Abstract
The control and eventual eradication of human malaria is considered one of the most important global public health goals of the 21st Century. Malaria, caused by intraerythrocytic protozoan parasites of the genus Plasmodium, is by far the most lethal and among the most prevalent of the infectious diseases. Four species of Plasmodium (P. falciparum, P. malariae, P. ovale, and P. vivax) are known to be infectious to humans, and more recent cases of infection due to P. knowlesi also have been reported. These species cause approximately 300 million annual cases of clinical malaria resulting in around one million deaths mostly caused by P. falciparum. The rapid emergence of drug-resistant Plasmodium strains has severely reduced the potency of medicines commonly used to treat and block the transmission of malaria and threatens the effectiveness of combination therapy in the field. New drugs that target important parasite functions, which are not the target of current antimalarial drugs, and have the potential to act against multi-drug-resistant Plasmodium strains are urgently needed. Recent studies in P. falciparum have unraveled new metabolic pathways for the synthesis of the parasite phospholipids and fatty acids. The present review summarizes our current understanding of these pathways in Plasmodium development and pathogenesis, and provides an update on the efforts underway to characterize their importance using genetic means and to develop antimalarial therapies targeting lipid metabolic pathways.
Keywords: malaria, lipid, metabolism, Plasmodium, therapy
INTRODUCTION
The pathological stage of malaria occurs following the invasion and subsequent destruction of human red blood cells by Plasmodium species. During a 48-hour intraerythrocytic lifespan, a single P. falciparum parasite invades a human red blood cell and, as it grows, consumes most of its host hemoglobin and initiates several nuclear divisions to produce a syncytium of up to 36 nuclei. Subsequently, the parasite undergoes cytokinesis followed by cellularization, thereby enveloping each new daughter parasite with a plasma membrane. It then completes its feast by destroying the host cell, releasing the newly made parasites in a state competent to repeat the cycle. This rapid growth and multiplication is fueled by precursors supplied by the host. Of these, fatty acids, serine, ethanolamine, and choline are of particular importance as they are the major building blocks used by the parasite in the synthesis of its structural and regulatory phospholipids. In addition to the transport of exogenous fatty acids, the parasite expresses all the enzymes for synthesis of fatty acids. Out of the biochemical, genetic, and pharmacological studies performed in P. falciparum and P. berghei, it became evident that the metabolic pathways of the synthesis of phospholipids and fatty acids play a crucial role in the growth and proliferation of Plasmodium species during the various stages of their life cycle.
BIOSYNTHESIS OF THE MAJOR PHOSPHOLIPIDS
Plasmodium infection is followed by a marked increase in the phospholipid content and a significant change in the lipid composition of the infected erythrocyte [Vial and Ben Mamoun, 2005], a phenomenon that is consistent with their need for large amounts of new membranes to achieve successful growth and proliferation. Like the majority of eukaryotes, phosphatidylcholine (PtdCho) is the major phospholipid in P. falciparum membranes. In most eukaryotes, including various protozoan parasites, phosphatidylcholine is synthesized by two routes. Synthesis can occur from choline via an enzymatic cascade (the de novo cytidine diphosphate (CDP)-choline pathway) involving three enzymes: choline kinase, CTP phosphocholine cytidylyltransferase, and choline/ethanolamine-phosphotransferase. The second route is from phosphatidylethanolamine (PtdEtn) via three transmethylation reactions that involve one or two phospholipid methyltransferases. Labeling studies in P. knowlesi-infected erythrocytes suggested that the methylation of PtdEtn into PtdCho occurs in this parasite [Moll et al., 1988]. Surprisingly, no phospholipid methyltransferase activity has been detected in P. falciparum extracts and no genes encoding homologs of phospholipid methyltransferases have been found in the P. falciparum genome [Pessi et al., 2004]. Consistent with these observations, analysis of the membrane composition of purified P. falciparum parasites revealed high levels of phosphatidylethanolamine (35–45%) in this parasite [Vial and Ancelin, 1998]. High levels of this phospholipid have also been reported in isolated merozoites of Babesia bovis, another apicomplexan parasite [Florin-Christensen et al., 2000]. However, the finding that P. falciparum is capable of normal growth when cultured in the absence of exogenous choline [Divo et al., 1985; Mitamura et al., 2000; Witola and Ben Mamoun, 2007] indicated the existence of an alternative pathway for the synthesis of phosphatidylcholine in this parasite.
CDP-Choline Pathway
The de novo CDP-choline (Kennedy) pathway for the synthesis of PtdCho (Fig. 1) initiates with the transport of choline from host serum into the infected erythrocyte, a process that involves the remnant erythrocytic choline carrier and the new permeation pathway induced by the parasite [Ancelin et al., 1991; Kirk et al., 1991]. A poly-specific organic cation transporter (OCT) functionally distinct from the known dedicated eukaryotic choline carriers, and whose gene has yet to be identified, mediates choline entry into the parasite [Biagini et al., 2004; Lehane et al., 2004]. Choline is then phosphorylated to phosphocholine by a parasite-specific choline kinase (PfCK), and subsequently coupled to CTP to generate CDP-Cho by a CDP-choline cytidylyltransferase (PfCCT) and further converted into PtdCho by a parasite CDP-diacylglycerol-cholinephosphotransferase (PfCEPT) in the presence of diacylglycerol. A similar de novo pathway exists in Plasmodium parasites for the synthesis of PtdEtn from ethanolamine [Vial and Ben Mamoun, 2005]. In mammalian cells, CCTs contain a conserved catalytic domain followed by a membrane-binding region and a phosphorylation site (Fig. 2). These enzymes are amphitropic, operating in an inactive soluble state and an active membrane-bound state [Attard et al., 2000; Davies et al., 2001]. In Plasmodium, CCT is the rate-limiting step enzyme of the PtdCho pathway. The P. falciparum enzyme consists of 899 amino acids and harbors an unusual duplication of the catalytic and membrane-binding domains (Fig. 2). Genetic studies in the rodent malaria parasite P. berghei failed to isolate knockout parasites lacking PbCK, PbCCT, PbECT, or PbECPT genes [Dechamps et al., unpublished data]. These findings suggest that these genes might play an essential function in P. berghei intraerythrocytic development and survival. Genetic studies in the human malaria parasites are needed to determine the importance of the genes involved in the CDP-choline pathway and validate them as possible drug targets.
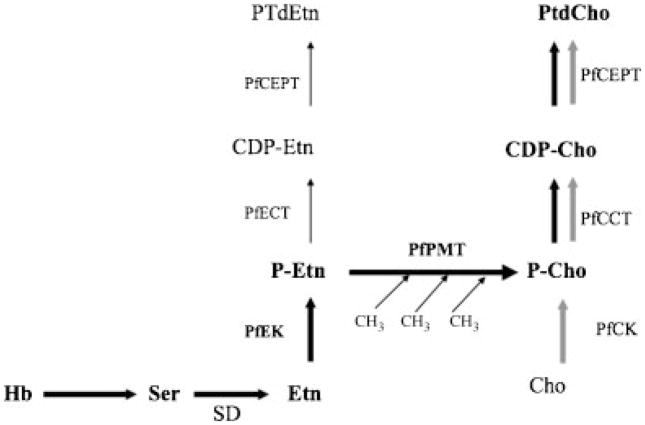
Fig. 1.
SDPM and CDP-choline pathways for phosphatidylcholine biosynthesis in P. falciparum. The cytidine diphosphate (CDP)-choline pathway is shown in gray. The SDPM pathway is represented in black. Cho: choline; HB: hemoglobin; Ser: serine; Etn: ethanolamine; CDP-Etn: CDP-ethanolamine; CDP-cho: CDP-choline; SD: serine decarboxylase; PfEK: P. falciparum ethanolamine kinase; PfCK: P. falciparum choline kinase; PfPMT: P. falciparum phosphoethanolamine methyltransferase; PtdEtn: phosphatidylethanolamine; PtdCho: phosphatidylcholine; PfCEPT: P. falciparum choline/ethanolamine-phosphate transferase; PfECT: P. falciparum CTP: phosphoethanolamine cytidylyltransferase; PfCCT: P. falciparum CTP: phosphocholine cytidylyltransferase.
