SUMMARY
Expression of the INK4b/ARF/INK4a tumor suppressor locus in normal and cancerous cell growth is controlled by methylation of histone H3 at lysine 27 (H3K27me) as directed by the Polycomb group proteins. The antisense non-coding RNA ANRIL of the INK4b/ARF/INK4a locus is also important for expression of the protein-coding genes in cis, but its mechanism has remained elusive. Here we report that chromobox 7 (CBX7) within the Polycomb Repressive Complex 1 binds to ANRIL, and both CBX7 and ANRIL are found at elevated levels in prostate cancer tissues. In concert with H3K27me recognition, binding to RNA contributes to CBX7 function and disruption of either interaction impacts the ability of CBX7 to repress the INK4b/ARF/INK4a locus and control senescence. Structure-guided analysis reveals the molecular interplay between non-coding RNA and H3K27me as mediated by the conserved chromodomain. Our study suggests a new mechanism by which non-coding RNA participates directly in epigenetic transcriptional repression.
INTRODUCTION
Transcription of the INK4a/ARF locus is tightly controlled in part by Polycomb group (PcG) protein complexes that initiate and maintain the silenced state (Gil and Peters, 2006). Initially identified as regulators of Hox genes during development, PcG proteins and their activating counterparts, the Trithorax group (TrxG) proteins, play an important role in gene silencing, development, stem cell self-renewal and differentiation (Müller and Verrijzer, 2009; Ringrose and Paro, 2004; Schuettengruber et al., 2007; Sparmann and van Lohuizen, 2006). As negative regulators PcG proteins have been best characterized as two multi-protein complexes, Polycomb Repressive Complex 1 (PRC1) and 2 (PRC2). PRC2 contains EED, Suz12 and the lysine methyltransferase EZH2, and initiates long-term repression of target genes by trimethylating histone H3 at lysine 27 (H3K27). PRC1, which typically consists of Bmi1/Mel18, mPh1/2, Ring1a/b and Pc/Chromobox (CBX), has been referred to as the maintenance system of silenced chromatin; it is responsible both for mono-ubiquitination at histone H2AK119 by Ring1b, and recognition of methylated H3K27 via the Pc protein. Mammalian forms of PRC1 may achieve additional contextual specificity by substituting for one of the multiple homologs of Pc/CBX. Of the chromobox proteins, CBX7 has been characterized as a regulator of cellular lifespan by directly repressing the INK4a/ARF locus (Bernard et al., 2005; Gil et al., 2004). Consistent with this CBX7 is up-regulated in follicular lymphoma (Scott et al., 2007) and certain prostate cancers (Bernard et al., 2005), although its down-regulation or loss of expression in other carcinomas suggests a more complex scenario.
In recent years, non-coding RNAs (ncRNAs) have been recognized for its major role in gene expression (Bernstein and Allis, 2005; Lee, 2009; Morris, 2009; Ponting et al., 2009; Umlauf et al., 2008; Zaratiegui et al., 2007), believed to control gene silencing by recruiting histone methyltransferases, such as by Air (Nagano et al., 2008) and Kcnq1ot1 (Pandey et al., 2008), or by creating a repressed nuclear compartment to which target genes are relocated, such as by Xist (Chaumeil et al., 2006). Both short and long ncRNAs can contribute to the recruitment of PcG complexes to target genes and the regulation of PcG protein expression (Hekimoglu and Ringrose, 2009; Khalil et al., 2009). In mammals and Drosophila, RNA-directed regulation may be dependent on direct interactions with PcG proteins such as EZH2, Suz12 and CBX (Bernstein et al., 2006; Pandey et al., 2008; Rinn et al., 2007; Zhao et al., 2008). Further, long ncRNAs can be transcribed adjacent to or within PcG target genes of which the expression of HOTAIR (Rinn et al., 2007) and Kcnq1ot1 (Pandey et al., 2008) is correlated with target silencing.
Given that CBX7 (and other CBX homologs) bind RNA in vitro (Bernstein et al., 2006), we postulated that both H3K27me and RNA binding activities might mediate CBX7 function, although it is unclear whether these two modes of regulation are analogous or cooperative. ANRIL is a long non-coding antisense RNA transcript overlapping the INK4b/ARF/INK4a locus (Pasmant et al., 2007) (Figure 1a) with estimates of the transcript spanning over 30–40 kilobases in length (Mattick et al., 2009). This transcript promotes and maintains the epigenetic state of the CDKN2B (INK4b) gene and is an anti-sense transcript to INK4b (Yu et al., 2008) and single nucleotide polymorphisms associated with ANRIL have been linked to enhanced risk of atherosclerosis (Broadbent et al., 2008; Samani et al., 2008). To determine whether ANRIL is a signature transcript for docking PcG proteins, we sought to characterize repression at the INK4b/ARF/INK4a locus by Polycomb CBX7 in the context of H3K27me and ANRIL interactions. We show that in vivo, CBX7 is specifically associated with ANRIL, and levels of both are elevated in prostate cancer tissues; in vitro, CBX7 employs overlapping yet distinct regions within its chromodomain for binding to H3K27me and RNA. Introduction of antisense transcripts to ANRIL, or structure-guided mutations of CBX7 of PRC1 that disrupt H3K27me or RNA binding affect cellular lifespan. Our results demonstrate how PcG proteins integrate the regulation of the INK4b/ARF/INK4a locus by two epigenetic factors – histone modification and ncRNA.
