Abstract
Expressed prostatic secretions (EPS) contain proteins of prostate origin that may reflect the health status of the prostate and be used as diagnostic markers for prostate diseases including prostatitis, benign prostatic hyperplasia, and prostate cancer. Despite their importance and potential applications, a complete catalog of EPS proteins is not yet available. We, therefore, undertook a comprehensive analysis of the EPS proteome using 2-D micro-LC combined with MS/MS. Using stringent filtering criteria, we identified a list of 114 proteins with at least two unique-peptide hits and an additional 75 proteins with only a single unique-peptide hit. The proteins identified include kallikrein 2 (KLK2), KLK3 (prostate-specific antigen), KLK11, and nine cluster of differentiation (CD) molecules including CD10, CD13, CD14, CD26, CD66a, CD66c, CD 143, CD177, and CD224. To our knowledge, this list represents the first comprehensive characterization of the EPS proteome, and it provides a candidate biomarker list for targeted quantitative proteomics analysis using a multiple reaction monitoring (MRM) approach. To help prioritize candidate biomarkers, we constructed a protein–protein interaction network of the EPS proteins using Cytoscape (www.cytoscape.org), and overlaid the expression level changes from the Oncomine database onto the network.
Keywords: Biomarker, Expressed prostatic secretions, Mass spectrometry, Prostate cancer
1 Introduction
Expressed prostatic secretions (EPS) are body fluids directly derived from prostate and should contain proteins of prostate origin, and therefore, may reflect the health status of prostate. Despite its importance and potential applications as a rich source of prostate disease biomarkers, the proteome of EPS, i.e., a complete catalog of proteins in the EPS, has not been established. Only a few proteins have been identified in the EPS from sporadic studies. In 1986, Lee et al. [1] observed, by 2-DE, a total of 57 major protein groups from specimens of pooled prostatic fluid collected by rectal massage from men under 50 years of age with no apparent prostatic disorders. However, due to limitation of the technologies available at that time, they were only able to identify a few prostate-associated proteins such as prostatic acid phosphatase (PAP), prostatic-specific antigen (PSA), and prostate-binding protein (PBP) [1].
The proteome of many body fluids such as nipple aspirate fluid [2], cerebrospinal fluid [3], cervical vaginal fluid [4], saliva, bronchoalveolar lavage fluid, synovial fluid, tear fluid, and amniotic fluid (reviewed by [5]) were cataloged previously; and their protein catalogs have served as a resource for biomarker discoveries for cancers and other human diseases. As far as we know, an analysis of the EPS proteome by current proteomics technology has not been done yet. We have therefore planned a study to identify candidate bio-markers from EPS. Our initial experimental design was to compare EPS from benign prostatic hyperplasia (BPH) patients or normal individuals to that from prostate cancer patients by ICAT (isotopic coded affinity tags) labeling coupled with MS/MS analysis; however, we found that it was difficult to get enough EPS from prostate cancer patients and that the amount we obtained from prostate cancer patients was not enough for comparative proteomics analysis by ICAT/MS/MS. In more than 30% of the prostate cancer patients the prostate is hardened, and the gland structure is greatly damaged, both making it difficult to get sufficient EPS.
Therefore, we had to lower our aim to only cataloging the proteins in the EPS, which we believed, nonetheless, would still be interesting to the scientific community. In this cataloging experiment, we have analyzed EPS from normal individuals, BPH patients, and prostate cancer patients. We undertook a comprehensive analysis using 2-D micro-LC combined with MS/MS. Using stringent filtering criteria, we identified a list of 114 proteins with at least two unique-pep-tide hits and additional 75 proteins with only a single unique-peptide hit. The proteins identified include kallikrein 2 (KLK2), KLK3 (PSA), KLK11, and nine cluster of differentiation (CD) molecules including CD10, CD13, CD14, CD26, CD66a, CD66c, CD 143, CD177, and CD224. To our knowledge, this list represents the first comprehensive characterization of the EPS proteome. Gene ontology (GO) analysis of the identified EPS proteins revealed that they are enriched for proteins in the GO categories such as immunity and defense, proteolysis, transport, and lipid and fatty acid transport under the category of biological processes.
Recently, Malmstrom et al. [6] heralded a targeted quantitative proteomics approach for biomarker discovery and validation to complement the shotgun proteomics approach that has numerous limitations including limited sensitivity, limited coverage, a high degree of redundancy, and under sampling, etc. In the targeted quantitative proteomics approach, candidate proteins are first identified and then quantified in multiple repeated analyses (i.e., large amount of samples) using a multiple reaction monitoring (MRM) approach. Candidate biomarkers can be derived from an analysis of the literature, from specific biological knowledge or from focused proteomics analysis such as our analysis of the EPS proteome. Integrative analysis of our EPS protein list with the prostate cancer microarray data from Oncomine (www.Oncomine.org) [7] revealed a list of 48 EPS proteins which 50% of the array data support as being upregulated in prostate cancer. Furthermore, we identified 26 EPS proteins that are prostate-specific/enriched compared with 33 massively parallel signature sequencing (MPSS) data generated by a collaboration with Solexa [8] using 33 normal tissues. Comparison of our EPS proteome with that of the proteome of prostasomes, which we previously characterized [9] through Entrez Gene identifiers, resulted in a common set of 34 proteins. This set includes CD10, CD224, CD13, PSA, PAP, Annexin A1, enolase 1, Ras-related proteins Rab-3D and Rab-27A, alpha-2-glycoprotein 1, etc. Finally, using Cytoscape (www.cytoscape.org), we were able to construct a protein–protein interaction network of the EPS proteins, suggesting that many EPS proteins form a network working in concert. We have also overlaid the expression changes of the mapped genes from the Oncomine database onto this map. This network will help us pinpoint key genes in the network that play important roles in prostate diseases and will also help us identify a composite panel of markers from different regions of the map that better reflect the biological state of the prostate and, therefore, constitute better bio-markers for prostate diseases.
2 Experimental procedures
2.1 EPS collection
EPS was collected with patients’ consent and an institutional review board (IRB) approval. EPS was collected by prostatic massage. About 30–100 µL of EPS was collected from each individual. The EPS was then diluted with 100 µL of cold PBS and centrifuged at 2000 rpm for 5 min to remove any possible cellular contamination. The EPS was then stored at −80°C until analysis.
EPS was collected from three confirmed prostate cancer patients, four BPH patients, and one normal individual. Their clinical information such as PSA levels is listed in Table 1.
Table 1.
Clinical information of the patients
| Patient no. | Diagnosis | PSA (ng/mL) | Gleason scores |
|---|---|---|---|
| 1 | CaP | 10.55 | 2 + 2 |
| 3 | CaP | > 100 | 4 + 3 |
| 4 | CaP | 62.3 | 5 + 4 |
| 5 | BPH | 3.62 | NA |
| 6 | BPH | 3.98 | NA |
| 7 | BPH | 4.71 | NA |
| 8 | BPH | 2.18 | NA |
| 9 | Normal prostate | 2.1 | NA |
2.2 Trypsin digestion
The EPS samples were treated with DTT followed by iodoacetamide before tryptic digestion. A standard trypsin digestion protocol (www.biochem.uwo.ca/wits/bmsl/in_solution_digestion.pdf) was followed using Promega sequencing grade modified trypsin (V511A). The EPS samples were also run on 1-D polyacrylamide gels, and four individual bands that showed differences between samples were cut out and trypsinized. The In-Gel Tryptic Digestion Kit (Pierce Biotechnology) was used for in-gel trypsin digestion.