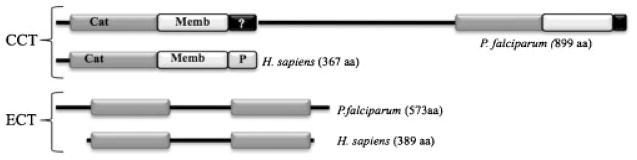
Fig. 2.
Comparison between P. falciparum and human CCT and ECT sequences. Cat: catalytic domain; Memb: membrane-binding domain; P: phosphorylation domain of the human CCT. Only Plasmodium CCT is duplicated, while all ECTs described so far are duplicated.
Phosphatidylcholine Biosynthesis From Serine/Ethanolamine in P. falciparum
The mechanism of P. falciparum phosphatidylcholine biosynthesis from non-choline precursors was demonstrated by lipid analysis following labeling of parasite-infected erythrocytes with radiolabeled ethanolamine or serine [Pessi et al., 2004]. Analysis of parasite-extracted lipid fractions revealed the formation of both phosphatidylethanolamine and phosphatidylcholine, whereas analysis of the soluble metabolites revealed the formation of phosphocholine but not choline from these precursors [Pessi et al., 2004]. Furthermore, addition of S-adenosylmethionine (SAM) and phosphoethanolamine to parasite protein preparations resulted in the production of phosphocholine [Pessi et al., 2004]. The combined results of these studies suggested that P. falciparum possesses an alternative pathway for the synthesis of phosphatidylcholine from serine and ethanolamine, which involves the methylation of phosphoethanolamine to form phosphocholine (Fig. 1). This pathway, which was termed the serine decarboxylase–phosphoethanolamine methyltransferase (SDPM) pathway [Pessi and Ben Mamoun, 2006], is also found in plants and nematodes [Bolognese and McGraw, 2000; Brendza et al., 2007; Charron et al., 2002; Nuccio et al., 2000b; Palavalli et al., 2006], but is absent in mammals, thus making it an excellent target for the development of new antimalarial drugs. Ethanolamine can be obtained in limited amounts from plasma and in larger quantities following serine decarboxylation [Elabbadi et al., 1997] by a parasite-encoded serine decarboxylase from serine either transported from the host or obtained from degradation of host hemoglobin. Ethanolamine formed via this reaction is subsequently phosphorylated into phosphoethanolamine, which serves as a substrate for PtdEtn biosynthesis, or is converted into phosphocholine and incorporated into PtdCho via the SDPM pathway.
Isolation and Functional Characterization of P. falciparum Phosphoethanolamine Methyltransferase
The P. falciparum enzyme responsible for the synthesis of phosphocholine from phosphoethanolamine (PfPMT) was identified by searching the P. falciparum genome database for proteins that contained a SAM-binding domain and shared sequence homology with plant phosphoethanolamine methyltransferases (PMTs). PfPMT, the gene encoding this enzyme, was cloned from P. falciparum genomic DNA. Cloning and characterization of PfPMT cDNA indicated that it encodes a polypeptide of 266 amino acids [Pessi et al., 2004]. PfPMT is expressed throughout the intraerythrocytic life cycle of the parasite with maximum expression detected during the trophozoite stage, at the peak of membrane biosynthesis [Pessi et al., 2004]. Genome sequencing efforts have thus far identified homologs of PfPMT in many species of plants, two species of African clawed frogs (Xenopus laevis and X. tropicalis), nematodes (Caenhorhabditis elegans and C. briggsae), zebrafish (Danio rerio), the florida lancelet (Brachiostoma floridae), and two other Plasmodium parasites (P. vivax and P. knowlesi). Based on their primary structure and distribution of their predicted catalytic domains, these enzymes can be divided into 4 classes: Class I includes the malarial enzymes, which are 264 (P. vivax and P. knowlesi) to 266 (PfPMT) amino acids in length and contain a single SAM-dependent catalytic domain; Class II is comprised of bipartite enzymes containing between 450–580 amino acids and harboring two SAM-dependent catalytic domains. This class includes PMTs from plants, frogs, zebrafish, and Florida lancelets; Class III and IV include enzymes from C. elegans and C. briggsae that are twice the size of the malarial enzymes but contain a single SAM-dependent catalytic domain located either at the N-terminal (Class III) or C-terminal (Class IV) end (Fig. 3). The amino acid sequence of PfPMT shows no specific organelle targeting signals, and no recognizable transmembrane domains. Analysis of the available genomes of various Plasmodium species indicates the presence of orthologs of PfPMT in P. vivax and P. knowlesi, and their absence in the rodent malaria parasites P. berghei and P. yoelii. Importantly, no homologs of PfPMT have been found in human or other mammalian databases, and PfPMT does not share homology with eukaryotic phosphatidylethanolamine methyltransferase proteins. Immunoelectron and immunofluorescence microscopy studies revealed that PfPMT localizes to the Golgi apparatus of the parasite [Witola et al., 2006]. This is the first enzyme in this pathway to be localized, and it is consistent with at least the transmethylation reaction occurring in the Golgi apparatus. The subcellular localization of the rest of the enzymes involved in the phosphatidylcholine/phosphatidylethanolamine synthesis pathways remains to be determined. When expressed in E. coli, recombinant PfPMT was shown to catalyze the conversion of phosphoethanolamine into phosphocholine using SAM as a methyl donor [Pessi et al., 2004]. Neither ethanolamine nor phosphatidylethanolamine acted as substrates for PfPMT, implying that phosphoethanolamine is the primary methyl acceptor of this enzyme [Pessi et al., 2004].
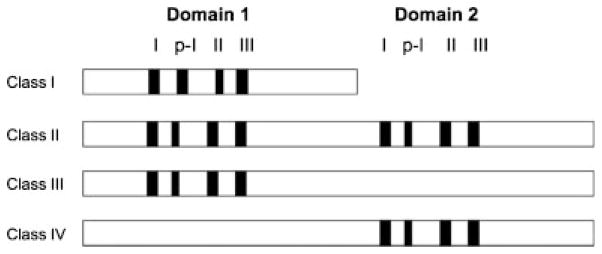
Fig. 3.
Schematic representation of the structure of the four classes of PMT enzymes. The four motifs (I, p-I, II, and III) of each PMT catalytic domain(s) are indicated as black boxes.
Functional analysis of PfPMT activity in vivo was determined using yeast as a surrogate system. Wild-type yeast cells inherently lack phosphoethanolamine methyltransferase activity. Expression of a codon-optimized PfPMT gene in yeast conferred phosphoethanolamine methylation activity on these cells [Pessi et al., 2005]. Furthermore, the expression of PfPMT in pem1Δpem2Δ yeast, which lack the ability to convert phosphatidylethanolamine to phosphatidylcholine and hence are auxotrophic for choline [Kodaki and Yamashita, 1987; Summers et al., 1988], restored the ability of these cells to grow in the absence of choline [Pessi et al., 2005; Reynolds et al., 2008]. Analysis of the phospholipid content revealed that, unlike wild-type yeast cells, PfPMT-expressing pem1Δpem2Δ yeast cells failed to synthesize the intermediates of the methylation of PtdEtn (monomethyl- and dimethyl-phosphatidylethanolamine). The growth of these complemented cells was ameliorated by the addition of choline, and required an active CDP-choline pathway. Altogether, these findings are consistent with phosphatidylcholine being synthesized via the CDP-choline pathway following the PfPMT-dependent production of phosphocholine from phosphoethanolamine [Pessi et al., 2005], and suggest that the in vivo activity of PfPMT is directly coupled to the CDP-choline pathway.
