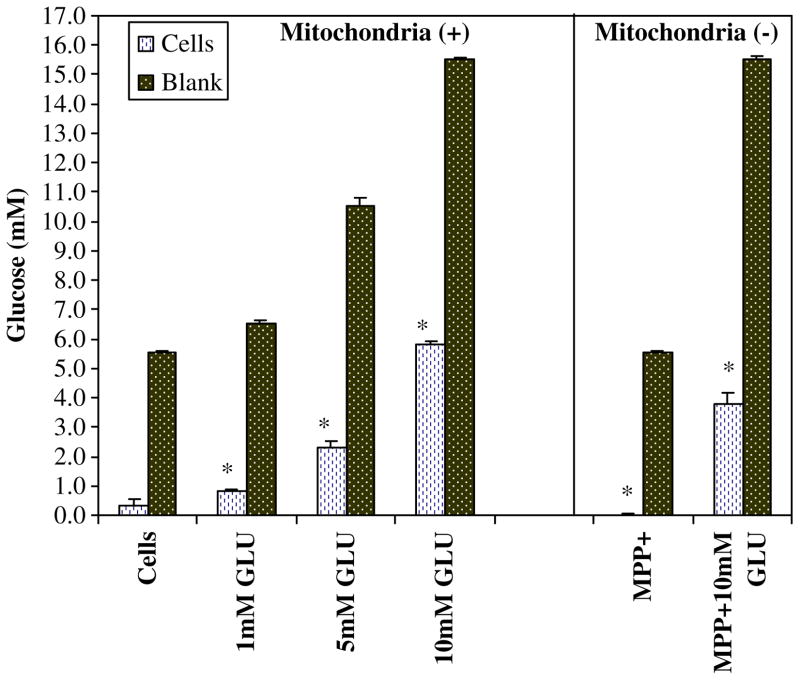
N-2A malignant neuroblastoma clearly display higher metabolic rates with increasing concentrations of glucose (1–10 mM), where high levels of glucose also prevent cell death incurred by the mitochondrial toxin MPP+ (500μM; Fig. 1). The attenuation of cell death appears to be the result of providing enough glucose to sustain energy supplies through driving glycolysis when anaerobic mitochondrial respiration is halted. Previously, we have confirmed that 500μM of MPP+ was adequate to block mitochondrial respiration by inhibiting complex I and IV in these particular cells (Mazzio and Soliman 2004). Figure 2 shows an acceleration of glucose utilization patterns of cells where oxidative phosphorylation (OXPHOS) is compromised with MPP+. In this instance, sustaining glucose concentrations required to maintain ATP produced through anaerobic glycolysis corresponds to the attenuation of cell death as in Fig. 1.
Fig. 1.
Metabolic rate was assessed in N-2A cells after treatment with variation in glucose concentrations in the presence or absence of MPP+ for 24 h at 37°C. The data represent cell metabolism (percent control) and are expressed as the mean±SEM (N=4). Statistical differences from controls were determined by a one-way ANOVA followed by Tukey post hoc test and Student’s t test for the MPP+-treated groups. *P<0.05
Fig. 2.
Variation in glucose utilization patterns by N-2A cells was examined after treatment with glucose (1–10 mM) in the presence or absence of MPP+ for 24 h at 37°C. Glucose concentrations within the experimental plating media were 1,000 mg/L. The data represent millimolar glucose as determined by glucose oxidase peroxidase-linked reaction. The data are expressed as the mean ± SEM (N=4), and statistical differences between glucose were utilized by [cells controls vs. MPP+] and [10 mM GLU vs 10mM GLU + MPP+] were determined by Student’s t test, *P<0.05
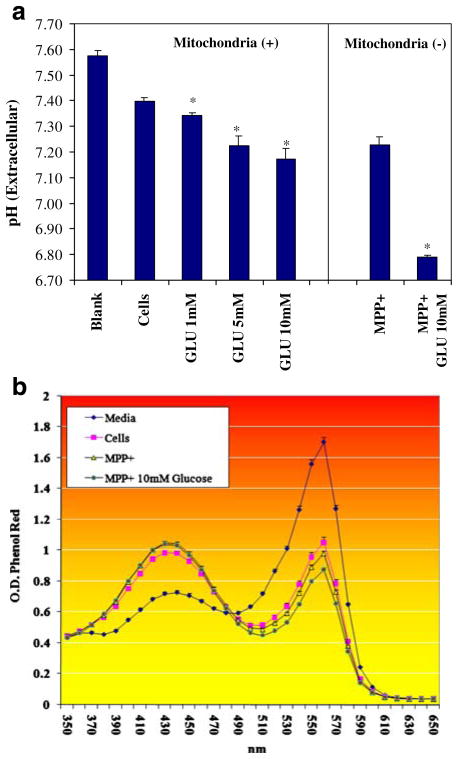
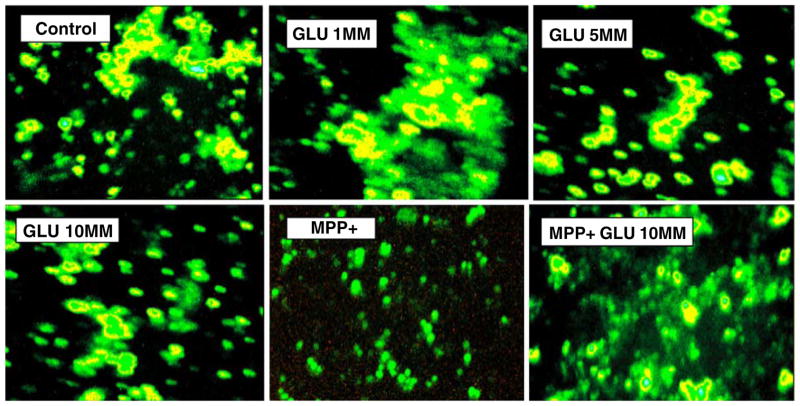