2.3 MS/MS analysis
Microcapillary high pressure LC nanoelectrospray IT MS/MS was carried out using an LTQ linear IT mass spectrometer (Thermo Fisher, San Jose, CA) with an in-house fabricated microelectrospray source and an HP1100 solvent delivery system (Agilent, Palo Alto, CA). Samples were automatically delivered by a FAMOS autosampler (LC Packings, San Francisco, CA) to a 100 µm internal diameter fusedsilica capillary precolumn packed to a 2 cm length with 200 Å pore-size Magic C18A™ material (Michrom Bioresources, Auburn, CA). Once on the precolumn, the samples were washed with solvent A (0.1% formic acid, 5% ACN) and then eluted with a gradient of 10–35% solvent B (100% ACN) over 30 min to a 75 µm × 10 cm fused-silica capillary column packed with 100 Å pore-size Magic C18AQ material (Michrom) and then into the mass spectrometer at a constant column tip flow-rate of ~300 nL/min. Eluting peptides were analyzed by µLC-MS and data-dependent µLC-MS/MS acquisition which selected three precursor ions for MS with a dynamic exclusion of one.
Each EPS sample was injected only once for the MS/MS analysis due to the limited amount of material. Thermo Finnegan’s Xcalibur Qual Browser (3.0) was used to generate the MS peak list. The ICIS peak-detection algorithm was used. The SEQUEST algorithm (Thermo Fisher) was used to search the ipi.HUMAN.v3.21.fasta database that contains 60 824 protein entries. A detailed SEQUEST search parameter is provided in Supporting Information Document 1. In brief, mass tolerance was set at 3 Da and fragment ion tolerance at 0 Da. A one-enzyme terminus search was used, and two amino acid modification options were used, one for STY phosphorylation (+80) and one for methionine oxidation (+16). A cysteine modification of +57 Da was used for the EPS proteins from in-solution digestion as they were alkylated by iodoacetamide. The PeptideProphet™ [10] and Protein-Prophet™ [11] programs were used for statistical analysis of the identified peptides and proteins. The Xinteract program was used to integrate the information. Detailed descriptions of these programs are available at the web site http://tools.proteomecenter.org/software.php.
2.4 Functional analysis
We used the Panther classification system (http://www.pantherdb.org/) to analyze our data in order to see which GO terms were enriched in the EPS proteins. Prostate cancer microarray datasets compiled at the Oncomine database (www.Oncomine.org) were used to search for EPS proteins that were differentially expressed. The one-sided t-test, as used in the Oncomine website, was used to calculate the p-values. For analyzing and determining prostate-specific genes, MPSS (http://mpss.licr.org/) datasets containing analyses of normal human tissue were downloaded [8]. Because liver was not present in these, we added normal liver MPSS data that we generated ourselves (data now shown). The specificity score was calculated using the formula listed in the paper by Jongeneel et al. [8], which was defined in our case as the log 2-transformed ratio of the expression of the gene in the prostate tissue divided by the sum of the expression levels in all other tissues plus 1 [8]. To categorize the EPS proteins as either classical or nonclassical secretory proteins, we used signal-peptide prediction servers such as SignalP 3.0, SecretomeP 2.0, and TMHMM (www.cbs.dtu.dk/services/).
To build a protein–protein interaction map of these EPS proteins in prostate, we extracted the protein–protein interaction data from the HPRD and the NCBI databases (www.hprg.org; www.ncbi.nlm.nih.gov/). We only extended one layer out to make the connections. We used Cytoscape (www.cytoscape.org) to display the result.
3 Results
3.1 A catalog of the proteins identified in the expressed prostatic secretion
We analyzed the proteome of EPS that was collected from three confirmed prostate cancer patients, four BPH patients, and one normal individual by micro-LC MS/MS analysis. After filtering the mass spectrum quality by PeptideProphet™ scores [10] of ≥0.9 and removing peptides corresponding to trypsin, we obtained a total of 2109 peptides (Supporting Information Document 2). These 2109 peptides can be reduced to 988 unique peptides corresponding to 288 unique proteins of which 135 are single-peptide hit proteins. To identify proteins that have the highest confidence, we further analyzed the data by ProteinProphet™ [11]. In the Protein-Prophet™ algorithm, peptides that correspond to more than a single protein in the sequence database are apportioned among all corresponding proteins, and a minimal protein list sufficient to account for the observed peptide assignments is derived using the expectation–maximization algorithm [11]. We reduced the number of proteins identified using the ProteinProphet™ algorithm to 206 unique proteins. We then filtered the protein list using the criterion that a protein must have two or more unique peptides identified (133 proteins). We also manually combined various Ig entries (e.g., heavy chain, light chain, etc.) into one entry and removed keratin, which we think is derived from contamination during EPS preparation and handling. We were left with a confidently identified list of 114 proteins (Table 2). To our knowledge, this list represents the first comprehensive characterization of the EPS proteome. We also identified an additional 75 proteins with only a single unique-peptide hit (Supporting Information Document 3).
Table 2.
Proteins identified and their MS identification information
| IPI protein IDs |
Protein probabilitya) |
Percent coverage |
No. of unique peptides |
Total no. of peptides |
Description | GO symbol |
|---|---|---|---|---|---|---|
| IPI00434943 | 1 | 16.1 | 2 | 4 | KLK2 | KLK2 |
| IPI00010858 | 1 | 54 | 27 | 49 | PSA | KLK3 |
| IPI00414609 | 1 | 28.1 | 6 | 13 | Isoform PSP94 of beta-microseminoprotein | MSMB |