Figure 1. Non-coding RNA ANRIL is Up-regulated in Prostate Cancer Tissues and Coincides with CBX7 and EZH2 Expression.
a, Schematic representation of the genomic organization of the INK4a/ARF gene cluster. Three genes in the INK4a/ARF locus are color-coded. Transcripts antisense to ANRIL (blue) used to inhibit ANRIL interactions (see Figure 2) are indicated above ANRIL. Roman numerals denote sites of quantitative ChIP as shown in Figure 3. b, Normalized expression levels of ANRIL, p16, CBX7 and EZH2 in normal prostate epithelium cells, and preneoplastic PIN and PCa prostate cancer cells. Each plotted point represents a separate tissue sample. c, Levels of ANRIL transcript relative to HPRT as detected in normal prostate epithelium and PC3 prostate cancer cells. d, RNA-FISH (fluorescence in situ hybridization) of ANRIL as detected in normal prostate and prostate cancer (PCa) cells. The housekeeping genes GAPDH and HPRT are shown as input controls. e, Relative enrichment of RNA polymerase II, EZH2 and CBX7 associated with loci spanning the INK4a/ARF region in the IMR90 fibroblasts (red) or PC3 prostate carcinoma (green) background. Standard ChIP analysis was performed at selected points overlapping transcript starts identified within the UCSC Genome Browser (chr9: 21,710,518-2,301,141), based on RefSeq, Uniprot, GeneBank, CCDS, and Comparative Genomics and plotted using Genome Graphs (http://genome.ucsc.edu). ChIP primers used are listed in Table S2. See also Figure S1.
RESULTS AND DISCUSSION
ANRIL RNA and CBX7 are Up-regulated in Prostate Cancer Tissues
Previous studies have shown that CBX7 is over-expressed in some prostate carcinomas (Bernard et al., 2005), which may collaborate with the over-expression of EZH2 (Varambally et al., 2002). Therefore, we analyzed several tissue specimens from prostate using quantitative PCR (qPCR) for expression of ANRIL non-coding RNA, and INK4a, CBX7 and EZH2 genes (Figure 1b). In comparison to preneoplastic (PIN) and untransformed (normal) prostate epithelium tissues, we observed elevated expression of ANRIL, CBX7 and EZH2 and a corresponding decrease in INK4a expression. Higher levels of ANRIL expression were seen in prostate cancer (PC3 and PCa) relative to normal prostate epithelial cells (Figure 1c) and other cancer tissues (data not shown). RNA-FISH (fluorescence in situ hybridization) imaging helps visualize the elevated expression of ANRIL in PCa, contrasting sharply with normal prostate cells where ANRIL is undetectable (Figure 1d). Additionally, we detected ANRIL coinciding with higher levels of occupancy of EZH2, CBX7 and RNA polymerase II (Pol II) near the promoter regions of INK4a and ARF in PC3 cells than in IMR90 diploid fibroblasts (Figure 1e). Collectively, these results argue that in addition to both being up-regulated in prostate cancer, CBX7 and ANRIL expression are likely correlated. Further, within the region of the locus separating the ANRIL and INK4a start sites, we observed an inverse correlation of Pol II occupation, suggesting a highly dynamic function of Pol II on the opposing DNA strand that may negatively impact INK4a transcription.
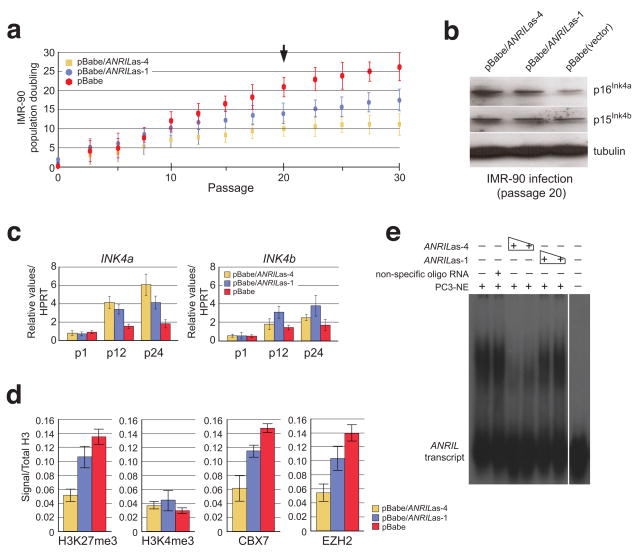
To investigate whether ANRIL influences INK4a transcription, we examined the effects of transcripts antisense to ANRIL on lifespan of IMR-90 diploid fibroblasts, which were transduced with retrovirus expressing each of two antisense cDNAs, one (ANRILas-4) targeting a site upstream to ARF and INK4a, and the other (ANRILas-1) upstream to INK4b (Figure 1a). Cell population doublings were measured and showed a 2- and 1.5-fold reduction, respectively, compared to the control at passage 20 (Figure 2a). Western blots of cells from passage 20 reveal that expression of ANRILas-4, and to a much lesser extent ANRILas-1, caused a marked increase in the expression of p16INK4a over cells infected with the vector alone. The effects on p15INK4b expression were much less profound and tubulin remained constant (Figure 2b). Notably, RNA transcript levels of INK4a and INK4b following passages showed patterns of changes similar to those of proteins (Figure 2c). Moreover, occupation of H3K27me3, CBX7, and EZH2 probed by chromatin immunoprecipitation (ChIP) at the transcription start site (position III) in the INK4a gene (Figure 1a) showed a similar reduction over the control, whereas occupation of H3K4me3 exhibited little change if not a slight increase (Figure 2d). Finally, we observed that ANRILas-4 competes effectively against in vitro transcribed ANRIL binding to nuclear proteins in a concentration-dependent manner in an RNA electrophoretic mobility shift assay (EMSA) (Figure 2e). Taken together, these results strongly support the existence of a direct role of ANRIL and ANRIL-associated factors that reinforce transcriptional control and are critical in the decision to repress the INK4b/ARF/INK4a locus.
Figure 2. Competitive inhibition of ANRIL transcript interactions results in loss of INK4a repression and limitation of cellular lifespan.
a, Transcript antisense to the ANRIL long non-coding transcript relative to the INK/ARF locus (as indicated by arrows above ANRIL in Figure 1a) influence lifespan of normal IMR-90 diploid fibroblasts, transduced with retrovirus expressing each of the antisense cDNA as indicated, and quantified through population doublings following each passage (n=3). Points equal mean; bars equal standard deviation. b, Levels of p16INK4a, p15INK4b and tubulin were evaluated from retrovirally-transduced IMR-90 cell extracts following passage 20 by immunoblots. c, RNA transcript levels of INK4a and INK4b following passages as indicated. d, Deposition and occupation of histone methylation marks, H3K27me3 and H3K4me3, CBX7, and EZH2 at the transcription start site (position III) of the INK4a locus as measured by ChIP analysis. e, Competition study using antisense transcripts to ANRIL were generated by in vitro transcription and analyzed from nuclear proteins bound by RNA-EMSA.