Anaerobic glucose metabolism initiates a drop in extracellular pH(ex) as anticipated through production of lactic acid. Figure 3a (pH(ex)), b (phenol red OD) indicates a shift toward an extracellular acidic environments with higher concentration of glucose, further exacerbated by blocking mitochondrial OXPHOS with MPP+. While a reduction in pH(ex) is evident with anaerobic metabolism, viable cells appear to regulate pH(i) (Fig. 4) within narrow limits regardless of the changes in pH(ex). BCECF-AM is the most commonly used fluorescent probe to measure pH(i) in cells, where it is readily transported through intact plasma membrane and hydrolyzed intracellularly to BCECF—a green fluorescein derivative. The test is usually used to visualize intracellular pH(i) based on a rate of increased fluorescence intensity and subsequent ratiometric monitoring of cellular pH. However, in this application, Fig. 4 demonstrates that malignant neuroblastoma exposed to various GLU concentrations or 10 mM GLU + MPP+ exhibits stability of internal pH(i) (spots of heightened intensity in yellow) relative to controls, where the only evidence for a reduction in pH(i) or lower fluorescence intensity is in cell samples incubated with MPP+ without additional glucose. This was likely due to a drop in ATP as a result of glucose deprivation (Fig. 2) and cell death (Fig. 1) incurred by MPP+, leading to loss of cell membrane potential and infiltration of extracellular acids into the intracellular environment. This pattern of cellular regulation of pH(i) in cancer cells has been previously reported where melanoma cells will continue to maintain pH(i) to suit cell survival, while extracellular tumor pH(ex) drops below 7.0 as a result of heightened expression of monocarboxylate transporters, which likely transport organic acids to the extracellular microenvironment (Wahl et al. 2002).
Fig. 3.
a Extracellular pH(ex) was evaluated in N-2A cells after treatment with variation in glucose concentration in the presence or absence of MPP+ after 24 h at 37°C. The data represent pH and are expressed as the mean ± SEM (N=4). Statistical differences from controls were determined by a one-way ANOVA followed by a Tukey post hoc test and Student’s t test for the MPP+-treated groups. *P<0.05. b Extracellular pH (ex) was evaluated in N-2A cells after treatment with variation in glucose ±MPP+ after 24 h at 37°C. The data represent the OD of phenol red indicator dye over 350–650 nm. The data are expressed as the mean±SEM (N=4)
Fig. 4.