Regulation of the SDPM Pathway
Unlike other organisms, the regulation of phospholipid metabolism in P. falciparum has only started to be elucidated. Studies by Elabbadi and colleagues indicated that distinct pools of PtdEtn are synthesized by P. falciparum from different precursors and via the de novo pathway from ethanolamine or following PtdSer decarboxylation, suggesting a possible compartmentalization of these metabolic pathways. However, it remains to be determined whether the genes encoding enzymes of the CDP-ethanolamine pathway or the PtdSer decarboxylase gene are regulated by their precursors. Evidence for the regulation of synthesis of PtdCho by its precursors has been established using wild-type and transgenic P. falciparum parasites. It was shown that exogenous choline leads to repression of transcription of PfPMT as well as the induction of its proteasomal degradation [Witola and Ben Mamoun, 2007]. Addition of exogenous choline to cultures of wild-type parasites produced a dose-dependent reduction in the amount of both PfPMT transcript and protein [Witola and Ben Mamoun, 2007]. The choline-mediated transcriptional response was not evident in transgenic parasites expressing PfPMT under the transcriptional control of a heterologous promoter, whereas PfPMT protein degradation persisted in these parasites. These findings suggest that the promoter of PfPMT may contain elements important for transcriptional regulation by choline (or its phosphorylated form), and that this substrate may also regulate PfPMT expression posttranscriptionally. Interestingly, the proteasome inhibitor, bortezomib, inhibited choline-induced repression of the PfPMT protein, indicating that proteasome activity is responsible for PfPMT degradation in the presence of choline [Witola and Ben Mamoun, 2007]. Studies to dissect the promoter of PfPMT are underway and may shed light on the mechanism of transcriptional regulation of this gene.
Inhibition of PfPMT Activity
The complete absence of phosphoethanolamine methyltransferase activity in mammalian cells bolsters PfPMT as a possible target for development of antimalarial compounds. Initial studies on PfPMT suggested that the enzyme activity was modulated by its own reaction product, phosphocholine [Pessi et al., 2004]. This finding intimated that PfPMT may also be inhibited by phosphocholine analogs. Indeed, hexadecylphosphocholine (miltefosine) inhibits PfPMT activity by 50% when present at a concentration of 50 μM and by 90% in the presence of 100 μM hexadecylphosphocholine [Pessi et al., 2004]. Parasite proliferation assays showed that hexadecylphosphocholine is capable of inhibiting parasite growth, with an IC50 value of ~80 μM [Pessi et al., 2004]. However, whether the inhibition of PfPMT by hexadecylphosphocholine accounts for its antimalarial activity remains to be determined. Structure-function studies in yeast taking advantage of the ability of PfPMT to complement the choline auxotrophy of the pem1Δpem2Δ mutant [Pessi et al., 2005] identified three residues (Asp-61, Gly-83, and Asp-128) in and near the catalytic domain that play essential roles in PfPMT activity [Reynolds et al., 2008]. Efforts are underway to determine the structure of PfPMT by NMR. These studies will aid in the understanding of the importance of these residues in PfPMT activity and will help in the rational design of specific PfPMT inhibitors.
PfPMT Plays an Important Role in P. falciparum Development and Multiplication
To determine the functional role of PfPMT in intact parasites, the PfPMT locus was disrupted to create a pfpmtΔ null mutant lacking PfPMT activity. Parasites lacking PfPMT display delay growth, altered DNA replication, reduced multiplication rate, and increased cell death [Witola et al., 2008]. The viability of pfpmtΔ knockout parasites is most likely due to the availability of residual choline in human red blood cells, which allows synthesis of phosphatidylcholine via the de novo pathway from choline. Interestingly, whereas choline is important for the survival of pfpmtΔ, its addition up to 10-times its physiological concentration did not complement the growth, replication, and multiplication defects of these knockout parasites [Witola et al., 2008]. This suggests that although the SDPM and CDP-choline pathways provide the same initial precursor (phosphocholine) for the synthesis of phosphatidylcholine, their functions are not completely redundant. These studies also suggest that inhibition of the initial steps of phosphatidylcholine biosynthesis would require compounds that inhibit both PfPMT activity and choline transport or phosphorylation. Screening of various chemical libraries is currently in progress to identify further compounds that efficiently inhibit PfPMT.
Arguments for and Against PtdEtn Transmethylation in Malaria Parasites
Biochemical and genetic studies in different species [Aktas and Narberhaus, 2009; Arondel et al., 1993; Kanipes et al., 1998; Keogh et al., 2009; Nuccio et al., 2000a; Vance et al., 2007] revealed the presence of PtdEtn methyltransferases capable of converting PtdEtn into PtdCho. In Plasmodium, such an activity has been proposed to exist based on a study in P. knowlesi-infected erythrocytes, which showed that radiolabeled PtdEtn (introduced by phospholipid-transfer proteins) can be converted into PtdCho [Moll et al., 1988]. However, the corresponding genes coding for such a PEMT activity have not been identified in any Plasmodium species. Furthermore, deletion of the PfPMT gene in P. falciparum parasites abolishes the incorporation of ethanolamine into PtdCho [Witola et al., 2008], suggesting that either PtdEtn transmethylation does not occur in P. falciparum, or that if such a reaction exists, it is catalyzed by PfPMT or requires a functional PfPMT enzyme. Thus far, biochemical and genetic analyses all appear to indicate that PfPMT does not catalyze the transmethylation of PtdEtn. First, unlike yeast extracts, P. falciparum extracts used in a PtdEtn transmethylation reaction in vitro failed to catalyze such a reaction. Second, purified recombinant PfPMT was found to catalyze the methylation of phosphoethanolamine but not PtdEtn. Third, using yeast as a model system it was shown that PfPMT complementation of pem1Δpem2Δ mutant growth defect in the absence of choline is ameliorated by ethanolamine supplementation and requires an active CDP-choline pathway. Thus, in P. falciparum available data do not support the existence of a direct methylation of PtdEtn to form Ptdcho. As to the study by Moll and colleagues [Moll et al., 1988], further biochemical and genetic studies in P. knowlesi are needed to determine whether the PMT ortholog can catalyze PtdEtn transmethylation and to identify a putative PEMT gene.
FATTY ACID BIOSYNTHESIS
Malaria parasites were thought to acquire all of the fatty acids required for blood stage growth through scavenging [Holz, 1977; Scheibel and Sherman, 1988; Vial et al., 1990]. This view came into question when sequencing of chromosome 2 from Plasmodium falciparum revealed the genes encoding two proteins typically associated with fatty acid biosynthesis in prokaryotes [Gardner et al., 1998, 1999]. These proteins, ACP (acyl carrier protein) and KASIII (β-ketoacyl-ACP synthase III), were subsequently shown to be targeted to the apicoplast, implicating this organelle as a possible site for de novo fatty acid biosynthesis [Waller et al., 2000]. By the time the P. falciparum genome was completed in 2002 [Gardner et al., 2002], genes encoding five other fatty acid biosynthesis enzymes had been identified and it appeared as though the parasites contained a complete Fatty Acid Synthase (FAS) capable of generating fatty acids from simple precursors.
The seven proteins found in the P. falciparum genome comprise a dissociated type II FAS. This type of FAS pathway is found in microorganisms and in the endosymbiont organelles of some eukaryotes (such as plant chloroplasts) [Harwood, 1996; Magnuson et al., 1993; Rock and Cronan, 1996]. The central hub of a type II FAS is the small, soluble protein ACP (Fig. 4). Nascent fatty acids are covalently linked to ACP through a thioester bond and are further modified by the FAS enzymes in this form. In a typical type II FAS pathway, MCAT (malonyl-coenzyme A:ACP transacylase) transfers a malonyl group from malonyl-CoA to ACP. Malonyl-ACP is then the substrate for KASIII, which catalyzes the decarboxylative condensation of the malonyl group with an acetyl group donated by acetyl-CoA. The product of KASIII is then reduced to acyl-ACP by the sequential action of KAR (β-ketoacyl-ACP reductase), HAD (β-hydroxyacyl-ACP dehydratase), and ENR (enoyl-ACP reductase). Further elongation of the acyl chain requires KASII (β-ketoacyl-ACP synthase II) followed by KAR, HAD, and ENR. These four enzymes form an elongation cycle that extends the acyl-ACP product by two carbons at a time and consumes malonyl-ACP (produced by MCAT). Substrate specificity, particularly of the ketoacyl-ACP synthases, ultimately determines the chain length of fatty acid produced by a type II FAS [Magnuson et al., 1993].