ANRIL RNA is Associated with Polycomb Protein Recruitment to the INK4a/ARF Locus
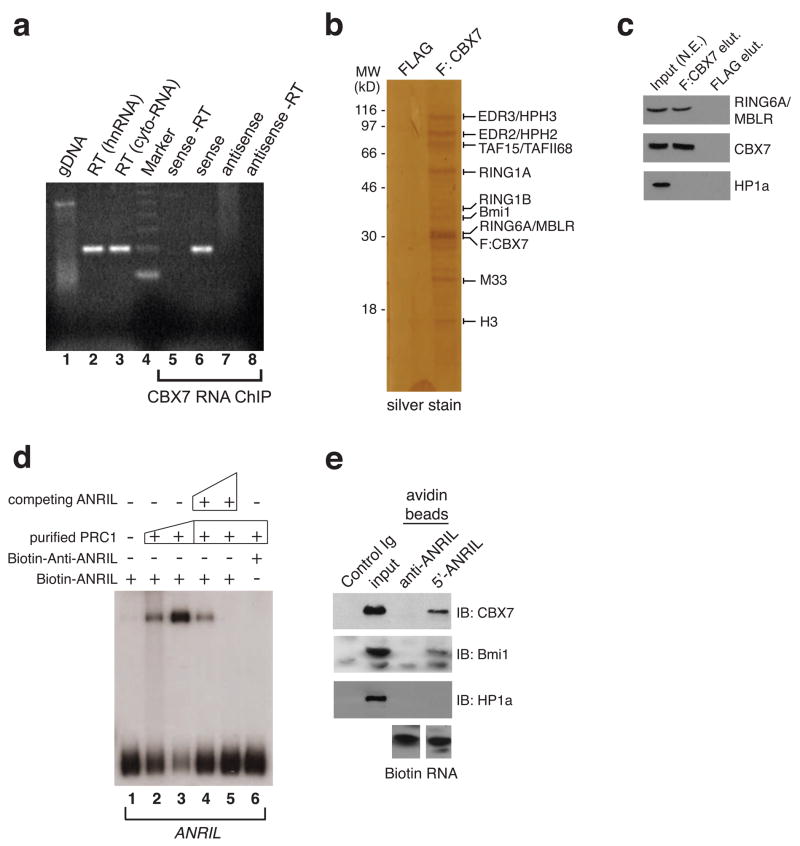
Using a modified RNA-ChIP method we confirmed that CBX7 forms a stable association with endogenous ANRIL in vivo in PC3 cells (Figure 3a). To ensure contextual RNA binding by CBX7 in the PRC1 complex, we purified PRC1 from the nuclear extracts of PC3 cells transfected with FLAG-tagged-CBX7, and confirmed CBX7 association with endogenous PRC1 by MALDI-TOF mass spectrometry and Western blot analyses (Figure 3b, c). An RNA EMSA using 5′-biotinylated ANRIL transcript further demonstrates that the purified PRC1 forms a stable complex with ANRIL but not an anti-ANRIL transcript (Figure 3d, lanes 2,3 versus 6; Figure 3e); the former can be competed off by a non-biotinylated ANRIL (Figure 3d, lanes 4,5). Based on genomic mapping of ncRNA association with CBX7, EZH2 and Pol II with RNA ChIP (Figure 1e), we detected that the ANRIL transcript initiates at start sites upstream of the INK4a and ARF genes on the opposing strand, preceding previously reported start sites by several hundred bases (Pasmant et al., 2007; Sharpless and DePinho, 2007; Yu et al., 2008). Mapping of the start site of ANRIL transcription by 5′ RACE confirms its proximity to the promoter of INK4a, which is also supported by the RNA ChIP analysis (Figure S1). These results confirm that ANRIL interacts with CBX7 of the PRC1 complex at the INK4b/ARF/INK4a locus.
Figure 3. ANRIL ncRNA Associates Specifically with Members of the PRC1 Complex.
a, Recovery of ANRIL transcripts by CLIP analysis of CBX7 in chromatin from asynchronously growing PC3 cells. Lane 1, genomic DNA prior to DNA hydrolysis from nuclei recovered; reverse transcription PCR (rtPCR) of heteronuclear RNA (lane 2) and cytoplasmic RNA (lane 3) recovered from PC3 cell nuclei; lane 4, DNA molecular ladder marker; RNA-ChIP of CBX7-bound RNA and rtPCR using antisense primer to detect sense strand of ANRIL transcript (lanes 5, 6) or sense primer to detect anti-sense strand (lanes 7, 8) in the absence or presence of AMV reverse transcriptase, respectively. b and c, FLAG-tagged CBX7 purified from human PC3 cells binds to the ANRIL transcript. Silver staining was used to visualize the human CBX7 complex purified from the PC3 cells. Identification of CBX7/PRC1 components was performed by MALDI-TOF mass spectrometry analysis and specific protein components confirmed (in c) by immunoblotting with antisera directed against RING6A/MBLR, CBX7, and HP1α. d, Band shift of the sense and anti-sense ANRIL transcript by the homogenous CBX7/PRC1 complex by RNA EMSA assay. 50 and 200 ng of purified PRC1 proteins were used to visualize the RNA binding potential of the CBX7-associated complex for the segment of the ncRNA transcript generated by in vitro transcription (lanes 2, 3). Competitive (50x and 1000x excess) unlabeled ncRNA transcript was used as shown to compete for ANRIL (lanes 4, 5). Note that anti-sense ANRIL does not form a stable complex with the purified PRC1 (lane 6). e, Total nuclear extract from PC3 cells was used to evaluate by Western blot analysis the association of PRC1 proteins CBX7, Bmi1 and HP1α with the biotinylated ANRIL transcript generated in d. See also Figure S2.