Intracellular pH(i) was assessed in N-2A cells treated with variation in GLU±of MPP+ after 24 h at 37°C. Heightened intensity of BCECF-AM represents a shift toward alkaline pH
Metabolic products corresponding to the drop in pH were investigated (Table 1). The data show that N2A cells utilize glucose (Fig. 2) and pyruvate within the experimental media concurrent to accumulation of lactate, acetate, fumarate, and α-ketoglutarate (Table 1). Coelution of malic acid and pyruvate at the same retention time (RT) prevented determination or analysis to examine malic acid production as a cellular metabolite. Anaerobic conditions set forth by the addition of MPP+ + GLU (10 mM) resulted in greater accumulation of acetate and lactate, with no significant changes in fumarate and a slight rise in α-ketoglutarate. It is possible that other acids are being produced and more so under anaerobic conditions as evidenced by the rise in two unidentified peaks parallel to the rise in lactic acid. Future analysis will be required to elucidate the identity of these metabolic analytes which could shed further light into anaerobic pathways. Acetaldehyde, ketones, acetoin, β-hydroxybutyric acid, and alcohols were also quantified relative to standard curves prepared in the same manner (Table 1). The data indicate no evidence for accumulation of these compounds both under aerobic or anaerobic conditions. However, there were trace levels of ethanol quantified below the LOD (3× STD) in the media after 24-h incubation. The data in general negate various types of fermentation reactions as a process involved with anaerobic energy metabolism in cancer cells.
Table 1.
Quantification of organic acids, amino acids, aldehydes, ketones, or alcohols produced by N-2A cells after treatment with variation in GLU ± MPP+ for 24 h at 37°C
| Blank | Aerobic Cells | Aerobic 10 mM GLU | Anaerobic 10 mM GLU + MPP+ | |
|---|---|---|---|---|
| Organic acids (mM) | ||||
| Oxalic acid | ND | ND | ND | ND |
| B-OH butyric acid | ND | ND | ND | ND |
| cis-Aconitate | ND | ND | ND | ND |
| Oxaloacetate | ND | ND | ND | ND |
| Citric acid | ND | ND | ND | ND |
| Isocitric acid | ND | ND | ND | ND |
| A-ketoglutarate | ND | 0.006±0.001* | 0.008±0.001* | 0.02±0.003* |
| Glyoxylate | ND | ND | ND | ND |
| Isobutyric acid | ND | ND | ND | ND |
| Malate/pyruvate | 2.6±0.31 | 0.33±0.031* | 0.54±0.046* | 0.85±0.093* |
| Malonic acid | ND | ND | ND | ND |
| Succinic acid | ND | ND | ND | ND |
| Glycolic acid | ND | ND | ND | ND |
| Lactic acid | ND | 7.5±0.48* | 12.4±1.25* | 18.1±1.8* |
| Formic acid | ND | ND | ND | ND |
| Acetic acid | ND | 0.83±0.137* | 1.3±0.147* | 2.7±0.390* |
| Fumaric acid | ND | .006±.001* | .006±.001* | .005±.001* |
| Aldehydes, ketones, alcohols, and acetoin | ||||
| Acetaldehyde/aldehydes | ND | ND | ND | ND |
| Ketones | ND | ND | ND | ND |
| Acetoin | ND | ND | ND | ND |
| Butanol, isopropanol | ND | ND | ND | ND |
| Ethanol | ND | <LOD | <LOD | <LOD |
| Amino acids (μM) | ||||
| Alanine | ND | 1,677.62±52.56* | 1,527.29±68.91* | 525.78±45.25* |
| Arginine | 322.45±2.00 | 322.25±3.31 | 260.64±12.49* | 283.37±6.57* |
| Asparagine | ND | 202.15±2.13* | 181.93±7.75* | 58.15±7.12* |
| Aspartic acid | ND | 84.82±4.91* | 52.88±6.18* | 35.07±4.32* |
| Cysteine/phenylalanine | 14.41±5.34 | 43.04±10.23* | 4.90±2.45* | 7.06±2.55* |
| Cysteine | ND | ND | ND | ND |
| GABA | ND | ND | ND | ND |
| Glutamate | ND | 285.51±9.71 | 220.99±8.38* | 111.23±9.59* |
| Glutamine | 2,868.20±32.79 | 697.50±36.42* | 845.05±33.73* | 2,370±91.73* |
| Glycine/threonine | 524.10±15.40 | 899.32±7.23* | 724.25±58.92* | 510.58±46.15 |
| Histidine | ND | ND | ND | ND |
| Isoleucine | ND | ND | ND | ND |
| Lysine | ND | ND | ND | ND |
| Methionine | ND | ND | ND | ND |
| Serine | 327.93±7.70 | 54.51±4.78* | 250.58±14.61* | 390.29±15.78* |
| Taurine | ND | ND | ND | ND |
| Tryptophan | ND | ND | ND | ND |
| Tyrosine | 447.00±13.45 | 492.10±18.15* | 473.96±16.78* | 398.21±12.48* |
The data represent concentration of metabolic products and are expressed as the mean ± SEM (N=3). Statistical differences from the blank media controls (incubated under the same conditions) and cell-treated samples were determined by a one-way ANOVA followed by a Tukey post hoc test
P<0.05
In contrast, routine cell metabolism was associated with accumulation of L-alanine both in the presence and absence of MPP+ (Table 1). Anaerobic metabolism rendered less utilization of L-glutamine and L-serine but maintained accumulation of end products L-asparagine, L-aspartate, and L-glutamate. These cells also produced L-glycine or L-threonine; however, coelution of these amino acids prevented determination as to the specific analyte. In summary, these findings indicate that the pyruvate–alanine transami-nase pathway may be involved with tumor cell energy metabolism in conjunction with glycolysis.