| IPI00023020 | 1 | 27.5 | 21 | 42 | Semenogelin-1 | SEMG1 |
| IPI00022429 | 1 | 25.4 | 4 | 6 | Alpha-1-acid glycoprotein 1 | ORM1 |
| IPI00465248 | 1 | 5.3 | 2 | 3 | Enolase 1 | ENO1 |
| IPI00007752 | 1 | 9.2 | 3 | 3 | Tubulin beta-2C chain | TUBB2C |
| IPI00396434 | 1 | 40.7 | 31 | 52 | PAP | ACPP |
| IPI00025415 | 1 | 26.1 | 23 | 35 | Semenogelin-2 | SEMG2 |
| IPI00025447 | 1 | 7.3 | 2 | 2 | Eukaryotic translation elongation factor 1 alpha-like 3 | EEF1A1 |
| IPI00010471 | 1 | 2.9 | 2 | 2 | Plastin-2 | LCP1 |
| IPI00456429 | 1 | 32.9 | 3 | 11 | Ubiquitin and ribosomal protein L40 | UBA52 |
| IPI00022418 | 1 | 21.4 | 60 | 76 | Isoform 1 of fibronectin | FN1 |
| IPI00027230 | 1 | 16.8 | 8 | 22 | Endoplasmin | TRA1 |
| IPI00027462 | 1 | 69.3 | 10 | 20 | Protein S100-A9 | S100A9 |
| IPI00021440 | 1 | 24 | 6 | 6 | Actin, cytoplasmic 2 | ACTG1 |
| IPI00219682 | 1 | 12.2 | 2 | 2 | Erythrocyte band seven integral membrane protein | STOM |
| IPI00247063 | 1 | 3.6 | 2 | 3 | Neprilysin (CD10) | MME (CD10) |
| IPI00177728 | 1 | 6.3 | 2 | 2 | Cytosolic nonspecific dipeptidase | CNDP2 |
| IPI00219018 | 1 | 23.1 | 7 | 14 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | GAPDH |
| IPI00479186 | 1 | 4.1 | 2 | 3 | Pyruvate kinase 3 isoform 1 | PKM2 |
| IPI00745161 | 1 | 9.2 | 2 | 2 | STEAP family member 4 | STEAP4 |
| IPI00553177 | 1 | 39.2 | 15 | 36 | Alpha-1-antitrypsin | SERPINA1 |
| IPI00018953 | 1 | 6.8 | 5 | 8 | Dipeptidyl peptidase 4 (CD26) | DPP4(CD26) |
| IPI00022977 | 1 | 6.3 | 3 | 3 | Creatine kinase B-type | CKB |
| IPI00306543 | 1 | 18.2 | 5 | 9 | Growth\differentiation factor 15 | GDF15 |
| IPI00299086 | 1 | 9.9 | 2 | 2 | Syntenin-1, syntenin isoform 2 | SDCBP |
| IPI00031461 | 1 | 6.5 | 3 | 4 | Rab GDP dissociation inhibitor beta | GDI2 |
| IPI00027223 | 1 | 6.8 | 3 | 3 | Isocitrate dehydrogenase (NADP) cytoplasmic | IDH1 |
| IPI00026259 | 1 | 7.5 | 2 | 4 | N(4)-(beta-N-acetylglucosaminyl)-l-asparaginase | AGA |
| IPI00022431 | 1 | 6.3 | 2 | 4 | Alpha-2-HS-glycoprotein | AHSG |
| IPI00741408 | 1 | 25.5 | 3 | 7 | Similar to neutrophil defensin 1 | DEFA1 |
| IPI00298497 | 1 | 14.5 | 4 | 4 | Fibrinogen beta chain | FGB |
| IPI00555812 | 1 | 17.9 | 4 | 4 | Vitamin D-binding protein | GC |
| IPI00032328 | 1 | 13.8 | 4 | 7 | Isoform HMW of kininogen-1 | KNG1 |
| IPI00236556 | 1 | 6.8 | 4 | 7 | Isoform H7 of myeloperoxidase | MPO |
| IPI00032179 | 1 | 13.5 | 3 | 5 | Antithrombin III variant | SERPINC1 |
| IPI00018901 | 1 | 6.5 | 3 | 3 | Gamma-glutamyltranspeptidase 1 (CD224 antigen) | GGT1(CD224) |
| IPI00293276 | 1 | 17.5 | 2 | 5 | Macrophage migration inhibitory factor | MIF |
| IPI00022974 | 1 | 61.6 | 18 | 29 | Prolactin-inducible protein | PIP |
| IPI00298547 | 1 | 22.2 | 2 | 2 | Protein DJ-1 | PARK7 |
| IPI00032220 | 1 | 6.4 | 2 | 2 | Angiotensinogen | AGT |
| IPI00022229 | 1 | 1.5 | 4 | 4 | Apolipoprotein B-100 | APOB |
| IPI00298828 | 1 | 16.5 | 7 | 9 | Beta-2-glycoprotein 1 | APOH |
| IPI00215983 | 1 | 15 | 3 | 4 | Carbonic anhydrase 1 | CA1 |
| IPI00218414 | 1 | 13.5 | 2 | 2 | Carbonic anhydrase 2 | CA2 |
| IPI00219713 | 1 | 19.7 | 10 | 13 | Gamma-A of fibrinogen gamma chain | FGG |
| IPI00478493 | 1 | 39.5 | 11 | 16 | Haptoglobin | HP |
| IPI00514159 | 1 | 11.4 | 3 | 3 | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 |
| IPI00655954 | 1 | 14.3 | 2 | 2 | Platelet-derived growth factor A chain | PDGFA |
| IPI00004573 | 1 | 11 | 8 | 12 | Polymeric-Ig receptor | PIGR |
| IPI00022432 | 1 | 41.5 | 10 | 15 | Transthyretin | TTR |
| IPI00643525 | 1 | 20.4 | 31 | 44 | Complement component 4A | C4A |
| IPI00219825 | 1 | 10.9 | 5 | 10 | PSAP | PSAP |
| IPI00513699 | 1 | 13.8 | 2 | 2 | Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 6 |
SERPINB6 |
| IPI00021841 | 1 | 21.3 | 7 | 7 | Apolipoprotein A-I | APOA1 |
| IPI00021854 | 1 | 42 | 6 | 7 | Apolipoprotein A-II | APOA2 |
| IPI00414018 | 0.91 | 4.5 | 2 | 4 | Complement component C8 alpha chain | C8A |
| IPI00465436 | 1 | 6.3 | 3 | 3 | Catalase | CAT |
| IPI00410714 | 1 | 88.7 | 58 | 97 | Hemoglobin subunit alpha | HBA2 |
| IPI00022488 | 1 | 23.8 | 9 | 10 | Hemopexin | HPX |
| IPI00022371 | 0.91 | 3.8 | 2 | 2 | Histidine-rich glycoprotein | HRG |
| IPI00401776 | 1 | 10.6 | 15 | 18 | Mucin 6, gastric isoform 1 | MUC6 |
| IPI00301579 | 1 | 34.4 | 5 | 5 | Epididymal secretory protein E1 | NPC2 |
| IPI00759575 | 1 | 7.1 | 3 | 6 | Cytoplasmic of glutathione reductase | GSR |
| IPI00549413 | 1 | 25.5 | 3 | 3 | Annexin A1 | ANXA1 |
| IPI00221224 | 1 | 6.6 | 6 | 13 | Aminopeptidase N (CD13) | ANPEP(CD13) |
| IPI00166729 | 1 | 44.6 | 28 | 93 | Alpha-2-glycoprotein 1, zinc | AZGP1 |
| IPI00295386 | 1 | 9.8 | 2 | 3 | Carbonyl reductase (NADPH) 1 | CBR1 |
| IPI00291262 | 1 | 16.7 | 11 | 13 | Clusterin, clusterin isoform 1 | CLU |
| IPI00032808 | 1 | 19.2 | 2 | 4 | Ras-related protein Rab-3D | RAB3D |
| IPI00219858 | 1 | 15 | 3 | 6 | Ras-related protein Rab-27A | RAB27A |
| IPI00216691 | 1 | 37.4 | 4 | 6 | Profilin-1 | PFN1 |
| IPI00294653 | 1 | 1.9 | 2 | 2 | Protein C21orf5 | DOPEY2 |
| IPI00000816 | 1 | 28.2 | 4 | 4 | 14-3-3 Protein epsilon | YWHAE |
| IPI00759663 | 1 | 36 | 3 | 5 | Peroxiredoxin-5 | PRDX5 |
| IPI00003865 | 1 | 3.7 | 2 | 2 | Heat shock cognate 71 kDa protein | HSPA8 |
| IPI00478003 | 1 | 19.5 | 23 | 40 | Alpha-2-macroglobulin | A2M |
| IPI00437751 | 1 | 1.6 | 2 | 2 | Angiotensin-converting enzyme, somatic isoform (CD143) |
ACE(CD143) |
| IPI00745872 | 1 | 72.6 | 145 | 435 | Serum albumin | ALB |
| IPI00029260 | 1 | 9.9 | 2 | 2 | Monocyte differentiation antigen CD14 | CD14 |
| IPI00641900 | 1 | 4.1 | 2 | 2 | Carcinoembryonic antigen-related cell adhesion molecule 1 isoform 2 (CD66a) |
CEACAM1(CD66a) |
| IPI00027412 | 1 | 13.7 | 3 | 4 | Carcinoembryonic antigen-related cell adhesion molecule 6 (CD66c) |
CEACAM6(CD66c) |
| IPI00639937 | 1 | 3.7 | 2 | 2 | Complement factor B | CFB |