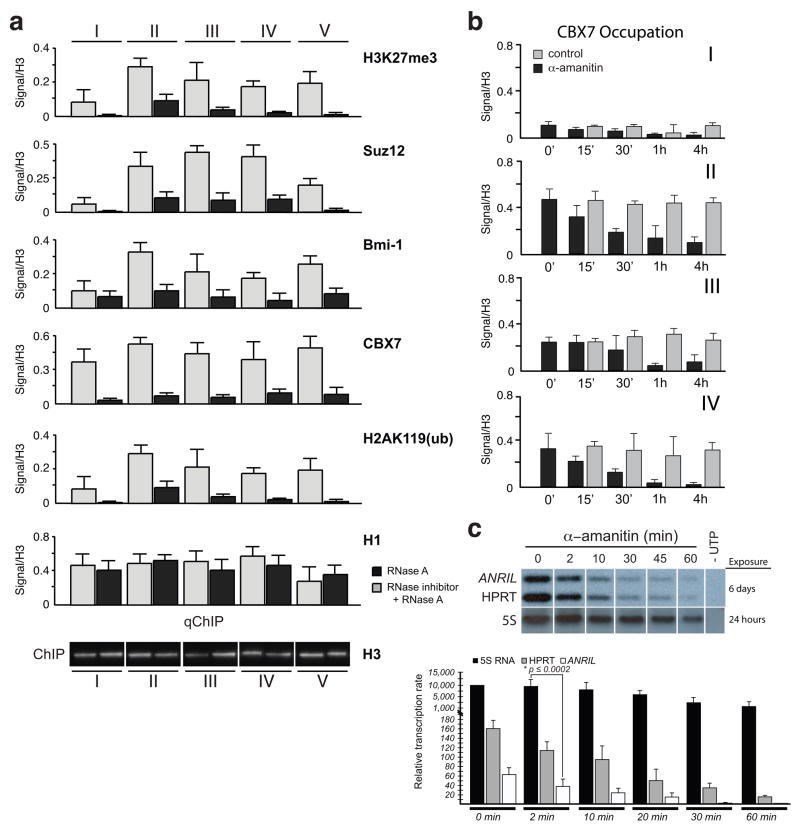
We then evaluated occupancy of CBX7 and other PRC proteins on the INK4b/ARF/INK4a locus by performing ChIP (Bracken et al., 2007) (Figure 1a). After treatment of PC3 cell nuclei with ribonuclease A (RNase A), site occupation by CBX7, PHC2 and Bmi1 of PRC1 as well as Suz12 of PRC2 was dramatically reduced while total levels of H1 and H3 remained about the same (Figure 4a). Levels of trimethylated H3K27 and ubiquitinated H2AK119, known as histone marks associated with PcG-mediated repression, were also greatly reduced at the corresponding CBX7 occupation sites. Moreover, RNAi knockdown of genes encoding the key PRC1 or PRC2 proteins individually or in combination with EZH2 results in a reduction of transcriptional repression of the INK4b/ARF/INK4a locus (Figure S2a,b,c). Reinstallation of H3K27 methylation in the EZH2 knocked down PC3 cells with controlled expression of a H3K27 methyltransferase vSET (Mujtaba et al., 2008) confirms the occupancy of CBX7, and to a lesser extent CBX8 but little if any CBX4 at the different sites in the INK4a/ARF locus, as revealed by ChIP analysis (Figure S2d). Finally, the depleted levels of CBX7 upon whole cell treatment of α-amanitin, a peptide inhibitor of RNA Pol II, support the notion that the RNA associating with CBX7 is a Pol II nascent transcript and is sensitive to Pol II inhibition more so at INK4a than INK4b (Figure 4b, I–IV). This result is consistent with in vitro transcription analysis of whole cell nuclei isolated from PC3 cells treated with α-amanitin, confirming that ANRIL is a nascent transcript generated by Pol II (Figure 4c). Collectively, these data illuminate a possible role of the ANRIL transcript in PcG recruitment and control of the INK4a/ARF locus.
Figure 4. Recruitment of Polycomb Proteins to the INK4a/ARF Locus is Mediated by RNA Polymerase II Transcripts.
a, Quantitative ChIP (qChIP) analysis from whole nuclei recovered from 3×109 PC3 cells following RNase A treatment, with or without RNase inhibitor added, at the sites noted in Figure 1a in Roman numerals (I to V) indicates the presence of CBX7, Bmi1, H3K27me3, PHC2 (polyhomeotic-like 2), Suz12 (Suppressor of Zeste 12 protein homolog), H2AK119Ub and histone H1. Controls are of mock treatment and levels of H3 are shown below. b, CBX7 occupation relative to histone H3 were measured with ChIP at the different sites along the INK4a/ARF locus and time points after PC3 cells treated with α-amanitin (10 μg/ml). c, In vitro transcription of ANRIL hnRNA transcripts from PC3 whole nuclei shows sensitivity to α-amanitin (upper panel). Exposure times for the transcripts are shown. Quantitative measurements of [α-32P]-UTP incorporation into nascent transcripts of ANRIL, HPRT and 5S RNA are plotted based on densitometry and liquid scintillation measurements of the [32P]-labeled transcripts (lower panel).
CBX7 Chromodomain Employs Structurally Distinct Modes to Bind RNA and H3K27me
To confirm that CBX7 directly associates with RNA we first examined the in vitro interaction between purified CBX7 and a 26-nt RNA identified from a modified RNA ChIP protocol using antisera against CBX7 in HeLa cells. RNA EMSA shows that purified CBX7 binds to this RNA, as does the chromodomain alone (CBX7CD) (Figure 5a). Because CBX7CD binds RNA in a manner similar to the full-length CBX7, and it is also the methyl-lysine recognition module of PRC1, we continued our in vitro studies with the chromodomain. CBX7CD binds RNA but not DNA (Figure 5a), but does not exhibit preference for different sequences of non-ANRIL RNA that we tested in vitro (data not shown) despite the observed contextual binding specificity for ANRIL by CBX7 (Figure 3a). Nevertheless, the CD of HP1γ, which shares approximately 50% sequence similarity to CBX7CD, does not bind RNA, while the CD of CBX8, the homolog most similar in sequence (~82%) to CBX7, binds RNA more weakly than CBX7CD, as indicated by the reduced intensity of band shifts (Figure 5a, right panel, lane 6 vs. lanes 7,8). An EMSA dually stained for RNA (SYBR Green) and protein (SYPRO Ruby) illuminates the RNA/CBX7CD species in yellow in a concentration dependent manner (Figure 5b). We observed increased viscosity of the protein/RNA complex at CBX7CD concentrations >0.8 mM, leading to decreased ability of the complex to migrate through the gel. However, CBX7CD/RNA binding is detected by reduction of free RNA signal (green dye at the bottom of the gel) as a function of protein concentration. The affinity for CBX7CD/RNA binding is estimated to be ~51 μM.