Discussion
Hypoxic and acid microenvironments propel aggressive malignancies where heightened anaerobic metabolism is associated with tumor progression, metastasis, and proliferation (Airley and Mobasheri 2007). Cancer cells require abnormally high concentration of glucose because ATP is inefficiently produced through SLP rather than mitochondrial oxidative phosphorylation (OXPHOS). This heavy reliance on the glycolytic pathway is a hallmark feature of tumor tissue often visualized using 18F-2-fluorodeoxyglucose positron emission tomography which shows distinct differences in the rate of glucose metabolism between the tumor and the host. While both use the glycolytic pathway, the disparity occurs at the point of pyruvate catabolism. In the host, pyruvate is largely converted to acetyl-CoA by mitochondrial PDH complex rendering reducing equivalents through Krebs cycle activity which drives the electron transport potential for production of ATP. Pyruvate is also the central molecule for diverse fermentation reactions in anaerobes, and in tumor cells pyruvate serves as substrate for LDH where NADH/NAD+ oxidation–reduction reactions drive the synthesis of ATP through SLP at phosphoglycerate kinase and pyruvate kinase (Armstrong 1983).
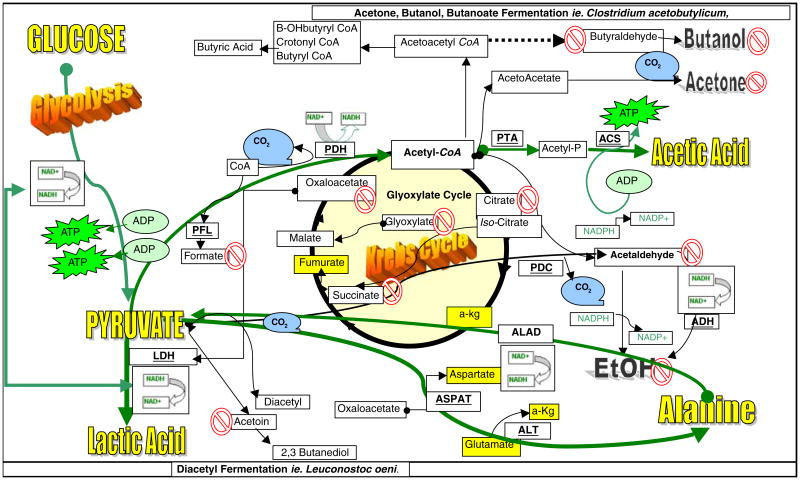
The goal of this study was to investigate for potential evidence to support other fermentation pathways that drive energy processes in tumor cells by examining accumulation of metabolic products. The data from this study exclude any conclusive evidence to support the following anaerobic microbial processes: alcohol fermentation (absence of acetaldehyde, ethanol), butanoate fermentation (absence of aldehydes, butanol, β-hydroxybutyric acid), glyoxylate cycle/hydroxypropionate cycle (absence of glyoxylic acid or accumulation of succinate, malic acid), diacetyl/acetoin/butanedione pathway (absence of acetoin, ketones), or acetone/isopropanol fermentations (absence of accumulated ketones or isopropanol). Figure 5 provides a summary of the findings in this study.
Fig. 5.