| IPI00029739 | 1 | 3.1 | 2 | 2 | Complement factor H | CFH |
| IPI00017601 | 1 | 11.5 | 8 | 14 | Ceruloplasmin | CP |
| IPI00032293 | 1 | 17.8 | 2 | 2 | Cystatin C | CST3 |
| IPI00032325 | 1 | 26.5 | 2 | 5 | Cystatin A | CSTA |
| IPI00242956 | 1 | 4.5 | 8 | 20 | Fc fragment of IgG binding protein | FCGBP |
| IPI00654755 | 1 | 97.3 | 47 | 94 | Hemoglobin subunit beta | HBB |
| IPI00473011 | 1 | 90.4 | 23 | 33 | Hemoglobin subunit delta | HBD |
| IPI00220706 | 1 | 24 | 2 | 2 | Hemoglobin subunit gamma-1 | HBG1 |
| IPI00014540 | 1 | 4 | 2 | 4 | Homeobox protein Hox-B4 | HOXB4 |
| IPI00743064 | 1 | 27.8 | 7 | 10 | Lipocalin 2 | LCN2 |
| IPI00023673 | 1 | 14.5 | 9 | 26 | Galectin-3-binding protein | LGALS3BP |
| IPI00298860 | 1 | 44 | 33 | 103 | Growth-inhibiting protein 12 | LTF |
| IPI00022255 | 1 | 8.6 | 3 | 4 | Olfactomedin 4 | OLFM4 |
| IPI00011571 | 1 | 7 | 2 | 5 | Protein disulfide-isomerase A2 | PDIA2 |
| IPI00642548 | 1 | 5.8 | 3 | 3 | Phosphoglycerate dehydrogenase, | PHGDH |
| IPI00000874 | 1 | 18.1 | 3 | 3 | Peroxiredoxin-1, 19 kDa protein | PRDX1 |
| IPI00218130 | 1 | 8.2 | 4 | 7 | Glycogen phosphorylase, muscle form | PYGM |
| IPI00007047 | 1 | 35.5 | 8 | 19 | Protein S100-A8 | S100A8 |
| IPI00550991 | 1 | 24.3 | 11 | 18 | Alpha-1-antichymotrypsin | SERPINA3 |
| IPI00007221 | 1 | 28.8 | 8 | 10 | Plasma serine protease inhibitor | SERPINA5 |
| IPI00027444 | 1 | 9.2 | 3 | 4 | Leukocyte elastase inhibitor | SERPINB1 |
| IPI00022463 | 1 | 31.5 | 27 | 79 | Serotransferrin | TF |
| IPI00032292 | 1 | 14.7 | 2 | 2 | Metalloproteinase inhibitor 1 | TIMP1 |
| IPI00746832 | 1 | 12.5 | 2 | 6 | Similar to triosephosphate isomerase (TIM) (Triosephosphate isomerase) isoform 2 |
TPI1 |
| IPI00022895 | 1 | 20.2 | 8 | 9 | Alpha-1B-glycoprotein | A1BG |
| IPI00164623 | 1 | 19.9 | 37 | 59 | Complement componebt C3 | C3 |
| IPI00297444 | 1 | 9.6 | 4 | 5 | Isoform 1 of CD177 antigen | CD177 |
| IPI00736872 | 1 | 39.8 | 20 | 54 | Lactotransferrin | LTF |
| IPI00784830 | 1 | 24.8 | 2 | 5 | Ig | |
| IPI00747115 | 1 | 39.5 | 2 | 3 | Similar to beta globin | HBB |
by the ProteinProphet™ program.
Previously, using MS/MS analysis, we cataloged the proteome of prostasomes, which are prostate-derived organelles in seminal plasma [9]. We reran the SEQUEST search using the same database and filtered the protein list with the same filtering criteria to make it comparable to the EPS data. We identified a list of 91 prostasome proteins that have two or more unique-peptide hits (Supporting Information Document 4). Comparing our EPS proteome with that of the prostasomes using Entrez Gene identifiers, we identified a common set of 31 proteins (Table 3 and Supporting Information Document 5). The common proteins include CD10, CD224, CD13, PSA, PAP, Annexin A1, enolase 1, Ras-related proteins Rab-3D and Rab-27A, alpha-2-glycoprotein 1, and others.
Table 3.
Integrative analysis of 26 EPS proteins with other datasets
| Protein (IPI number) |
GO symbol | Found in prostasomes |
Identified as downregulated in oncomine |
Identified as upregulated in oncomine |
Prostate enrichment scores |
SignalP prediction |
|---|---|---|---|---|---|---|
| IPI00434943 | KLK2 | No | 2 | 3 | 9.24 | Yes |
| IPI00010858 | KLK3 | No | 3 | 4 | 8.79 | Yes |
| IPI00414609 | MSMB | No | 7.25 | Yes | ||
| IPI00023020 | SEMG1 | Yes | 6.41 | Yes | ||
| IPI00022429 | ORM1 | No | 6.02 | Yes | ||
| IPI00465248 | ENO1 | Yes | 0 | 1 | 5.29 | No |
| IPI00007752 | TUBB2C | Yes | 5.25 | No | ||
| IPI00396434 | ACPP | Yes | 5.12 | Yes | ||
| IPI00025415 | SEMG2 | Yes | 0 | 2 | 4.91 | Yes |
| IPI00025447 | EEF1A1 | No | 4.32 | No | ||
| IPI00010471 | LCP1 | Yes | 1 | 6 | 3.91 | No |
| IPI00456429 | UBA52 | No | 2 | 4 | 3.46 | No |
| IPI00022418 | FN1 | No | 3.32 | Yes | ||
| IPI00027230 | TRA1 | No | 3.32 | Yes | ||
| IPI00027462 | S100A9 | No | 3.17 | No | ||
| IPI00021440 | ACTG1 | Yes | 0 | 3 | 3.00 | No |
| IPI00219682 | STOM | No | 2.81 | Signal anchor | ||
| IPI00247063 | MME (CD10) | Yes | 2.32 | Signal anchor | ||
| IPI00177728 | CNDP2 | Yes | 0 | 1 | 2.00 | No |
| IPI00219018 | GAPDH | Yes | 2.00 | No | ||
| IPI00479186 | PKM2 | Yes | 2.00 | No | ||
| IPI00745161 | STEAP4 | No | 1.74 | No | ||
| IPI00553177 | SERPINA1 | No | 0 | 1 | 1.58 | Yes |
| IPI00018953 | DPP4 (CD26) | Yes | 1.19 | Yes | ||
| IPI00022977 | CKB | Yes | 1.17 | No |
3.2 EPS proteins as putative prostate cancer biomarkers
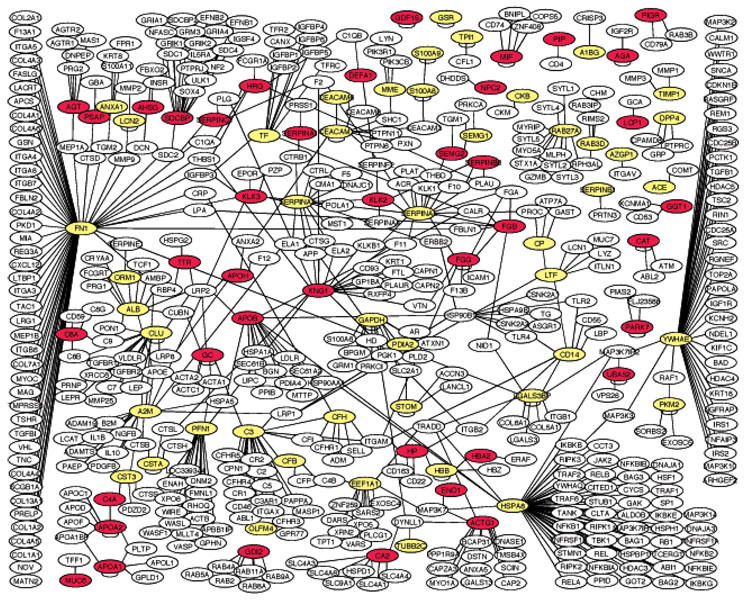
In order to assess whether these EPS proteins have any potential roles in prostate carcinogenesis and to assess whether any of these may be potential cancer markers, we compared the EPS protein list that we generated with the transcriptomics data on prostate cancer collected at the Oncomine database which collects, standardizes, analyzes, and delivers published cancer gene expression data to the research community (www.Oncomine.org) [7]. There are 11 prostate cancer microarray datasets that contain normal and cancer tissue type comparisons. Using p<0.05 as a cut-off score, we tabulated the number of times that a protein was identified as significantly up- or downregulated. As a protein could be identified as upregulated in one dataset, but down-regulated in another dataset, we calculated the percentage of times that a protein was identified as upregulated in these 11 prostate cancer datasets. In the end, we identified a list of 48 EPS proteins that had more than 50% of the array data in support of their being upregulated in prostate cancer. We included those genes that were identified as upregulated only in the one array study in the database. The information was added to Table 3 and Supporting Information Document 5. We also mapped these 48 proteins onto the protein–protein network (Fig. 1), thus creating a map reflecting the disease state (prostate cancer) of the network.