Figure 5. Molecular Interplay between ANRIL-derived RNA and H3K27me as mediated by the CBX7 chromodomain.
a, EMSA showing CBX7 full-length (CBX7FL, left) and chromodomain (CBX7CD, right) binding to RNA. Increasing concentrations of CBX7FL are shown in the presence of a 26-nt RNA; CBX7CD and other chromodomains are shown in the presence of a 13-nt ssDNA, 13-nt RNA (RNA13) or 26-nt RNA (RNA26). b, Fluorescence-based EMSA of titration of CBX7CD (red) into a constant concentration of RNA (green) shows an increasing amount of CBX7CD-RNA complex formation (yellow). The binding curve (KD = ~51 μM) based on complex overlay signal is shown at right. c, [1H,15N]-HSQC spectra of CBX7CD in the presence of H3K27me2 peptide, 14-nt ssDNA, 14-nt loop RNA and RNA after treatment with RNase A. The resonances of several residues not observed in the HSQC underwent severe line broadening and were confirmed by 3D experiments. d, Binding curves for CBX7CD binding to fluorescein-labeled ANRIL loopC RNA (black), H3K27me3 (red) or H3K27me2 (blue) peptide, as monitored by fluorescence anisotropy. e, H3K27me3/RNA competition experiments, in which unlabeled competing ligand is titrated against constant concentrations of CBX7CD and fluorescein-labeled ligand. f, Binding curves for CBX7CD binding to fluorescein-labeled ANRIL loop C in the presence (black) or absence (red) of saturating amounts of H3K27me3 peptide. The KD values for these curves are 75.9 ± 11.2 μM and 56.3 ± 6.8 μM, respectively, yielding a negative cooperativity factor α of 1.3. All anisotropy data were measured in triplicate. See also Figures S1, S3 and S4.
We next examined CBX7CD binding to RNA and methylated H3K27 in 2D [1H,15N]-HSQC (heteronuclear single quantum correlation) NMR spectra. Upon titration of an H3K27me3 peptide some protein amide peaks shift slightly while others undergo line broadening (Figure 5c, left panel); the latter effect was less severe with an H3K27me2 peptide (Figure S3a). Because of this, we carried out the detailed structural and biochemical binding studies (see below) using the H3K27me2 peptide. Based on the extent of peptide binding-induced NMR resonance perturbations, we observed a slight preference of CBX7CD for H3K27me3 over H3K27me2, and H3K27me3 over H3K9me3 (Figure S3a–c). Biological significance for CBX7CD/H3K9me3 binding, also reported in an in vitro study (Bernstein et al., 2006), remains unclear. Consistent with the RNA EMSA, CBX7CD binds RNA but not DNA (Figure 5c, middle panel), resulting in severe NMR line broadening at molar stoichiometry of CBX7CD/RNA as low as 1:0.1, which is more profound than that by the H3K27me peptide. Such an effect was noted for RNAs of different lengths (data not shown), and the HSQC spectrum was restored upon addition of RNase A (Figure 5c, right panel), indicating that the CBX7CD/RNA interaction is specific.
In an attempt to establish how this specificity is achieved we further analyzed a fragment of the target region of ANRILas-4 anti-sense RNA, which was used to competitively inhibit ANRIL transcripts (Figure 2). Secondary structure prediction of this target sequence revealed a number of potential hairpins (Figure S4a), three of which were assessed for binding by fluorescence anisotropy using fluorescein-labeled RNA (loops A–C, Figure S4b). The binding affinities observed for these potential RNA loops (14-16-nt in length; KD = 13.7 ± 1.3 μM, 30.2 ± 3.7 μM, 14.6 ± 2.7 μM, respectively) were notably stronger than that for the 26-nt RNA identified from the HeLa ChIP assay (KD = 121.1 ± 22.1 μM) (Table S1).
We measured fluorescence anisotropy of fluorescein-labeled H3K27me3 or H3K27me2 peptide upon binding to CBX7CD, and determined comparable affinities of KD = 59.0 ± 4.7 μM and 108.4 ± 7.4 μM, respectively (Figure 5d), which are consistent with NMR measurements. We observed a stronger binding affinity for H3K9me3 (KD = 16.5 ± 0.8 μM, Figure S3d) by fluorescence anisotropy compared with that obtained by NMR, likely due to differences in assay and peptide conditions. We next assessed a possible mutual influence between CBX7CD binding to methyl-lysine and to ANRIL RNA loop C (Figure 5e). In the presence of 50 μM CBX7CD (~55% saturation), titration of non-labeled RNA results in a rise in H3K27me3 anisotropy signal, suggesting possible formation of a ternary complex. The non-labeled RNA (up to 500 μM) does not bind to the H3K27me peptide alone. We observed a similar increase in ANRIL loop C RNA anisotropy signal with the titration of non-labeled H3K27me3 peptide. However, at higher peptide concentrations (>500 μM) the signal starts to decrease, indicating that the RNA is being competed off of CBX7CD. This competing effect appears consistent with our EMSA observation of a decrease of PC3-isolated PRC1/in vitro translated ANRIL RNA complex formation upon titration with H3K27me peptide from 50–250 μM, but not with H3K4me (Figure S1e). By measuring the binding affinity of fluorescein-labeled ANRIL loop C RNA for CBX7CD in the presence of saturating concentrations of H3K27me3 peptide, we established that a slight negative cooperativity exists between H3K27me3 and loop C (α = 1.3, see Figure 5f). Therefore, the three components (CBX7CD, H3K27me3 peptide and ANRIL loop RNA) are able to form a ternary complex (Figure 5e), but the presence of one ligand weakens the affinity of CBX7CD for the other, confirming that at very high peptide concentrations it is able to compete with RNA for binding to CBX7CD (Figure 5e, right). Taken together, our data suggest that the binding mode of CBX7 may be dictated by the relative concentration of RNA and H3K27me3 at the locus.