Carbohydrate metabolism and fermentation in tumor cells. Glucose metabolism in malignant neuroblastoma occurs primarily through glycolysis where pyruvate is converted to lactate via LDH with NADH reduced to NAD+. NAD+ drives substrate-level phosphorylation through phosphoglycerate kinase and pyruvate kinase producing energy in the form of ATP through glycolysis (Armstrong 1983). While tumor cells have a mitochondria, its function is expendable, resulting in meager amount of pyruvate converted to acetyl-CoA through mitochondrial pyruvate dehydrogenase (PDH). An absence of [formic acid] in this study indicates that conversion of pyruvate to acetyl-CoA + formate through pyruvate formate-lyase (PFL) is not likely. An absence of accumulated [isocitrate], [succinate], or [glyoxylate] suggests a lack of a functional glyoxylate cycle. Absence of [acetaldehyde] accumulation indicates that pyruvate conversion to acetaldehyde through pyruvate decarboxylase (PDC) or acetaldehyde to [alcohol] through alcohol dehydrogenase (ADH) is unlikely events required for anaerobic fermentation. A lack of [acetoin] indicates that conversion of pyruvate through the diacetyl/acetoin biosynthetic pathway is unlikely, and a lack of quantifiable [aldehydes and/or ketones] in general suggests that butanoate fermentation is not likely involved with energy metabolism as well. The quantifiable accumulation of [L-alanine] in conjunction with several amino acids (yellow highlighted) such as [aspartate], [glutamate], and [α-ketoglutarate] suggests potential involvement of pyruvate–alanine transaminase (ALT) or L-alanine/oxaloacetate aminotransferase (ASPAT) pathway. L-alanine could be reconverted to pyruvate through L-alanine dehydrogenase (ALAD) to regenerate NADH to drive LDH. Evidence also suggests that pyruvate may be condensing with coenzyme A to form acetyl-CoA which is converted to acetyl phosphate by phosphotransacetylase (PTA) and [acetate] by acetyl-CoA synthetase (ACS) which is a reaction that could contribute to a second source of ATP production
The data in this study suggest that carbohydrate metabolism in tumor cells is largely dependent upon the fate of pyruvate and could involve the pyruvate–alanine transaminase pathway which is inherent to Escherichia coli. The evidence to support this is based on accumulation of L-alanine in highest yield of the 19 amino acids quantified, along with nitrogen metabolites that appear to drive pyruvate–alanine transamination reactions such as α-ketoglutarate, L-glutamate, and L-aspartate. In this study, forced anaerobic conditions, using MPP+, initiated the simultaneous loss of L-alanine concentration, corresponding to high accumulation of L-glutamine. These findings suggest that deamination reactions, in particular involving L-alanine, could be in high demand during anaerobic metabolism in order to sustain energy needs, despite ample glucose supply. The relevance of these findings needs further investigation to determine exactly how or why a reversal of L-alanine conversion to pyruvate influence tumor cell anaerobic metabolism. Accumulation of L-alanine is also reported in ancestral bacterial anaerobes, as it is readily synthesized from glucose through reductive amination of pyruvate by alanine dehydrogenase (ALAD) [EC 1.4.1.1] as in the case with lactic acid bacteria (Kleerebezemab et al. 2000), E. coli (Lee et al. 2004) and sulfur-metabolizing strict anaerobes such as archaeon Archaeoglobus fulgidus (Gallagher et al. 2004). The known capability of anaerobes to produce L-alanine has broad application in pharmaceutical industry (Kleerebezemab et al. 2000; Wada et al. 2007) where it is manufactured from the carbon in glucose as a by-product of energy production in the absence of O2 (Hashimoto and Katsumata 1999). Briefly, pyruvate is converted to L-alanine through (1) alanine transaminase (ALT) [2.6.1.2] where pyruvate + L-glutamate <=> L-alanine + α-ketoglutarate or (2) L-alanine/oxaloacetate aminotransferase (ASPAT) [2.6.1.12] where pyruvate + L-aspartate <=> L-alanine + oxaloacetate, also a reversible reaction. L-Alanine can be reconverted to pyruvate through ALAD (1) L-ALAD [1.4.1.1] where L-alanine + H2O + NAD+<=> pyruvate + NH3 + NADH + H+. ALAD if present in cancer cells could provide a secondary means to sustain glycolytic flux during anaerobic metabolism in a similar manner to LDH, where it regenerates reducing equivalents (NADH+) to propel SLP in glycolysis.
Our findings corroborate previous reports of accumulated L-alanine, glutamate, L-alanine, and aspartate in malignant blastoma (Márquez et al. 1989). However, the role of a metabolic pathway involving these metabolites needs to be further explored because it may lead to identification of a therapeutic target to block energy production in the tumor cell without harm to the host. Similarly, ALAD antagonists are thought to have therapeutic value in the treatment of other anaerobic pathogens such as Mycobacterium tuberculosis where ALAD expression is heightened with both severity of disease and hypoxia (Davidow et al. 2005; Starck et al. 2004; Agren et al. 2008).