Figure 1.
A protein–protein interaction network of the EPS proteins. Colored (yellow and red) nodes indicate the EPS proteins that we identified by MS. Noncolored nodes are derived from the human protein–protein interaction databases of HPRD and NCBI. We only retrieved proteins that interact with an EPS protein, i.e., only one-step retrieval of the protein–protein interaction. We only showed the large network connecting most of the EPS proteins. Smaller networks and isolated nodes were not shown. The red color also indicates the nodes with microarray data from the Oncomine database suggesting that they are upregulated in prostate cancer cells.
We have been promoting the idea that organ-specific genes may be rich resources for identifying cancer bio-markers [12]. We have compared 33 MPSS data generated through a collaboration with Solexa [7] using 33 normal tissues and used the specificity score defined by Jongeneel et al. [8] to identify prostate-specific/enriched expressed genes. Using a log 2 enrichment score greater than 1 (meaning that more than 50% of the total combined transcript copy numbers come from the prostate tissue) as a cut-off score, we identified 26 EPS proteins that are prostate-specific/enriched, including well-known prostate-specific genes such as KLK2, PSA, and CD10. This information is also provided in Table 3.
3.3 Functional categorization of the EPS proteins
We used the Panther classification system (http://www.pantherdb.org/) to identify those functional categories that characterized the majority of the EPS proteins. Using NCBI’s Homo sapiens genes as reference lists and using the Bonferroni correction for multiple testing, we identified many enriched biological processes (p<0.05) as listed in Table 4. Biological processes such as immunity and defense, proteolysis, transport, and lipid and fatty acid transport are all enriched. Grouped by molecular function, the enriched categories (p<0.05) are transfer/carrier proteins, proteases and their inhibitors, defense/immunity proteins, apolipoproteins, and others. All of these data support and further illustrate the functions of the prostate.
Table 4.
Enriched GO terms in the EPS proteins
| NCBI: H. sapiens genes (REF) | EPS proteins | Expected | p-value | |
|---|---|---|---|---|
| Biological process | ||||
| Immunity and defense | 1263 | 35 | 5.88 | 1.03E-16 |
| Proteolysis | 893 | 23 | 4.16 | 3.31E-09 |
| Blood circulation and gas exchange | 87 | 9 | 0.41 | 1.28E-08 |
| Blood clotting | 84 | 9 | 0.39 | 4.49E-08 |
| Transport | 1237 | 22 | 5.76 | 0.00000172 |
| Protein metabolism and modification | 2858 | 29 | 13.31 | 0.00109 |
| Lipid and fatty acid transport | 114 | 6 | 0.53 | 0.00259 |
| Miscellaneous | 129 | 5 | 0.6 | 0.0115 |
| Complement-mediated immunity | 52 | 4 | 0.24 | 0.0166 |
| Molecular function | ||||
| Transfer/carrier protein | 305 | 19 | 1.42 | 1.16E-14 |
| Protease inhibitor | 118 | 14 | 0.55 | 1.14E-13 |
| Serine protease inhibitor | 72 | 9 | 0.34 | 1.62E-08 |
| Other transfer/carrier protein | 110 | 9 | 0.51 | 0.000000507 |
| Protease | 531 | 14 | 2.47 | 0.0000059 |
| Defense/immunity protein | 362 | 10 | 1.69 | 0.000241 |
| Serine protease | 175 | 8 | 0.82 | 0.000308 |
| Cysteine protease inhibitor | 20 | 4 | 0.09 | 0.000561 |
| Select regulatory molecule | 1152 | 17 | 5.37 | 0.00072 |
| Apolipoprotein | 26 | 4 | 0.12 | 0.00125 |
| Peroxidase | 28 | 4 | 0.13 | 0.00167 |
| Complement component | 46 | 4 | 0.21 | 0.0114 |
| Oxidoreductase | 582 | 9 | 2.71 | 0.0482 |
EPS proteins are secreted proteins. Secretory proteins can be divided into two categories: classical secretory proteins with signal peptides and nonclassical secretory proteins including those without signal peptides and those that are shed from cell-surface membrane proteins. To assess the distribution of the EPS proteins between these categories, we used the signal-peptide prediction servers SignalP 3.0 and SecretomeP 2.0 (www.cbs.dtu.dk/services/). We found that 67 EPS proteins have signal peptides as predicted by the SignalP program and that they are classical secreted proteins (Table 3 and Supporting Information Document 5). The rest are probably nonclassical secreted proteins or are shed proteins.
To understand the function of these EPS proteins in prostate, we retrieved the protein–protein interactions of the EPS proteins from the HPRD and the NCBI databases (www.hprg.org; www.ncbi.nlm.nih.gov/) and used Cytoscape (www.cytoscape.org) to display the network (Fig. 1). This network suggests that many EPS proteins are connected by protein–protein interactions and, therefore, function in concert in the prostate. This network will provide a basis for mapping disease genes or biomarkers onto the network and will help us to identify key therapeutic targets and a panel of biomarkers that reflects the broad physiological function of the prostate. Such a biomarker panel may better reflect the heath state of the prostate than any single marker alone.