CBX7 Chromodomain Uses Distinct Regions and Residues for Binding H3K27me or RNA
To understand the molecular interplay between CBX7CD binding to H3K27me and to RNA, we solved the three-dimensional solution structure of CBX7CD bound to an H3K27me2 peptide (Figure 6a, Table 1). We could not use an H3K27me3 peptide as it caused severe line broadening effects of protein NMR resonances, thus complicating resonance assignments and precluding structure determination. The structure is similar to that of other chromodomains bound to H3 peptides including mouse HP1βCD/H3K9me2 and Drosophila PcCD/H3K27me3 (Fischle et al., 2003; Jacobs and Khorasanizadeh, 2002; Min et al., 2003; Nielsen et al., 2002). The dimethyl group of H3K27 resides in an aromatic cage composed of Phe 11, Trp 32 and Trp 35 in a position similar to that of the trimethyl group of H3K27 in the PcCD structure (Figure S5). This supports the notion that the mode of interaction for the trimethyl group by CBX7CD is likely the same. A higher binding affinity is expected by an additional methyl group, which should allow for stronger polar and van der Waals interactions with the protein (Hughes et al., 2007; Jacobs and Khorasanizadeh, 2002).
Figure 6. CBX7 Chromodomain Employs Distinct Regions and Residues for Binding H3K27me or RNA.
a, Representative 3D NMR structure of CBX7CD in complex with H3K27me2 peptide depicted as ribbon or surface colored by Poisson-Boltzmann electrostatic potential, with aromatic cage residues as shown. b, CBX7CD structure with residues as determined by NMR titration depicting severe line broadening (red) or moderate peak shifts (yellow) upon binding H3K27me2 peptide. c, Residues undergoing perturbation upon RNA addition at early (CBX7CD:RNA molar ratio 1:0.2, blue) and later (1:0.8, cyan) points during titration. d, Fluorescence-based EMSA depicting 26-nt RNA binding to different point mutants of CBX7CD based on the NMR titration in b and c. e, [1H,15N]-HSQC spectra of wild-type and corresponding point mutants showing affected H3K27me2 or RNA binding. See also Figures S5 and S6.
Table 1.
NMR and Structural Statistics of the CBX7CD/H3K27me2 Complex
| NMR distance and dihedral constraints | |
|---|---|
| Distance restraints | |
| Total NOE | 1,735 |
| Intra-residue | 879 |
| Inter-residue | 856 |
| Sequential | 298 |
| Non-sequential | 558 |
| Hydrogen bonds | 33 |
| Protein-peptide intermolecular | 25 |
| Total dihedral angle restraints | 78 |
| phi | 39 |
| psi | 39 |
| Structure statistics | ||
|---|---|---|
| Violations | ||
| Distance constraints (Å) | 0.037 ± 0.011 | |
| Dihedral angle constraints (°) | 0.222 ± 0.043 | |
| Maximum dihedral angle violation (°) | 0.344 | |
| Maximum distance constraint violation (°) | 0.054 | |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.003 ± 0.000 | |
| Bond angles (°) | 0.432 ± 0.017 | |
| Impropers (°) | 0.355 ± 0.020 | |
| Average pairwise r.m.s.d., 20 structures (Å) | ||
| CBX7CD (aa 13–64) | ||
| Heavy | 0.792 ± 0.055 | |
| Backbone | 0.295 ± 0.062 | |
| H3K27me2 peptide (aa 23–27) | ||
| Heavy | 1.083 ± 0.190 | |
| Backbone | 0.344 ± 0.155 | |
| Complex | ||
| Heavy | 0.843 ± 0.050 | |
| Backbone | 0.329 ± 0.059 | |
| Ramachandran plot statistics | All structured* | Secondary structure** |
| Most favored regions (%) | 78.4 | 85.3 |
| Additional allowed (%) | 21.6 | 14.7 |
| Generously allowed (%) | 0.0 | 0.0 |
| Disallowed regions (%) | 0.0 | 0.0 |
CBX7CD residues 13–64, H3K27me2 peptide residues 23–27.
CBX7CD residues 14–22, 25–32, 41–44, 51–65.
CBX7CD residues that show major backbone amide chemical shift changes are clustered in regions mostly surrounding the H3K27me2 binding site (Figure 6b), which was confirmed by our nuclear Overhauser effect (NOE) data. Indeed, NOEs were only observed for residues 23–29 of H3, indicating the flexible nature of the two ends of the peptide in the complex structure. We next mapped the residues undergoing the strongest line broadening at the earliest RNA titration points onto the structure (Figure 6c), and found that while common residues are also perturbed in peptide binding, a distinct and contiguous surface perturbed in RNA binding is somewhat distal to the peptide binding site.
We designed point mutations of CBX7CD to abolish either H3K27me (F11A, W32A, W35A) or RNA binding (E14A, R17A, K31A, K33A). These residues were chosen based on their location in the methyl-lysine binding site or their NMR resonance perturbation upon RNA binding (Figure 6c, dark blue). From comparative binding studies by EMSA (Figure 6d), NMR (Figure 6e) and fluorescence anisotropy (Table S1), an R17A mutation showed a nearly complete loss of RNA binding, but retained H3K27me binding at a level comparable to that of the wild-type. Conversely, a W35A mutation adversely affected binding to H3K27me (Figure 6e), but not RNA (Figure 6d, e). Moreover, an R17AW35A double mutant exhibited inability to bind either H3K27me or RNA (Figure S6a). We also evaluated RNA binding by CBX7 fusions to the DNA binding domain of the GAL4 transactivator (CBX7/GAL4) in nuclear extracts recovered from HeLa cells and observe strongly reduced RNA-protein complex mobility by both R17Q and R17QW35A mutants, but not by wild-type or W35A mutant (Figure S1d). Finally, we ruled out mutation effects on any major conformational changes in the protein, as the HSQC spectra of the mutants show spectral changes localized to the mutation sites (Figure 6e and Figure S6b).