While the results of this study negate involvement of a number of metabolic pathways such as alcohol fermentation, butanoate metabolism, or diacetyl fermentation, the results suggest potential involvement of acetic acid metabolism (Fig. 5). The observation of a coexistence of accumulated lactate, L-alanine, and acetic acid is not unique having been reported in organisms adept at surviving hypoxic environments including fly species resistant to hypoxia (Feala et al. 2007), anaerobic bacteria, and parasites which thrive in the absence of O2 (Ugurbil et al. 1978; Ravot et al. 1996; Darling et al. 1987; Kawanaka et al. 1989). Acetate is a typical fermentation product often seen in facultative anaerobes such as S. cerevisiae or E. coli under anaerobic conditions where its production is involved with SLP and its utilization to drive glyoxylate cycle biosynthetic reactions. Typically, in microbes, acetate is formed from pyruvate which condenses with CoA to form acetyl-CoA which is converted to acetyl phosphate by phosphotransacetylase, serving as a substrate for acetyl-CoA synthetase (ACS) [EC:6.2.1.1], which produces acetic acid + ATP (Guillermo 2005; Lee and Liao 2008). The ACS pathway is active in Candida albicans and is required not only for glucose metabolism but also propanoate metabolism, histone acetylation (also critical role in tumor cell transcriptional response to hypoxia), fixation of CO2 through the Wood–Ljungdahl (reductive acetyl-CoA pathway), and reductive TCA cycle (Carman et al. 2008; Johnson et al. 2008; Takahashi et al. 2006; Ragsdale 2008). And a recent report corroborates the existence of this metabolic avenue in cancer cells as evidenced by high cytosolic ACS expression which is required for tumor cell survival, although its function remains unknown (Yoshii et al. 2009). Although acetate is involved with ethanol fermentation reactions, the data show no evidence of this occurring in tumor cells.
In summary, we have assessed a diverse range of metabolic products commonly produced by a number of yeasts and lactic acid bacteria in association with reduction in pH(ex) (Panagou et al. 2008; Mugula et al. 2003). The findings in this study suggest that there may exist other anaerobic pathways that drive the production of ATP outside of the EMP pathway. Future research will be required to outline metabolic events and define potential therapeutic targets which could enable blocking of tumor energy metabolism without adverse effect on normal host respiration.
References
- Agren D, Stehr M, Berthold CL, Kapoor S, Oehlmann W, Singh M, et al. Three-dimensional structures of apo- and holo-L-alanine dehydrogenase from Mycobacterium tuberculosis reveal conformational changes upon coenzyme binding. J Mol Biol. 2008;377:1161–73. doi: 10.1016/j.jmb.2008.01.091. [DOI] [PubMed] [Google Scholar]
- Airley RE, Mobasheri A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: novel pathways and targets for anticancer therapeutics. Chemotherapy. 2007;53:233–56. doi: 10.1159/000104457. [DOI] [PubMed] [Google Scholar]
- Armstrong FB. Biochemistry. 2. New York: Oxford University Press; 1983. pp. 184–5.pp. 270–271.pp. 283pp. 285 [Google Scholar]
- Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38:3751–7. [PubMed] [Google Scholar]
- Bongaerts GP, van Halteren HK, Verhagen CA, Wagener DJ. Cancer cachexia demonstrates the energetic impact of gluconeogenesis in human metabolism. Med Hypotheses. 2006;67:1213–22. doi: 10.1016/j.mehy.2006.04.048. [DOI] [PubMed] [Google Scholar]
- Carman AJ, Vylkova S, Lorenz MC. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot Cell. 2008;7:1733–41. doi: 10.1128/EC.00253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling TN, Davis DG, London RE, Blum JJ. Products of Leishmania braziliensis glucose catabolism: release of D-lactate and, under anaerobic conditions, glycerol. Proc Natl Acad Sci U S A. 1987;84:7129–33. doi: 10.1073/pnas.84.20.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow A, Kanaujia GV, Shi L, Kaviar J, Guo X, Sung N, et al. Antibody profiles characteristic of Mycobacterium tuberculosis infection state. Infect Immun. 2005;73:6846–51. doi: 10.1128/IAI.73.10.6846-6851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Milito A, Fais S. Tumor acidity, chemoresistance and proton pump inhibitors. Future Oncol. 2005;1:779–86. doi: 10.2217/14796694.1.6.779. [DOI] [PubMed] [Google Scholar]