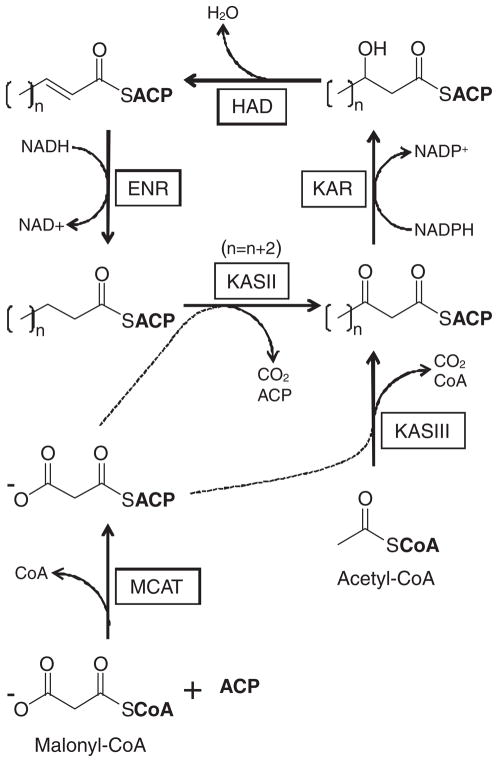
Fig. 4.
Type II FAS pathway as found in P. falciparum. MCAT catalyzes the production of malonyl-ACP, which is a substrate for both KASII and KASIII. KASIII catalyzes the condensation of malonyl-ACP with acetyl-CoA, forming acetoacetyl-ACP. This product enters an elongation cycle catalyzed by KAR, HAD, ENR, and KASII. The KASII reaction extends the carbon chain by 2 carbons, noted by increasing the number (n) of CH2 groups in the acyl chain by 2 (n+2). MCAT: malonyl-coenzyme A:ACP transacylase; KAS: β-ketoacyl-ACP synthase; KAR: β-ketoacyl-ACP reductase; HAD: β-hydroxyacyl-ACP dehydratase; ENR: enoyl-ACP reductase.
The Role of the Type II FAS in Malaria: Changing Paradigms
What is the role of fatty acid biosynthesis during the blood stages of malaria? This question has been unexpectedly difficult to answer. Early experiments led to the conclusion that P. falciparum does not synthesize fatty acids [Oaks et al., 1991]. However, the discovery of the type II FAS led to a revised hypothesis. The thought was that plasma lipids are scavenged (particularly the fatty acids palmitate and oleate) [Mitamura et al., 2000], but that key FAS products, such as lipoic acid, would have to be produced by the type II FAS pathway during the blood stages (lipoic acid was recently shown to be scavenged by P. falciparum during the blood stages) [Allary et al., 2007]. The biocide triclosan was initially used to validate the essential role of the type II FAS enzyme ENR. Triclosan was shown to inhibit ENR enzymatic activity, to inhibit the in vitro growth of P. falciparum in culture, and to inhibit the in vivo growth of the rodent malaria, P. berghei [Surolia and Surolia, 2001]. Importantly, triclosan was found to inhibit the incorporation of radiolabeled acetyl-CoA into newly synthesized fatty acids, demonstrating the apparent mechanism of antiparasitic activity [Surolia and Surolia, 2001]. Taken together, these studies were interpreted as pharmacological validation of FAS pathway enzymes as drug targets for blood-stage malaria.
Ultimately, it was pursuit of ENR as a drug target that led to a reexamination of the role that the type II FAS plays in malaria biology. The overall correlation between inhibition of enzyme activity and the inhibition of parasite growth proved to be poor, and an effort was made to genetically validate ENR as a target [Yu et al., 2008]. The P. falciparum enr gene was knocked out by double crossover homologous recombination without any observable growth defect, and no shift in the susceptibility of this strain for triclosan. Clearly, triclosan must have a different target in vivo. A key experiment demonstrated that the incorporation of radiolabeled acetyl-CoA into fatty acids was unaltered in the ENR knockout parasites [Yu et al., 2008]. This result indicated that some other metabolic pathway, such as fatty acid elongation, is probably responsible for the observed incorporation of acetyl-CoA [Spalding and Prigge, 2008].
Despite the presence of the type II FAS in malaria, the original conclusion that blood-stage parasites do not synthesize fatty acids may have been correct all along. Indeed, transcript levels of several type II FAS genes are very low during the blood stages. But even this conclusion requires a second look. A recent study examined the in vivo expression profiles of P. falciparum parasites collected from 43 patients in Senegal [Daily et al., 2007]. This study identified a population of parasites in which the genes encoding metabolic enzymes, including those comprising the type II FAS, were upregulated. The implications of this observation are still being discussed [Lemieux et al., 2009; Wirth et al., 2009], but it may be the case that there is a role for type II FAS in vivo that is not observed in culture conditions.
An unexpected result derived from recent genetic studies is that malaria type II FAS is critical for liver stage development. Vaughan and coworkers generated FAS gene knockouts in the rodent malaria parasite P. yoelii [Vaughan et al., 2009]. The behavior of pyhadΔ and pykasIIΔ strains was analyzed in different stages of the malaria life cycle. No phenotype was observed during the blood stages and mosquito stages; however, these strains were unable to complete liver stage development. Detailed examination showed that the knockout parasites are able to infect hepatocytes and develop normally until morphological abnormalities are observed during the second day of development [Vaughan et al., 2009]. Yu and coworkers conducted similar studies in another rodent parasite, P. berghei [Yu et al., 2008]. They observed a similar block in late liver stage development when analyzing the behavior of pbenrΔ parasites. However, pbenrΔ parasites were able to complete liver stage development in some cases, leading to blood stage infection, albeit with a significantly delayed patency [Yu et al., 2008]. It is not clear if breakthrough infections occur in pbenrΔ parasites and not in pyhadΔ and pykasIIΔ strains due to differences between the two parasite species, or due to differences attributable to the genes chosen for deletion. In any case, it now appears as though the type II FAS found in malaria parasites is critical for liver stage development, and that FAS enzymes may be appropriate targets for prophylactic drugs.
Type II FAS as an Antimalaria Drug Target
The discovery of the type II FAS in malaria generated considerable interest as a drug target because this pathway differs considerably from the type I FAS found in humans [Waller et al., 2003]. Within a few years after the initial report on triclosan activity, all seven proteins of the P. falciparum type II FAS had been biochemically characterized. Early work with pure recombinant proteins showed that ACP, MCAT, and KASIII function to initiate fatty acid biosynthesis and use acetyl-CoA and malonyl-CoA as carbon sources (Fig. 4) [Prigge et al., 2003; Waters et al., 2002]. Subsequently, HAD [Sharma et al., 2003] and KAR [Pillai et al., 2003] were shown to be active using acetoacetyl-CoA and β-hydroxybutyryl-CoA as surrogate substrates in lieu of the corresponding ACP-linked physiological substrates. Finally, KASII activity and substrate specificity for various acyl-ACP species were described [Lack et al., 2006]. In an effort to facilitate drug discovery efforts, crystal structures have been determined for ACP [Gallagher and Prigge, 2009], KAR [Wickramasinghe et al., 2006], HAD [Kostrewa et al., 2005; Swarnamukhi et al., 2006, 2007], and ENR [Freundlich et al., 2005, 2006, 2007; Muench et al., 2007; Perozzo et al., 2002; Pidugu et al., 2004].