3.4 MS coverage of the putative PSA peptides
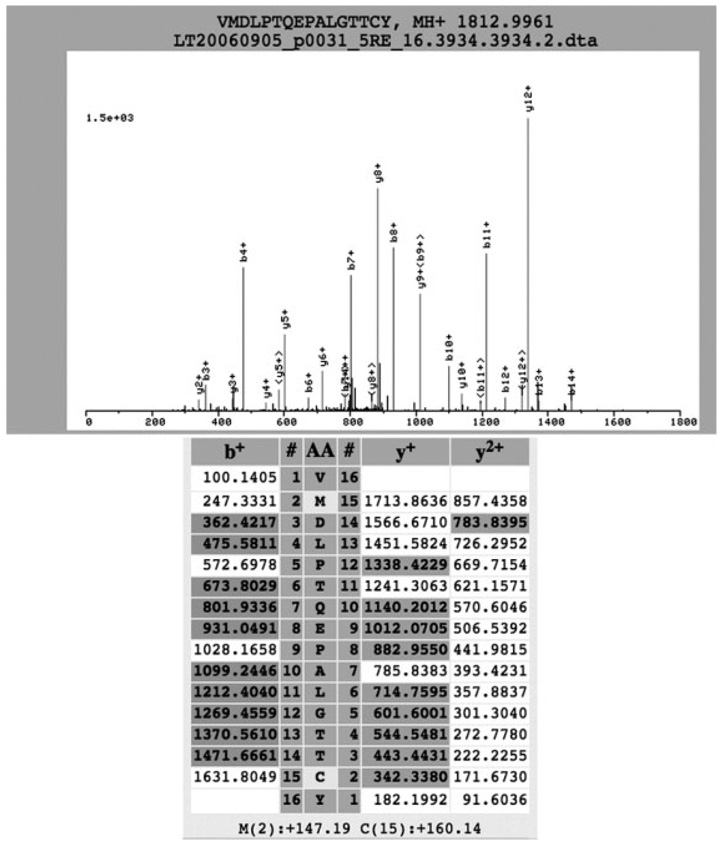
As PSA is currently the marker for prostate cancer diagnosis and as we have identified multiple peptides by MS that correspond to the PSA protein, we thought it might be interesting to study these PSA peptides in detail. In silico digestion of PSA produces 22 double-tryptic peptides, and about 13 of these peptides are in the MW range of 500–3500, detectable by the mass spectrometer with the settings that we used. We identified 26 peptides (Supporting Information Document 6) specific to the PSA protein (with an adjusted PeptideProphet™ p>0.7 and manual inspection of the spectra), which corresponds to a coverage of 54% (141 out of 261 amino acid residues). To our knowledge, this provides the best coverage in identification of PSA by MS. We identified five double-tryptic peptides and 21 single-tryptic peptides. The identification of many single-tryptic sites was probably caused by other proteases in the EPS that digest PSA prior to our trypsin digestion, as EPS is rich in proteases. We also identified oxidized methionine (+16) at positions 122 and 139 (Fig. 2 and Supporting Information Document 6) and a double-tryptic peptide WIKDTIVANP with an uncleaved tryptic site. This information may be useful in understanding the ionization properties of the PSA peptides and the potential native cleavage sites of PSA by endogenous enzymes.
Figure 2.
The tandem mass spectra of the PSA peptide VMDLPTQEPALGTTCY. The methionine is oxidized (+16) and the cysteine is carbamidated (+57). The top panel shows the MS/MS spectrum, and the bottom shows the corresponding identified b and y ions (highlighted).
4 Discussion
Using micro-LC ESI MS, we cataloged the proteome of EPS. Using stringent criteria, we identified a list of 114 proteins (Table 1) with at least two unique-peptide hits and an additional 75 proteins that have only a single unique-peptide hit. The PeptideProphet™ scores for these peptides are all greater than 0.9, and some of the single unique peptides were observed multiple times.
Proteins identified with only a single unique peptide but seen multiple times (Supporting Information Documents 2 and 3) may also represent true EPS proteins. For example, KLK11 was identified in the EPS by a single unique peptide that was observed twice. Diamandis et al. [13] showed that KLK11 existed in seminal plasma and further demonstrated that KLK11 is useful for discriminating between prostate cancer and BPH when used in combination with PSA [14]. Our list does not represent a complete list of the EPS proteins, as there may be scarce proteins that were not identified due to the limited dynamic range of our proteomics technology and due to our limited protein amount. As we have analyzed samples from eight individuals separately (i.e., eight micro-LC-MS/MS), we believe that we have identified most of the proteins in the detection range of our proteomics capabilities.
In this cataloging experiment, we have analyzed EPS from normal individual, BPH, and prostate cancer patients. We also used spectrum counting [15], a semiquantitative method for label-free quantitative proteomics analysis of complex samples. However, due to small sample sizes and the fact that we used as much protein as possible from what we got for each individual to get the maximum recovery, the protein amount was not normalized. Because of these concerns, we did not believe that the results were reliable and therefore have decided not to include them in this manuscript. Since we had EPS from only one normal individual but had EPS from multiple cancer patients and since the protein list was derived from the EPS of normal and disease samples, the analysis may be biased toward the diseased EPS.
We observed that the majority of the EPS proteins could be categorized under the GO biological processes of immunity and defense, proteolysis, transport, and lipid and fatty acid transport. EPS has been shown to have antibacterial activity which has been attributed to the high zinc concentration and to the existence of Igs [16–18]. Our identification of 35 proteins involved in immunity and defense extended our understanding of EPS’s role in antibacterial activity and immunity. These proteins may help to distinguish different types of chronic prostatitis (CP). It is difficult to diagnose and differentiate different types of CP such as chronic bacterial prostatitis, chronic nonbacterial prostatitis and recurrent nonspecific urethritis (NSU) without prostatitis; because the symptoms are ambiguous [19]. Treatments for CP are not effective and produce inconsistent results.
The EPS proteins are also enriched with proteins involved in proteolysis, transport, and lipid and fatty acid transport. Prostate contributes 25–30% of the proteins in human semen [20]. We identified 23 EPS proteins that are involved in proteolysis and 22 proteins involved in transport. These proteins may perform important functions in human semen including semen liquefaction and fertility.
Finding biomarkers directly from serum by MS-based proteomics technology has turned out to be extremely challenging due to the low dynamic detection range of current mass spectrometers and the extremely high dynamic range (up to 10–12 orders of magnitude) of serum proteins [21]. Finding biomarkers directly in blood is a difficult task as potential cancer biomarkers secreted from a tissue would be diluted in about 5 L of blood, the volume of blood in an average human adult. Body fluid that is in close proximity to a tissue or organ may have a higher concentration of proteins (biomarkers) that reflect the disease state, such as cancer, of the tissue or organ compared to serum or plasma. Therefore, identifying biomarker candidates from tissues and body fluids first and then validating them in serum is an approach that many researchers are taking now.
By comparing the proteins identified from EPS with the proteins identified from sera (data not shown) using direct proteomics analysis, we found that we were able to identify many more known cancer biomarkers from EPS than from serum samples. The proteins identified from EPS are also present in normal serum samples, albeit at lower concentrations. For example, without any abundant protein removal, we were able to identify three KLKs (PSA, KLK2, and KLK11) from EPS while we were not able to identify any of these KLKs from serum directly (data not shown). It is well-known that the abundant proteins prevent the identification of lowly expressed, possible bio-markers by MS. This is exactly the obstacle that makes it extremely difficult to successfully discover biomarkers directly from the blood, as blood proteins span a concentration range of at least ten orders of magnitude [21] and the 22 most abundant proteins constitute 99% of the total blood protein mass [22].
EPS proteins are directly derived from the prostate and changes in their protein expression patterns or levels may be used as diagnostic indicators of prostate diseases including prostatitis, BPH, and prostate cancer; and may be also used for disease classification such as differentiating different kinds of CP or different types or stages of prostate cancer. Such applications have already been used. Elevation of dipeptidylpeptidase IV (CD26) activity in prostatic secretions of men with prostate cancer was identified as a possible prostate cancer disease marker [23]. The prostate cancer molecular markers GSTP1 and hTERT in EPS were used as predictors of biopsy results [24]. Interleukin-1 beta (IL-1 β) and tumor necrosis factor alpha (TNF-α) in the EPS were used as markers for CP [25].