- Feala JD, Coquin L, McCulloch AD, Paternostro G. Flexibility in energy metabolism supports hypoxia tolerance in Drosophila flight muscle: metabolomic and computational systems analysis. Mol Syst Biol. 2007;3:99. doi: 10.1038/msb4100139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finklestein JZ, Tittle K, Meshnik R, Weiner J. Murine neuroblastoma: further evaluation of the C1300 model with single antitumor agents. Cancer Chemother Rep. 1975;59:975–83. [PubMed] [Google Scholar]
- Gallagher DT, Monbouquette HG, Schröder I, Robinson H, Holden MJ, Smith NN. Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin. J Mol Biol. 2004;342:119–30. doi: 10.1016/j.jmb.2004.06.090. [DOI] [PubMed] [Google Scholar]
- Guillermo G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate: sugar phosphotransferase system. Microb Cell Fact. 2005;4:14. doi: 10.1186/1475-2859-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto SI, Katsumata R. Mechanism of alanine hyperproduction by Arthrobacter oxydans HAP-1: metabolic shift to fermentation under nongrowth aerobic conditions. Appl Environ Microbiol. 1999;65:2781–3. doi: 10.1128/aem.65.6.2781-2783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann PC, Herrmann EC. Oxygen metabolism and a potential role for cytochrome c oxidase in the Warburg effect. J Bioenerg Biomembr. 2007;39:247–50. doi: 10.1007/s10863-007-9084-z. [DOI] [PubMed] [Google Scholar]
- Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat Res. 2008;640:174–9. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanaka M, Matsushita K, Kato K, Ohsaka A. Glucose metabolism of adult Schistosoma japonicum as revealed by nuclear magnetic resonance spectroscopy with D-[13C6] glucose. Physiol Chem Phys Med NMR. 1989;21:5–12. [PubMed] [Google Scholar]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezemab M, Holsa P, Hugenholtza J. Lactic acid bacteria as a cell factory: rerouting of carbon metabolism in Lactococcus lactis by metabolic engineering. Enzyme Microbial Tech. 2000;26:840–8. doi: 10.1016/s0141-0229(00)00180-0. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL Tumour Angiogenesis Research Group. Lactate dehydrogenase 5 expression in operable colorectal cancer: strong association with survival and activated vascular endothelial growth factor pathway—a report of the Tumour Angiogenesis Research Group. J Clin Oncol. 2006;24:4301–8. doi: 10.1200/JCO.2006.05.9501. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Bougioukas G, Sivridis E. Lung cancer: a comparative study of metabolism related protein expression in cancer cells and tumor associated stroma. Cancer Biol Ther. 2007;6:1476–9. doi: 10.4161/cbt.6.9.4635. [DOI] [PubMed] [Google Scholar]
- Lee SG, Liao JC. Control of acetate production rate in Escherichia coli by regulating expression of single-copy pta using lacQ in multicopy plasmid. J Microbiol Biotechnol. 2008;18:334–7. [PubMed] [Google Scholar]
- Lee M, Smith GM, Eiteman MA, Altman E. Aerobic production of alanine by Escherichia coli aceF ldhA mutants expressing the Bacillus sphaericus alaD gene. Appl Microbiol Biotechnol. 2004;65:56–60. doi: 10.1007/s00253-004-1560-3. [DOI] [PubMed] [Google Scholar]
- Márquez J, Sánchez-Jiménez F, Medina MA, Quesada AR, Núñez de Castro I. Nitrogen metabolism in tumor bearing mice. Arch Biochem Biophys. 1989;268(2):667–75. doi: 10.1016/0003-9861(89)90335-4. [DOI] [PubMed] [Google Scholar]
- Mazzio EA, Soliman KF. Effects of enhancing mitochondrial oxidative phosphorylation with reducing equivalents and ubiquinone on 1-methyl-4-phenylpyridinium toxicity and complex I–IV damage in neuroblastoma cells. Biochem Pharmacol. 2004;67(6):1167–84. doi: 10.1016/j.bcp.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Gunaratnam L, Lee S. Oxygen sensing by H+: implications for HIF and hypoxic cell memory. Cell Cycle. 2004;3:1027–9. [PubMed] [Google Scholar]