Inhibitors have been reported for several of the type II FAS enzymes from P. falciparum [Lu et al., 2005]. Compounds with low micromolar IC50 values were reported for KAR [Wickramasinghe et al., 2006] and HAD [Sharma et al., 2003]. However, most of the drug discovery effort has focused on KASIII and ENR. The natural product thiolactomycin (TLM) inhibits bacterial β-ketoacyl-ACP synthase enzymes (FabH, FabB, and FabF) and inhibited the growth of P. falciparum with an IC50 value of 50 μM [Waller et al., 1998]. Several groups evaluated TLM analogs and found compounds that inhibit parasite growth at low micromolar concentrations [Jones et al., 2004; Prigge et al., 2003; Waller et al., 2003]. The establishment of an enzyme-based screen yielded a collection of submicromolar KASIII inhibitors, 60% of which are also effective at inhibiting the growth of cultured P. falciparum parasites [Lee et al., 2009]. Drug discovery for ENR has largely focused on analogs of the biocide triclosan (TRC). Two groups showed that the growth of P. falciparum is inhibited by triclosan with an IC50 value of approximately 1 μM [McLeod et al., 2001; Surolia and Surolia, 2001], and this discovery led to extensive structure-based drug discovery efforts. Hundreds of TRC analogs were synthesized and their inhibitory properties were evaluated in enzyme-based and parasite-based assays [Chhibber et al., 2006; Freundlich et al., 2005, 2006, 2007; Kuo et al., 2003; Nicola et al., 2007; Perozzo et al., 2002]. Although very potent inhibitors of ENR activity and malaria growth were found, there was ultimately a poor correlation between these phenomena, leading to the realization that type II FAS is not essential for the growth of blood-stage malaria in vitro [Yu et al., 2008].
Unfortunately, the search for type II FAS inhibitors has so far focused on the erythrocytic stages of malaria [Lu et al., 2005]. Typically, potent inhibitors of malaria FAS enzymes have been subjected to further selection for activity against cultured blood-stage P. falciparum. Ironically, this selection process probably enriched the population of off-target inhibitors and diminished the pool of compounds capable of inhibiting ENR in vivo. As discussed above, type II FAS enzymes may be appropriate targets for liver-stage therapeutics. As this new hypothesis is being pursued, there is a strong rationale for retesting potent inhibitors of malaria FAS enzymes, this time in a liver-stage malaria model.
Targeting the Synthesis of Plasmodium Phospholipids
One of the fundamental goals in the study of Plasmodium membrane biogenesis is to discover new metabolic pathways and key steps that play an important function in parasite development, proliferation, differentiation, and pathogenesis, and are either absent in humans or different enough from their human counterparts to be targeted for the development of novel antimalarial drugs. The establishment of an in vitro culture system for P. falciparum by Trager and Jensen [1976] made it possible to identify nutrients that the parasite actively transports in order to synthesize new membranes. Compounds that mimic the structure of membrane precursors such as fatty acids, ethanolamine, serine, or choline were thus tested for their antimalarial activity and found to inhibit parasite proliferation with IC50 values in the low micromolar range [Vial et al., 1984]. Biochemical analyses revealed that these analogs are transported into the parasitized erythrocyte and incorporated into the parasite membranes. By doing so, these compounds dramatically alter the lipid composition and physico-chemical properties of the membrane leading to parasite death [Beaumelle and Vial, 1988]. Unquestionably, the most advanced approach targeting lipid metabolism in Plasmodium relates to the inhibition of PtdCho biosynthesis by large molecules possessing one or two quaternary ammoniums and are structural analogs of choline. The first compounds within this category that were tested against P. falciparum were commercially available and inhibited parasite proliferation with IC50 values of 0.7–10 μM [Ancelin et al., 1985]. Metabolic studies revealed that these compounds block the entry of choline into the infected erythrocyte and cause a specific decrease in the biosynthesis of PtdCho [Ancelin and Vial, 1986]. The excellent antimalarial activity of this class of compounds paved the way for a more rationale approach to design new structural analogs and optimize them in order to select derivatives with more potent antimalarial activity in vitro. These studies led to the discovery that a molecular variation involving duplication of pharmacophoric groups (“twin-drug”) considerably increased the antimalarial activity with IC50 values in the sub-nanomolar to nanomolar rangs. Bis-ammonium salts were generally 100-fold more active than mono-ammonium salts [Calas et al., 2000]. SAR studies highlighted the importance of the spacer that separates the 2 cationic heads and the role of steric hindrance and lipophilicity of the N-substitution. In a second generation, the pyrolidinium moiety of the intrinsically potent lead compound, G25, was substituted by a less toxic thiazolium group that is present in vitamin B1 [Hamze et al., 2005]. Proof of concept of realistic antimalarial pharmacology with potent antimalarial activity was obtained in rodent malaria and in non-human primate models under very severe conditions of parasitemia and short course treatment [Wengelnik et al., 2002]. An important feature of the biscationic choline analogs was their ability to accumulate by several hundred-fold in malaria-infected erythrocytes, which makes them potent and specific agents against hematozoan-infected erythrocytes including Babesia [Richier et al., 2006]. This accumulation does in part occur in the Plasmodium food vacuole, where the compound associates with heme. Heme binding was shown to be critical for drug accumulation and likely contributes to the antimalarial activity of these compounds [Biagini et al., 2003].
Thus far, all efforts to unravel the mechanism of action of these compounds at different stages of the intraerythrocytic life cycle highlighted the pathways for the synthesis of PtdCho as the main targets. The compounds were found to inhibit choline entry into infected erythrocytes. However, because choline is not essential for P. falciparum intraerythrocytic development and survival, the inhibition of choline uptake alone cannot account for the antimalarial activity of these compounds. Transcriptome profiling to characterize the global response of P. falciparum to the bisthiazolium choline analogue T4, demonstrates cell cycle arrest and a general induction of genes involved in gametocytogenesis but no apparent transcriptional changes in genes involved in the PtdCho biosynthetic pathways. On the other hand, proteomic analysis revealed a significant decrease in the level of the Cho/Eth-phosphotransferase (PfCEPT) involved in the final step of synthesis of PtdCho. This effect was further supported by metabolic studies [Le Roch et al., 2008]. Other enzymes of the CDP-choline and SDPM pathways have also been shown to be inhibited by these compounds albeit at higher concentrations. Recently, genetic studies in P. falciparum and P. berghei have been crucial in the validation of candidate genes as possible targets of specific antimalarial drugs. Such genetic strategies are needed to identify primary target(s) of choline analogs. Nevertheless, the possibility that choline analogs may have multiple targets may represent a major strength of these inhibitors, as it could help delay the development of resistance. The development of this exciting new class of compounds is currently being conducted by Sanofi-Aventis. Phase 2 clinical trials of the bisthiazoliums salts T3/SAR97276 [Vial et al., 2004] are under way for parenteral cure of severe malaria. This clinical candidate is structurally unrelated to existing antimalarial agents, and acts through new independent mechanisms of action. Its unique properties are of tremendous interest as anti-infectious agents.
CONCLUSIONS
The metabolic machineries for the synthesis of phospholipids and fatty acids have stimulated great interest as potential targets for the development of novel antimalarial drugs, largely due to their importance for the growth, proliferation, and pathogenesis of Plasmodium parasites. In addition, the enzymes comprising these pathways are either absent from humans, or markedly different from their human counterparts. With the advances made during the past few years in the genetic manipulation of different Plasmodium species, it is becoming possible to validate specific steps and networks in these metabolic pathways as targets for the development of new antimalarial therapeutic strategies. The success of the chemical approach that led to the synthesis of potent antimalarial quaternary ammonium compounds highlights the importance of these pathways as drug targets. Thus far, only a few genes encoding lipid metabolism enzymes have been genetically ablated to validate their role in parasite growth and survival. From these limited genetic analyses, we have learned about the importance of these metabolic machineries not only during the intraerythrocytic stage but also during sexual differentiation and development in the mosquito and in hepatocytes. Thus, combining biochemical and metabolic knowledge with more advanced genetic, genomic, and structural analyses will set the stage for the design of novel drugs or combination therapies to block both malaria infection and transmission.