Relative contribution of CBX7 interactions with H3K27me and RNA to senescence regulation
Consistent with our previous results, we observed that ANRIL expression declined during serial passage of IMR90 cells, correlating with increased levels of INK4a transcription as cells reached senescence (Figure 7a). To dissect the effect of CBX7 interaction with H3K27me and RNA on its function, we expressed wild type or mutant versions of CBX7 in MEFs (Figure 7) or IMR90 cells (Figure S7) and analyzed their effects on replicative lifespan and expression of the INK4b/ARF/INK4a locus. Expression of wild type Cbx7 (mouse) resulted in extended cellular lifespan and increased cell growth as observed in growth curves, colony formation assays (Figure 7b–c) or by quantifying BrdU incorporation at different passages (Figure S7d). However, expression of a Cbx7 W35A mutant, which affects Cbx7 ability to bind to H3K27me, resulted in premature growth arrest with characteristics of senescence, similar to the effect observed upon expression of oncogenic RasV12. We also tested a Cbx7 R17A mutant, which specifically affects Cbx7 ability to bind to RNA. Expression of this mutant resulted in extended lifespan of MEFs when compared with vector-infected controls, but cells did not grow as vigorously as those expressing Cbx7 wild type (Figure 7b, c and Figure S7d). The defects in growth of the RNA binding deficient Cbx7 R17A mutant were more obvious in colony formation assays (Figure 7c) and resulted in decreased BrdU incorporation when compared to Cbx7 wt (Figure S7d). Analysis of the effect of the different CBX7 mutants on the expression of the products of the Ink4b/ARF/Ink4a locus by Western blot or RT-PCR (Figure 7d,e) showed that the W35A mutation caused an up-regulation of p19Arf, p16Ink4a and to a lesser extent p15Ink4b. Conversely, expression of Cbx7 wild type repressed p19Arf, p16Ink4a and to a lesser extent p15Ink4b when compared with vector-infected control cells. Consistent with its effects on cell growth, the expression of a Cbx7 R17Q mutant resulted in less effective repression of p16Ink4a and p19Arf compared to Cbx7 wild type. Expression of p15Ink4b was similar in Cbx7 R17Q infected cells to that in the vector controls (Figure 7d,e). Similar results were observed in the IMR-90 cells (see Figure S7a–c). Although mutations affecting Cbx7 binding to H3K27me have a more dramatic effect on cellular lifespan than those affecting its ability to bind RNA, overall these data confirm that ablation of either H3K27me or RNA binding abilities of CBX7 compromises its capacity to repress the INK4b/ARF/INK4a locus and control senescence.
Figure 7. Relative contributions of CBX7 interactions with H3K27me and RNA to senescence regulation.
a, Comparison of transcript levels relative to HPRT of ANRIL and INK4a in IMR90 cells undergoing replicative senescence. b, MEFs were infected with the indicated vectors and after selection, growth curves were performed. c, Colony formation assay of MEFs infected with the indicated vectors. Cells were seeded at passage 4. d, Western blots assessing expression levels of p15Ink4b, p16Ink4a and p19Arf in MEFs infected with the indicated constructs. e, Quantitative RT-PCR showing levels of Ink4b, Ink4a and Arf in same cells as in d. f, Model of ncRNA-mediated gene repression of the INK4b/ARF/INK4a locus by the Polycomb group complexes. Schematic model illustrating molecular interplay between methylated H3K27 (by EZH2 of PRC2) and ncRNA transcripts ANRIL for silencing of the INK4b/ARF/INK4a locus by Polycomb CBX7. Both PRC1 and PRC2 complexes are retained at a repression site of a target gene through their association with the nascent ANRIL transcripts of Pol II. Note that the stem loop RNA structure is used for illustration only; RNA/CBX7 binding could result in dissociation of PRC1 from H3K27me, leading to a reversal of transcriptional repression of the target gene. See also Figure S7.
CONCLUDING REMARKS
Growing evidence argues that RNA, particularly long non-coding RNAs, is actively engaged in gene transcription (Hirota et al., 2008; Sessa et al., 2007; Wang et al., 2008; Zhao et al., 2008), but how ncRNAs function mechanistically in concert with histone modifications in gene regulation has remained elusive. Recently, PRC2, which methylates H3K27 through its component EZH2, has been suggested to be recruited to its target gene by virtue of its association with HOTAIR, a 2.2 kb ncRNA in the HOXC locus (Rinn et al., 2007), which could be a general mechanism for in trans recruitment of PRC complexes at non-HOX loci. Our identification of the ncRNA transcript ANRIL associated with the INK4a/ARF locus argues that the nascent ncRNA transcripts could be involved in in cis recruitment of PRC complexes for gene silencing. In this capacity, long ncRNA likely functions as an architectural matrix in addition to its instructional role to recruit and retain the PRC complexes at the target gene locus (Bracken et al., 2007; Wang et al., 2008) in conjunction with the spreading of chromatin structure compaction that is associated with gene repression. In principle, the nascent RNA produced by Pol II may also recruit a H3K4 methyltransferase, identified as mixed leukemia lineage (MLL) of the activating Trithorax complex (Trx) (Lee et al., 2007; Rinn et al., 2007), which in turn recruits a JmjC-family H3K27 demethylase leading to a reversal of gene repression. We therefore propose a general model, in which ncRNA, always present at the locus, recruits both PRC1 and PRC2 to the target gene for silencing; in balance with Trx, the modification, modification-mediated interactions, and de-modification of H3K27me is likely dynamically modulated by these multi-protein complexes (Jensen et al., 2009; Ringrose and Paro, 2004) along with ncRNA (Figure 7f).