- Montero VM, Wright LS, Siegel F. Increased glutamate, GABA and glutamine in lateral geniculate nucleus but not in medial geniculate nucleus caused by visual attention to novelty. Brain Res. 2001;916:152–8. doi: 10.1016/s0006-8993(01)02886-4. [DOI] [PubMed] [Google Scholar]
- Mugula JK, Nnko SA, Narvhus JA, Sørhaug T. Microbiological and fermentation characteristics of togwa, a Tanzanian fermented food. Int J Food Microbiol. 2003;80:187–99. doi: 10.1016/s0168-1605(02)00141-1. [DOI] [PubMed] [Google Scholar]
- Ozkan P, Mutharasan R. A rapid method for measuring intracellular pH using BCECF-AM. Biochim Biophys Acta. 2002;1572(1):143–8. doi: 10.1016/s0304-4165(02)00303-3. [DOI] [PubMed] [Google Scholar]
- Panagou EZ, Schillinger U, Franz CM, Nychas GJ. Microbiological and biochemical profile of cv. Conservolea naturally black olives during controlled fermentation with selected strains of lactic acid bacteria. Food Microbiol. 2008;25:348–58. doi: 10.1016/j.fm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Pastorekova S, Zatovicova M, Pastorek J. Cancer-associated carbonic anhydrases and their inhibition. Curr Pharm Des. 2008;14:685–98. doi: 10.2174/138161208783877893. [DOI] [PubMed] [Google Scholar]
- Rafailidis PI, Kapaskelis A, Christodoulou C, Galani E, Falagas ME. Concurrent M. tuberculosis, Klebsiella pneumoniae, and Candida albicans infection in liver metastasis of bowel carcinoma. Eur J Clin Microbiol Infect Dis. 2008;27:753–5. doi: 10.1007/s10096-008-0488-4. [DOI] [PubMed] [Google Scholar]
- Ragsdale SW. Enzymology of the wood–Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci. 2008;1125:129–36. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravot G, Ollivier B, Fardeau ML, Patel BK, Andrews KT, Magot M, et al. L-Alanine production from glucose fermentation by hyperthermophilic members of the domains bacteria and Archaea: a remnant of an ancestral metabolism? Appl Environ Microbiol. 1996;62:2657–9. doi: 10.1128/aem.62.7.2657-2659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva BM, Andrade PB, Mendes GC, Seabra RM, Ferreira MA. Study of the organic acids composition of quince (Cydonia oblonga Miller) fruit and jam. J Agric Food Chem. 2002;50:2313–7. doi: 10.1021/jf011286+. [DOI] [PubMed] [Google Scholar]
- Starck J, Källenius G, Marklund BI, Andersson DI, Akerlund T. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology. 2004;150:3821–9. doi: 10.1099/mic.0.27284-0. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleo-cytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Ugurbil K, Brown TR, den Hollander JA, Glynn P, Shulman RG. High-resolution 13C nuclear magnetic resonance studies of glucose metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:3742–6. doi: 10.1073/pnas.75.8.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Narita K, Yokota A. Alanine production in an H+-ATPase- and lactate dehydrogenase-defective mutant of Escherichia coli expressing alanine dehydrogenase. Microbiol Biotechnol. 2007;76:819–25. doi: 10.1007/s00253-007-1065-y. [DOI] [PubMed] [Google Scholar]
- Wahl ML, Owen JA, Burd R, Herlands RA, Nogami SS, Rodeck U, et al. Regulation of intracellular pH in human melanoma: potential therapeutic implications. Mol Cancer Ther. 2002;1:617–28. [PubMed] [Google Scholar]
- Walenta S, Schroeder T, Mueller-Klieser W. Lactate in solid malignant tumors: potential basis of a metabolic classification in clinical oncology. Curr Med Chem. 2004;11:2195–204. doi: 10.2174/0929867043364711. [DOI] [PubMed] [Google Scholar]
- Yoshii Y, Furukawa T, Yoshii H, Mori T, Kiyono Y, Waki A, et al. Cytosolic acetyl-CoA synthetase affected tumor cell survival under hypoxia: the possible function in tumor acetyl-CoA/acetate metabolism. Cancer Sci. 2009;100:821–7. doi: 10.1111/j.1349-7006.2009.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziello JE, Jovin IS, Huang Y. Hypoxia-inducible factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J Biol Med. 2007;80:51–60. [PMC free article] [PubMed] [Google Scholar]