The EPS proteins that we identified consist of many interesting ones. For example, we identified nine CD molecules including CD10, CD13, CD14, CD26, CD 66a, CD 66c, CD 143, CD177, and CD224. Many of these CD markers are cell-surface enzymes or adhesion molecules that play important functions in prostate development and prostate cancer. These genes may also be targets for prostate cancer therapies. Loss of CD10 (neutral endopeptidase) is a frequent and early event in human prostate cancer [26]. CD13 expression is absent in most tumor cells, and a decreased frequency of CD26 (DPP IV) expression was found in metastatic tumors [27]. Circulation of PSA/CD14 double-positive cells was found to be a biomarker indicating low risk for hematogenous metastasis of prostate cancer [28]. CD66a (CEACAM1) is an adhesion molecule of the carcinoembryonic antigen family, and its protein expression is lost in most prostate tumors [29]. Downregulation of CD66a (CEACAM1) in human prostate cancer is correlated with loss of cell polarity, increased proliferation rate, and a Gleason grade 3 to 4 transition [30].
Many of the EPS proteins that we identified have been implicated in prostate cancer. For example, KLK2 and KLK3 are well-known examples of prostate cancer biomarkers [31]. By 2-DE and MS analyses protein disulfide isomerase was shown to be over expressed in prostate cancer compared to its expression in benign tissues [32]. Calcium-binding proteins S100A8 and S100A9 were observed to be upregulated in prostatic intraepithelial neoplasia and preferentially expressed in high-grade adenocarcinomas by immunohistochemistry staining when compared to benign tissue that was negative for or showed weak expression of these proteins [33]. S100A9 serum levels were significantly elevated in cancer patients compared with BPH patients or healthy individuals [33]. Prosaposin (PSAP) was identified as a secreted protein expressed in androgen-independent (AI) prostate cancer cells, and its genomic region was shown to be amplified in the metastatic AI prostate cancer cell lines PC-3, DU-145, MDA-PCa 2b, M-12, and NCI-H660; in LuCaP 58; and in two lymph-node metastases [34]. In addition, PSAP mRNA was shown to be over expressed without evidence of genomic amplification in the AI metastatic prostate cancer cell lines C4-2B and IA8-ARCaP [34]. Annexin I protein expression is downregulated in high-grade prostatic intraepithelial neoplasia (HGPIN) and prostate cancer [35]. Clusterin expression is significantly enhanced in prostate cancer cells following androgen-withdrawal therapy, confers resistance to androgen ablation, and helps accelerate the progression to androgen independence [36]. Aberrant expression of cystatin C in prostate cancer is associated with neuroendocrine differentiation [37]. Some of the EPS proteins, although not well studied in prostate cancer, were studied in other types of cancers. For example, Zhang et al. [38] applied serum-proteomics analysis and identified apolipoprotein A1, transthyretin, and interalpha-trypsin inhibitor heavy chain H4 as markers for ovarian cancer.
We, as well as many other investigators, have taken the approach of first identifying candidate biomarkers using mRNA-expression profiling to compare mRNA expression between normal tissue and the diseased tissue and then assessing their potential as biomarkers in serum. However, which of these differentially expressed genes are likely to produce proteins that are secreted or shed into the serum? We applied the in silico prediction programs SecretomeP, TargetP, and SingalP (http://www.cbs.dtu.dk/services/), and a computationly predicted human secretome database was constructed [39]; however, direct evidence for secretion of a protein will come from proteome analysis of body fluids including the EPS which we have analyzed here.
A systematic comparison of the EPS proteome with the available microarray data of prostate tissues (cancer and normal) (www.Oncomine.org) reinforced our initial observation that EPS proteins are a rich source of putative biomarkers. We identified a list of 48 EPS proteins that have at least one microarray dataset which compares normal and cancer samples and have more than 50% of the array data corroborating their upregulation in prostate cancer. Integrated analysis with the transcriptomics data from normal human tissues identified 26 EPS proteins that are prostate-specific/enriched including well-known prostate-specific genes such as KLK2, PSA, and CD10. We identified six transmembrane epithelial antigen of the prostate 4 (STEAP4) as an EPS protein, the expression of which is enriched in prostate tissue (enrichment score of 1.74). Other genes in the STEAP family such as STEAP1 and STEAP2 have been identified in prostate tissues, and their expressions were related to prostate cancer [40, 41]. STEAP4 may be a good candidate marker for prostate cancer as well, and therefore it is worthwhile to investigate this possibility further. Unfortunately, there is no antibody available against STEAP4. We are currently generating antibodies against this gene and will further evaluate this gene as a marker for prostate cancer.
In summary, we have characterized the proteome of EPS proteins, which provides a starting point and a candidate list of biomarkers for prostate cancer. Integrative analysis with both existing data and our unpublished data suggested many potential candidate biomarkers. Network analysis revealed that many EPS proteins form a network working in concert. Changes in the network may reflect the biological state of the prostate. One approach is to use EPS directly for prostate cancer diagnosis, which offers a less invasive means than performing biopsies. Using EPS directly as a route for diagnosis may be more accurate than prostate biopsies as it probably represents the secretion from the whole prostate while biopsies often sample only a few locations in the prostate making it possible that certain cancerous locations are missed. However, it may be technically difficult to obtain enough EPS from prostate cancer patients based on our experience. Another strategy is to evaluate the EPS markers in serum samples as biomarkers for prostate cancer by using antibody-based approaches as well as by combining these with the use of MRM-based MS.
In conclusion, we believe that our list may provide a rich source of biomarkers for prostate diseases such as prostate cancer, BPH, and prostatitis. The potential biomarkers can be evaluated further by quantitative targeted proteomics, heralded recently by Malmstrom et al. [6] to complement the shotgun-proteomics approach.
Supplementary Material
Acknowledgments
This work was partially supported by grants 5P50GM076547 and 5U54CA119347 from NIH, USA, by a grant from the National Basic Research Program of China (2006CB504005) (R. Li), and a grant from the Science and Technology Commission of Shanghai Municipality (05dz22321) (B. Lin). We thank James White for critical reading of the manuscript.
Abbreviations
- BPH
benign prostatic hyperplasia
- CD
cluster of differentiation
- CP
chronic prostatitis
- EPS
expressed prostatic secretions
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GO
gene ontology
- KLK
kallikrein
- MPSS
massively parallel signature sequencing
- PAP
prostatic acid phosphatase
- PSA
prostate-specific antigen
- PSAP
prosaposin
- STEAP
six trans-membrane epithelial antigen of the prostate
Footnotes
The authors have declared no conflict of interest.