Acknowledgments
Grant sponsor: Department of Defense; Grant number: PR033005; Grant sponsor: Burroughs Wellcome Fund Award; Grant number: 1006267; Grant sponsor: European Union; Grant numbers: LSHP-CT-2004-503578; IP-018834; Grant sponsor: National Institutes of Health; Grant number: R01 AI065853; Grant sponsors: Johns Hopkins Malaria Research Institute; Bloomberg Family Foundation.
This research was supported by the Department of Defense Grant PR033005 and Burroughs Wellcome Fund Award 1006267 (to Choukri Ben Mamoun); the European Union (FP6 Network of Excellence BioMalPar LSHP-CT-2004-503578 and Integrated project Antimal, No. IP-018834 to Henri Vial); and the National Institutes of Health (R01 AI065853), the Johns Hopkins Malaria Research Institute, and the Bloomberg Family Foundation (to Sean Prigge). Choukri Ben Mamoun is a recipient of the Burroughs Wellcome Award, Investigators of Pathogenesis of Infectious Diseases. We thank past and present members of the Ben Mamoun, Prigge, and Vial laboratories for their contribution to the studies described in this review.
References
- Aktas M, Narberhaus F. In vitro characterization of the enzyme properties of the phospholipid N-methyltransferase PmtA from Agrobacterium tumefaciens. J Bacteriol. 2009;191:2033–2041. doi: 10.1128/JB.01591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allary M, Lu JZ, Zhu L, Prigge ST. Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol Microbiol. 2007;63:1331–1344. doi: 10.1111/j.1365-2958.2007.05592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin ML, Vial HJ. Quaternary ammonium compounds efficiently inhibit Plasmodium falciparum growth in vitro by impairment of choline transport. Antimicrob Agents Chemother. 1986;29:814–820. doi: 10.1128/aac.29.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin ML, Vial HJ, Philippot JR. Inhibitors of choline transport into Plasmodium-infected erythrocytes are effective antiplasmodial compounds in vitro. Biochem Pharmacol. 1985;34:4068–4071. doi: 10.1016/0006-2952(85)90390-9. [DOI] [PubMed] [Google Scholar]
- Ancelin ML, Parant M, Thuet MJ, Philippot JR, Vial HJ. Increased permeability to choline in simian erythrocytes after Plasmodium knowlesi infection. Biochem J. 1991;273:701–709. doi: 10.1042/bj2730701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arondel V, Benning C, Somerville CR. Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J Biol Chem. 1993;268:16002–16008. [PubMed] [Google Scholar]
- Attard GS, Templer RH, Smith WS, Hunt AN, Jackowski S. Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc Natl Acad Sci USA. 2000;97:9032–9036. doi: 10.1073/pnas.160260697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumelle BD, Vial HJ. Correlation of the efficiency of fatty acid derivatives in suppressing Plasmodium falciparum growth in culture with their inhibitory effect on acyl-CoA synthetase activity. Mol Biochem Parasitol. 1988;28:39–42. doi: 10.1016/0166-6851(88)90177-6. [DOI] [PubMed] [Google Scholar]
- Biagini GA, Richier E, Bray PG, Calas M, Vial H, Ward SA. Heme binding contributes to antimalarial activity of bis-quaternary ammoniums. Antimicrob Agents Chemother. 2003;47:2584–2589. doi: 10.1128/AAC.47.8.2584-2589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini GA, Pasini EM, Hughes R, De Koning HP, Vial HJ, O’Neill PM, Ward SA, Bray PG. Characterization of the choline carrier of Plasmodium falciparum: a route for the selective delivery of novel antimalarial drugs. Blood. 2004;104:3372–3377. doi: 10.1182/blood-2004-03-1084. [DOI] [PubMed] [Google Scholar]
- Bolognese CP, McGraw P. The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phosphoethanolamine N-methyltransferase. Plant Physiol. 2000;124:1800–1813. doi: 10.1104/pp.124.4.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza KM, Haakenson W, Cahoon RE, Hicks LM, Palavalli LH, Chiapelli BJ, McLaird M, McCarter JP, Williams DJ, Hresko MC, Jez JM. Phosphoethanolamine N-methyltransferase (PMT-1) catalyzes the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochem J. 2007;404:439–448. doi: 10.1042/BJ20061815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calas M, Ancelin ML, Cordina G, Portefaix P, Piquet G, Vidal-Sailhan V, Vial H. Antimalarial activity of compounds interfering with Plasmodium falciparum phospholipid metabolism: comparison between mono- and bisquaternary ammonium salts. J Med Chem. 2000;43:505–516. doi: 10.1021/jm9911027. [DOI] [PubMed] [Google Scholar]
- Charron JB, Breton G, Danyluk J, Muzac I, Ibrahim RK, Sarhan F. Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol. 2002;129:363–373. doi: 10.1104/pp.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber M, Kumar G, Parasuraman P, Ramya TN, Surolia N, Surolia A. Novel diphenyl ethers: design, docking studies, synthesis and inhibition of enoyl ACP reductase of Plasmodium falciparum and Escherichia coli. Bioorg Med Chem. 2006;14:8086–8098. doi: 10.1016/j.bmc.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, Levasseur K, Thomas E, Tamayo P, Dong C, Zhou Y, Lander ES, Ndiaye D, Wirth D, Winzeler EA, Mesirov JP, Regev A. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- Davies SM, Epand RM, Kraayenhof R, Cornell RB. Regulation of CTP: phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: an important role for stored curvature strain energy. Biochemistry. 2001;40:10522–10531. doi: 10.1021/bi010904c. [DOI] [PubMed] [Google Scholar]
- Divo AA, Geary TG, Davis NL, Jensen JB. Nutritional requirements of Plasmodium falciparum in culture. I. Exogenously supplied dialyzable components necessary for continuous growth. J Protozool. 1985;32:59–64. doi: 10.1111/j.1550-7408.1985.tb03013.x. [DOI] [PubMed] [Google Scholar]
- Elabbadi N, Ancelin ML, Vial HJ. Phospholipid metabolism of serine in Plasmodium-infected erythrocytes involves phosphatidyl-serine and direct serine decarboxylation. Biochem J. 1997;324:435–445. doi: 10.1042/bj3240435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin-Christensen J, Suarez CE, Florin-Christensen M, Hines SA, McElwain TF, Palmer GH. Phosphatidylcholine formation is the predominant lipid biosynthetic event in the hemoparasite Babesia bovis. Mol Biochem Parasitol. 2000;106:147–156. doi: 10.1016/s0166-6851(99)00209-1. [DOI] [PubMed] [Google Scholar]
- Freundlich JS, Anderson JW, Sarantakis D, Shieh HM, Yu M, Valderramos JC, Lucumi E, Kuo M, Jacobs WR, Jr, Fidock DA, Schieshser GA, Jacobus DP, Sacchettini JC. Synthesis, biological activity, and X-ray crystal structural analysis of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 1:4′-substituted triclosan derivatives. Bioorg Med Chem Lett. 2005;15:5247–5252. doi: 10.1016/j.bmcl.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Freundlich JS, Yu M, Lucumi E, Kuo M, Tsai HC, Valderramos JC, Karagyozov L, Jacobs WR, Jr, Schiehser GA, Fidock DA, Jacobus DP, Sacchettini JC. Synthesis and biological activity of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 2:2′-substituted triclosan derivatives. Bioorg Med Chem Lett. 2006;16:2163–2169. doi: 10.1016/j.bmcl.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Freundlich JS, Wang F, Tsai HC, Kuo M, Shieh HM, Anderson JW, Nkrumah LJ, Valderramos JC, Yu M, Kumar TR, Valderramos SG, Jacobs WR, Jr, Schiehser GA, Jacobus DP, Fidock DA, Sacchettini JC. X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy. J Biol Chem. 2007;282:25436–25444. doi: 10.1074/jbc.M701813200. [DOI] [PubMed] [Google Scholar]
- Gallagher JR, Prigge ST. Plasmodium falciparum acyl carrier protein crystal structures in disulfide-linked and reduced states and their prevalence during blood stage growth. Proteins. 2009 doi: 10.1002/prot.22582. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, Koonin EV, Shallom S, Mason T, Yu K, Fujii C, et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Smith HO, Fraser CM, Venter JC, Hoffman SL. The malaria genome sequencing project: complete sequence of Plasmodium falciparum chromosome 2. Parasitologia. 1999;41:69–75. [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:49–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamze A, Rubi E, Arnal P, Boisbrun M, Carcel C, Salom-Roig X, Maynadier M, Wein S, Vial H, Calas M. Mono- and bis-thiazolium salts have potent antimalarial activity. J Med Chem. 2005;48:3639–3643. doi: 10.1021/jm0492608. [DOI] [PubMed] [Google Scholar]
- Harwood JL. Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta. 1996;1301:7–56. doi: 10.1016/0005-2760(95)00242-1. [DOI] [PubMed] [Google Scholar]
- Holz GG. Lipids and the malarial parasite. Bull World Health Org. 1977;55:237–248. [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Urch JE, Brun R, Harwood JL, Berry C, Gilbert IH. Analogues of thiolactomycin as potential anti-malarial and anti-trypanosomal agents. Bioorg Med Chem. 2004;12:683–692. doi: 10.1016/j.bmc.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Kanipes MI, Hill JE, Henry SA. The Schizosaccharomyces pombe cho1 + gene encodes a phospholipid methyltransferase. Genetics. 1998;150:553–562. doi: 10.1093/genetics/150.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MR, Courtney PD, Kinney AJ, Dewey RE. Functional characterization of phospholipid N-methyltransferases from Arabidopsis and soybean. J Biol Chem. 2009;284:15439–15447. doi: 10.1074/jbc.M109.005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk K, Wong HY, Elford BC, Newbold CI, Ellory JC. Enhanced choline and Rb+ transport in human erythrocytes infected with the malaria parasite Plasmodium falciparum. Biochem J. 1991;278:521–525. doi: 10.1042/bj2780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- Kostrewa D, Winkler FK, Folkers G, Scapozza L, Perozzo R. The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci. 2005;14:1570–1580. doi: 10.1110/ps.051373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MR, Morbidoni HR, Alland D, Sneddon SF, Gourlie BB, Staveski MM, Leonard M, Gregory JS, Janjigian AD, Yee C, Musser JM, Kreiswirth B, Iwamoto H, Perozzo R, Jacobs WRJ, Sacchettini JC, Fidock DA. Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data. J Biol Chem. 2003;278:20851–20859. doi: 10.1074/jbc.M211968200. [DOI] [PubMed] [Google Scholar]
- Lack G, Homberger-Zizzari E, Folkers G, Scapozza L, Perozzo R. Recombinant expression and biochemical characterization of the unique elongating beta-ketoacyl-acyl carrier protein synthase involved in fatty acid biosynthesis of Plasmodium falciparum using natural and artificial substrates. J Biol Chem. 2006;281:9538–9546. doi: 10.1074/jbc.M509119200. [DOI] [PubMed] [Google Scholar]
- Lee PJ, Bhonsle JB, Gaona HW, Huddler DP, Heady TN, Kreishman-Deitrick M, Bhattacharjee A, McCalmont WF, Gerena L, Lopez-Sanchez M, Roncal NE, Hudson TH, Johnson JD, Prigge ST, Waters NC. Targeting the fatty acid biosynthesis enzyme, beta-ketoacyl-acyl carrier protein synthase III (PfKASIII), in the identification of novel antimalarial agents. J Med Chem. 2009;52:952–963. doi: 10.1021/jm8008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane AM, Saliba KJ, Allen RJ, Kirk K. Choline uptake into the malaria parasite is energized by the membrane potential. Biochem Biophys Res Commun. 2004;320:311–317. doi: 10.1016/j.bbrc.2004.05.164. [DOI] [PubMed] [Google Scholar]
- Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, Day F, Kyes SA, Conway DJ, Holmes CC, Newbold CI. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc Natl Acad Sci USA. 2009;106:7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Ahiboh H, Chung DW, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates JR, Mamoun CB, Winzeler EA, Vial H. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics. 2008;9:513. doi: 10.1186/1471-2164-9-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Lee PJ, Waters NC, Prigge ST. Fatty acid synthesis as a target for antimalarial drug discovery. Comb Chem High Throughput Screen. 2005;8:15–26. doi: 10.2174/1386207053328192. [DOI] [PubMed] [Google Scholar]
- Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod R, Muench SP, Rafferty JB, Kyle DE, Mui EJ, Kirisits MJ, Mack DG, Roberts CW, Samuel BU, Lyons RE. Triclosan inhibits the growth of Plasmodium falciparum and Toxoplasma gondii by inhibition of Apicomplexan Fab I. Int J Parasitol. 2001;31:109–113. doi: 10.1016/s0020-7519(01)00111-4. [DOI] [PubMed] [Google Scholar]
- Mitamura T, Hanada K, Ko-Mitamura EP, Nishijima M, Horii T. Serum factors governing intraerythrocytic development and cell cycle progression of Plasmodium falciparum. Parasitol Int. 2000;49:219–229. doi: 10.1016/s1383-5769(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Moll GN, Vial HJ, Ancelin ML, Op den Kamp JA, Roelofsen B, van Deenen LL. Phospholipid uptake by Plasmodium knowlesi infected erythrocytes. FEBS Lett. 1988;232:341–346. doi: 10.1016/0014-5793(88)80765-8. [DOI] [PubMed] [Google Scholar]
- Muench SP, Prigge ST, McLeod R, Rafferty JB, Kirisits MJ, Roberts CW, Mui EJ, Rice DW. Studies of Toxoplasma gondii and Plasmodium falciparum enoyl acyl carrier protein reductase and implications for the development of antiparasitic agents. Acta Crystallogr D Biol Crystallogr. 2007;63:328–338. doi: 10.1107/S0907444906053625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola G, Smith CA, Lucumi E, Kuo MR, Karagyozov L, Fidock DA, Sacchettini JC, Abagyan R. Discovery of novel inhibitors targeting enoyl-acyl carrier protein reductase in Plasmodium falciparum by structure-based virtual screening. Biochem Biophys Res Commun. 2007;358:686–691. doi: 10.1016/j.bbrc.2007.04.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J Biol Chem. 2000a;275:14095–14101. doi: 10.1074/jbc.275.19.14095. [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J Biol Chem. 2000b;275:14095–14101. doi: 10.1074/jbc.275.19.14095. [DOI] [PubMed] [Google Scholar]
- Oaks SC, Mitchell VS, Pearson GW, Carpenter CCJ, editors. Malaria: obstacles and opportunities. Washington, DC: National Academy Press; 1991. p. 309. [PubMed] [Google Scholar]
- Palavalli LH, Brendza KM, Haakenson W, Cahoon RE, McLaird M, Hicks LM, McCarter JP, Williams DJ, Hresko MC, Jez JM. Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry. 2006;45:6056–6065. doi: 10.1021/bi060199d. [DOI] [PubMed] [Google Scholar]
- Perozzo R, Kuo M, Sidhu AbS, Valiyaveettil JT, Bittman R, Jacobs WR, Jr, Fidock DA, Sacchettini JC. Structural elucidation of the specificity of the antibacterial agent triclosan for malarial enoyl acyl carrier protein reductase. J Biol Chem. 2002;277:13106–13114. doi: 10.1074/jbc.M112000200. [DOI] [PubMed] [Google Scholar]