References
- 1.Lee C, Tsai Y, Sensibar J, Oliver L, Grayhack JT. Two-dimensional characterization of prostatic acid phosphatase, prostatic specific antigen and prostate binding protein in expressed prostatic fluid. Prostate. 1986;9:135–146. doi: 10.1002/pros.2990090204. [DOI] [PubMed] [Google Scholar]
- 2.Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, et al. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin. Cancer Res. 2004;10:7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- 3.Ramstrom M, Ivonin I, Johansson A, Askmark H, et al. Cerebrospinal fluid protein patterns in neurodegenerative disease revealed by liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:4010–4018. doi: 10.1002/pmic.200400871. [DOI] [PubMed] [Google Scholar]
- 4.Cottingham K. The cervical-vaginal fluid proteome and possible biomarkers of preterm birth. J. Proteome. Res. 2007;6:1241–1242. doi: 10.1021/pr070737o. [DOI] [PubMed] [Google Scholar]
- 5.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malmstrom J, Lee H, Aebersold R. Advances in proteomic workflows for systems biology. Curr. Opin. Biotechnol. 2007;1S:378–384. doi: 10.1016/j.copbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhodes DR, Yu J, Shanker K, Deshpande N, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jongeneel CV, Delorenzi M, Iseli C, Zhou D, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome. Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utleg AG, Yi EC, Xie T, Shannon P, et al. Proteomic analysis of human prostasomes. Prostate. 2003;56:150–161. doi: 10.1002/pros.10255. [DOI] [PubMed] [Google Scholar]
- 10.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 11.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 12.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 13.Diamandis EP, Okui A, Mitsui S, Luo LY, et al. Human kallikrein 11: A new biomarker of prostate and ovarian carcinoma. Cancer Res. 2002;62:295–300. [PubMed] [Google Scholar]
- 14.Nakamura T, Scorilas A, Stephan C, Jung K, et al. The usefulness of serum human kallikrein 11 for discriminating between prostate cancer and benign prostatic hyperplasia. Cancer Res. 2003;63:6543–6546. [PubMed] [Google Scholar]
- 15.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 16.Fair WR, Parrish RF. Antibacterial substances in prostatic fluid. Prog. Clin. Biol. Res. 1981;75A:247–264. [PubMed] [Google Scholar]
- 17.Fowler JE, Jr, Kaiser DL, Mariano M. Immunologic response of the prostate to bacteriuria and bacterial prostatitis. I. Immunoglobulin concentrations in prostatic fluid. J. Urol. 1982;128:158–164. doi: 10.1016/s0022-5347(17)52810-4. [DOI] [PubMed] [Google Scholar]
- 18.Fowler JE., Jr Secretory immunity of the prostate gland. Infection. 1991;19:S131–S137. doi: 10.1007/BF01643682. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez-Cruz JF, Broseta-Rico E. Classification, etiology, diagnosis and treatment of prostatitis. Other types of prostatitis. Acute and chronic prostatitis. Enferm. Infecc. Microbiol. Clin. 2005;23:47–56. doi: 10.1157/13091448. [DOI] [PubMed] [Google Scholar]
- 20.Mandal A, Bhattacharyya AK. Biochemical composition of washed human seminal coagulum in comparison to sperm-free semen from the same donors. J. Reprod. Fertil. 1990;88:113–118. doi: 10.1530/jrf.0.0880113. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, et al. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Wilson MJ, Haller R, Li SY, Slaton JW, et al. Elevation of dipeptidylpeptidase iv activities in the prostate peripheral zone and prostatic secretions of men with prostate cancer: Possible prostate cancer disease marker. J. Urol. 2005;174:1124–1128. doi: 10.1097/01.ju.0000168621.84017.5c. [DOI] [PubMed] [Google Scholar]
- 24.Crocitto LE, Korns D, Kretzner L, Shevchuk T, et al. Prostate cancer molecular markers GSTP1 and hTERT in expressed prostatic secretions as predictors of biopsy results. Urology. 2004;64:821–825. doi: 10.1016/j.urology.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Ye L, Jiang H, Zhou J, et al. Clinical evaluation of IL-1beta and TNF-alpha in prostatic secretions for chronic prostatitis. Zhonghua Nan Ke Xue. 2004;10:449–450. 454. [PubMed] [Google Scholar]
- 26.Freedland SJ, Seligson DB, Liu AY, Pantuck AJ, et al. Loss of CD10 (neutral endopeptidase) is a frequent and early event in human prostate cancer. Prostate. 2003;55:71–80. doi: 10.1002/pros.10202. [DOI] [PubMed] [Google Scholar]
- 27.Bogenrieder T, Finstad CL, Freeman RH, Papandreou CN, et al. Expression and localization of aminopeptidase A, aminopeptidase N, and dipeptidyl peptidase IV in benign and malignant human prostate tissue. Prostate. 1997;33:225–232. doi: 10.1002/(sici)1097-0045(19971201)33:4<225::aid-pros1>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Brandt B, Griwatz C, Brinkmann O, Zanker KS. Circulating prostate-specific antigen/CD14 double-positive cells: A biomarker indicating low risk for hematogeneous metastasis of prostate cancer. J. Natl. Cancer Inst. 1997;89:174. doi: 10.1093/jnci/89.2.174. [DOI] [PubMed] [Google Scholar]
- 29.Luo W, Tapolsky M, Earley K, Wood CG, et al. Tumor-suppressive activity of CD66a in prostate cancer. Cancer Gene Ther. 1999;6:313–321. doi: 10.1038/sj.cgt.7700055. [DOI] [PubMed] [Google Scholar]
- 30.Busch C, Hanssen TA, Wagener C, O’Brink B. Down-regulation of CEACAM1 in human prostate cancer: Correlation with loss of cell polarity, increased proliferation rate, and Gleason grade 3 to 4 transition. Hum. Pathol. 2002;33:290–298. doi: 10.1053/hupa.2002.32218. [DOI] [PubMed] [Google Scholar]
- 31.Henttu P, Vihko P. Prostate-specific antigen and human glandular kallikrein: Two kallikreins of the human prostate. Ann. Med. 1994;26:157–164. doi: 10.3109/07853899409147884. [DOI] [PubMed] [Google Scholar]
- 32.Lexander H, Palmberg C, Auer G, Hellstrom M, et al. Proteomic analysis of protein expression in prostate cancer. Anal. Quant. Cytol. Histol. 2005;27:263–272. [PubMed] [Google Scholar]
- 33.Hermani A, Hess J, De Servi B, Medunjanin S, et al. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin. Cancer Res. 2005;11:5146–5152. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 34.Koochekpour S, Zhuang YJ, Beroukhim R, Hsieh CL, et al. Amplification and overexpression of prosaposin in prostate cancer. Genes Chromosomes Cancer. 2005;44:351–364. doi: 10.1002/gcc.20249. [DOI] [PubMed] [Google Scholar]
- 35.Kang JS, Calvo BF, Maygarden SJ, Caskey LS, et al. Dysregulation of annexin I protein expression in high-grade prostatic intraepithelial neoplasia and prostate cancer. Clin. Cancer Res. 2002;8:117–123. [PubMed] [Google Scholar]
- 36.July LV, Akbari M, Zellweger T, Jones EC, et al. Clusterin expression is significantly enhanced in prostate cancer cells following androgen withdrawal therapy. Prostate. 2002;50:179–188. doi: 10.1002/pros.10047. [DOI] [PubMed] [Google Scholar]
- 37.Jiborn T, Abrahamson M, Gadaleanu V, Lundwall A, Bjartell A. Aberrant expression of cystatin C in prostate cancer is associated with neuroendocrine differentiation. BJU Int. 2006;98:189–196. doi: 10.1111/j.1464-410X.2006.06345.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Bast RC, Jr, Yu Y, Li J, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Yu P, Luo J, Jiang Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome. 2003;14:859–865. doi: 10.1007/s00335-003-2296-6. [DOI] [PubMed] [Google Scholar]
- 40.Porkka KP, Helenius MA, Visakorpi T. Cloning and characterization of a novel six-transmembrane protein STEAP2, expressed in normal and malignant prostate. Lab. Invest. 2002;82:1573–1582. doi: 10.1097/01.lab.0000038554.26102.c6. [DOI] [PubMed] [Google Scholar]
- 41.Hubert RS, Vivanco I, Chen E, Rastegar S, et al. STEAP: A prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc. Natl. Acad. Sci. USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.