- Pessi G, Ben Mamoun C. Pathways for phosphatidylcholine biosynthesis: targets and strategies for antimalarial drugs. Future Med, Future Lipidol. 2006;1:173–180. [Google Scholar]
- Pessi G, Kociubinski G, Mamoun CB. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc Natl Acad Sci USA. 2004;101:6206–6211. doi: 10.1073/pnas.0307742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Choi JY, Reynolds JM, Voelker DR, Mamoun CB. In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J Biol Chem. 2005;280:12461–12466. doi: 10.1074/jbc.M414626200. [DOI] [PubMed] [Google Scholar]
- Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K. Structural basis for the variation in triclosan affinity to enoyl reductases. J Mol Biol. 2004;343:147–155. doi: 10.1016/j.jmb.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Pillai S, Rajagopal C, Kapoor M, Kumar G, Gupta A, Surolia N. Functional characterization of beta-ketoacyl-ACP reductase (FabG) from Plasmodium falciparum. Biochem Biophys Res Commun. 2003;303:387–392. doi: 10.1016/s0006-291x(03)00321-8. [DOI] [PubMed] [Google Scholar]
- Prigge ST, He X, Gerena L, Waters NC, Reynolds KA. The initiating steps of a type II fatty acid synthase in Plasmodium falciparum are catalyzed by pfACP, pfMCAT, and pfKASIII. Biochemistry. 2003;42:1160–1169. doi: 10.1021/bi026847k. [DOI] [PubMed] [Google Scholar]
- Reynolds JM, Takebe S, Choi JY, El Bissati K, Witola WH, Bobenchik AM, Hoch JC, Voelker DR, Mamoun CB. Biochemical and genetic analysis of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum. J Biol Chem. 2008;283:7894–7900. doi: 10.1074/jbc.M709869200. [DOI] [PubMed] [Google Scholar]
- Richier E, Biagini GA, Wein S, Boudou F, Bray PG, Ward SA, Precigout E, Calas M, Dubremetz JF, Vial HJ. Potent antihematozoan activity of novel bisthiazolium drug T16: evidence for inhibition of phosphatidylcholine metabolism in erythrocytes infected with Babesia and Plasmodium spp. Antimicrob Agents Chemother. 2006;50:3381–3388. doi: 10.1128/AAC.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- Scheibel LW, Sherman IW. Plasmodial metabolism and related organellar function during various stages of the life-cycle: proteins, lipids, nucleic acids and vitamins. In: Wernsdorfer W, McGregor I, editors. Principles and practice of malariology. New York: Churchill Livingstone; 1988. pp. 219–252. [Google Scholar]
- Sharma SK, Kapoor M, Ramya TN, Kumar S, Kumar G, Modak R, Sharma S, Surolia N, Surolia A. Identification, characterization, and inhibition of Plasmodium falciparum beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) J Biol Chem. 2003;278:45661–45671. doi: 10.1074/jbc.M304283200. [DOI] [PubMed] [Google Scholar]
- Spalding MD, Prigge ST. Malaria pulls a FASt one. Cell Host Microbe. 2008;4:509–511. doi: 10.1016/j.chom.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surolia N, Surolia A. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nature Med. 2001;7:167–173. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- Swarnamukhi PL, Sharma SK, Bajaj P, Surolia N, Surolia A, Suguna K. Crystal structure of dimeric FabZ of Plasmodium falciparum reveals conformational switching to active hexamers by peptide flips. FEBS Lett. 2006;580:2653–2660. doi: 10.1016/j.febslet.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Swarnamukhi PL, Sharma SK, Padala P, Surolia N, Surolia A, Suguna K. Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum. Acta Crystallogr D Biol Crystallogr. 2007;63:458–464. doi: 10.1107/S0907444907003228. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Vance DE, Li Z, Jacobs RL. Hepatic phosphatidylethanolamine N-methyltransferase, unexpected roles in animal biochemistry and physiology. J Biol Chem. 2007;282:33237–33241. doi: 10.1074/jbc.R700028200. [DOI] [PubMed] [Google Scholar]
- Vaughan AM, O’Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial H, Ancelin M. Malarial Lipids. In: Sherman I, editor. Malaria: parasite biology, biogenesis, protection. Washington, DC: ASM Press; 1998. pp. 159–175. [Google Scholar]
- Vial HJ, Ben Mamoun C. Plasmodium lipids: metabolism and Function. In: Sherman IW, editor. Molecular approaches to malaria. Washington, DC: ASM Press; 2005. pp. 327–352. [Google Scholar]
- Vial HJ, Thuet MJ, Ancelin ML, Philippot JR, Chavis C. Phospholipid metabolism as a new target for malaria chemotherapy. Mechanism of action of D-2-amino-1-butanol. Biochem Pharmacol. 1984;33:2761–2770. doi: 10.1016/0006-2952(84)90693-2. [DOI] [PubMed] [Google Scholar]
- Vial HJ, Ancelin ML, Philippot JR, Thuet MJ. Biosynthesis and dynamics of lipids in Plasmodium-infected mature mammalian erythrocytes. Blood Cells. 1990;16:531–555. discussion 556–561. [PubMed] [Google Scholar]
- Vial HJ, Wein S, Farenc C, Kocken C, Nicolas O, Ancelin ML, Bressolle F, Thomas A, Calas M. Prodrugs of bisthiazolium salts are orally potent antimalarials. Proc Natl Acad Sci USA. 2004;101:15458–15463. doi: 10.1073/pnas.0404037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Ralph SA, Reed MB, Su V, Douglas JD, Minnikin DE, Cowman AF, Besra GS, McFadden GI. A type II pathway for fatty acid biosynthesis presents drug targets in Plasmodium falciparum. Antimicrob Agents Chemother. 2003;47:297–301. doi: 10.1128/AAC.47.1.297-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. Embo J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters NC, Kopydlowski KM, Guszczynski T, Wei L, Sellers P, Ferlan JT, Lee PJ, Li Z, Woodard CL, Shallom S. Functional characterization of the acyl carrier protein (PfACP) and beta-ketoacyl ACP synthase III (PfKASIII) from Plasmodium falciparum. Mol Biochem Parasitol. 2002;123:85–94. doi: 10.1016/s0166-6851(02)00140-8. [DOI] [PubMed] [Google Scholar]
- Wengelnik K, Vidal V, Ancelin ML, Cathiard AM, Morgat JL, Kocken CH, Calas M, Herrera S, Thomas AW, Vial HJ. A class of potent antimalarials and their specific accumulation in infected erythrocytes. Science. 2002;295:1311–1314. doi: 10.1126/science.1067236. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe SR, Inglis KA, Urch JE, Muller S, van Aalten DM, Fairlamb AH. Kinetic, inhibition and structural studies on 3-oxoacyl-ACP reductase from Plasmodium falciparum, a key enzyme in fatty acid biosynthesis. Biochem J. 2006;393:447–457. doi: 10.1042/BJ20050832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth D, Daily J, Winzeler E, Mesirov JP, Regev A. In vivo profiles in malaria are consistent with a novel physiological state. Proc Natl Acad Sci USA. 2009;106:E70. doi: 10.1073/pnas.0904478106. author reply E71–E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola WH, Ben Mamoun C. Choline induces transcriptional repression and proteasomal degradation of the malarial phosphoethanolamine methyltransferase. Eukaryot Cell. 2007;6:1618–1624. doi: 10.1128/EC.00229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola WH, Pessi G, El Bissati K, Reynolds JM, Mamoun CB. Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus. J Biol Chem. 2006;281:21305–21311. doi: 10.1074/jbc.M603260200. [DOI] [PubMed] [Google Scholar]
- Witola WH, El Bissati K, Pessi G, Xie C, Roepe PD, Mamoun CB. Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J Biol Chem. 2008;283:27636–27643. doi: 10.1074/jbc.M804360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]