It is interesting to note that in Drosophila, PRC1 can be recruited to PREs independent of the H3K27 methylation induced by PRC2, and that repression might require H3K27me in regions flanking, rather than within PREs (Jensen et al., 2009). It is unclear, however, whether ncRNA is directly required for repression in this case, or that the effects observed are due to compromised recruitment to the locus. This argues that the contextual specific binding of CBX7 to ANRIL we observed, which may be manifested through RNA structure-mediated interactions with the CBX chromodomain, is likely dictated by other subunits of CBX7/PRC1 capable of binding RNA or even other RNA binding proteins, which together provide greater complexity to the organization of ncRNAs in regulating chromatin structure (Wilusz et al., 2009). This notion is supported by the purification of PRC1 through CBX7 that identifies multiple RNA binding proteins (Figure 3b). Further studies are required to determine their role in augmenting RNA-binding specificity. In conclusion, our study reported here provides a new molecular mechanism of how ncRNA transcripts functionally coordinate with the chromatin-associated factors that modify and interact with H3K27 methylation, a repressive mark for gene transcription.
EXPERIMENTAL PROCEDURES
RNA Fluorescent in situ Hybridization (RNA-FISH)
Prostate specimens received as a generous resource from the NYU Langone Medical Center’s Department of Pathology (Drs. Eva Hernando and Jonathan Melamed) were obtained with prior IRB approval following all HIPAA guidelines. 10 mm sections were fixed in 4% formaldehyde, 5% acetic acid and 150 mM NaCl prior to drying. Anti-sense RNA probe preparation, probe denaturation, hybridizations, and detection were performed using the FISH Tag™ RNA Red Kit (Invitrogen).
RNA Recovery and qPCR of ANRIL Transcripts From Human Prostate Specimens
Frozen specimens recovered from dissected tissue surrounding normal prostate epithelium, prostate intraepithelial neoplasias, and malignant prostate duct tumors as described above were prepared as total RNA using QuantiTect Whole Transcriptome Kit (Qiagen). qPCR was performed with TaqMan probes and quantitation of ANRIL transcripts by SYBR Green labeling.
Genomic Mapping of CBX7, EZH2 and RNA Polymerase II Across the INK4a/ARF Locus
Using standardized ChIP methodology and the MagnaChIP kit (Millipore), the genomic mapping of CBX7, EZH2 and RNA polymerase II was performed at the positions relative to key transcript start sites overlapping the position of human chr 9 (chr9:21,710,518-22,301,141). DNA isolated from ChIP experiments was subjected to quantitation by real time PCR using Brilliant SYBR green master mix (Stratagene). Primers were identified using Geneious Pro 4.0 for primer alignment of genomic regions.
RNA-Chromatin Immunoprecipitation (CLIP) Assay
PC3 cells were cultured under normal growth conditions (Li et al., 2005). Following UV cross-linking isolation of chromatin from the treated PC3 cells was performed using standard nuclear chromatin isolation techniques (Kuo and Allis, 1999). CLIP was performed using a previously established procedure (Ule et al., 2005) with modifications.
RNA electrophoretic mobility shift assay (RNA EMSA)
The fragment of ANRIL cDNA generated by reverse transcription PCR was cloned into pTOPO into the correct orientation. Radiolabeled transcripts were generated by in vitro transcription using the Riboprobe® System with T7 RNA Polymerase (Promega) and [32P]-UTP (UTP, [α-32P]-3000Ci/mmol 10mCi/ml; Perkin-Elmer). Biotin UTP-labeled RNA transcripts were generated using in vitro transcription-translation (Promega) for detection by the LightShift kit (Pierce).
5′ Primer Extension (5′-RACE) Analysis of ANRIL Transcripts
Primer extension analysis was carried out with RNA isolated from nuclei of PC3 cells and using the primer extension system with AMV reverse transcriptase (Promega). Isolation of nuclear RNA (hnRNA) from PC3 cells was carried out using a cytoplasmic/nuclear RNA separation and preparation system (Norgen Biotek Inc.).
In vitro Transcription Analysis of ANRIL From Whole Nuclei
Whole nuclei from 5 × 107 PC3 cells were collected as described previously (Walsh et al., 1987) and kept on ice. Nuclei numbers were obtained by staining with 4′, 6-diamidino-2-phenylindole (DAPI), and counted with a hemacytometer using fluorescence microscopy.
NMR Structure Determination
The three-dimensional solution structure of the CBX7CD/H3K27me2 complex (1:2 molar ratio) was determined in PBS buffer, pH 7.4, containing 5 mM DTT, 90% H2O/10% 2H2O at 25°C, using established 2D and 3D NMR methods (Clore and Gronenborn, 1994). Distance restraints were obtained from 3D 15N and/or 13C-edited, as well as 15N/13C- filtered/edited NOESY spectra. Dihedral angle and hydrogen bond restraints were derived from TALOS analysis (Cornilescu et al., 1999), and the structures of the complex were calculated using ARIA/CNS (Brunger et al., 1998; Rieping et al., 2007).
Retrovirus Production, Infection and Proliferation Assays
Mouse CBX7 was cloned into the MSCV-iresGFP vector to express Cbx7-GFP fusion protein. Retroviral production, infection, growth curves, colony formation assays, BrdU incorporation, western blot and qPCR analysis of members of the INK4b/ARF/INK4a locus were performed as described (Barradas et al., 2009; Gil et al., 2004). Growth kinetics and p16 expression in normal diploid fibroblasts were performed using established methods.
Supplementary Material
Acknowledgments
We wish to acknowledge the use of the NMR Facility at the New York Structural Biology Center. We thank S. Hearn of the Cold Spring Harbor Laboratory Core Microscopy Facility for advice and help with fluorescence microscopy and imaging, G. Gerona-Navarro for fluorescein-labeled peptide and A. Kelly for help with data analysis. This work was supported by a Terry Fox Foundation postdoctoral fellowship from the National Cancer Institute of Canada (K.L.Y.) and grants from the Empire State Stem Cell Trust Fund (NYSTEM) and the National Institutes of Health (M.-M.Z and M.J.W.), MRC UK (J.G.) and the National Science Foundation (M.-M.Z.).
Footnotes
ACCESSION NUMBERS
Accession numbers for the coordinates of the CBX7CD/H3K27me2 complex structure and the NMR resonance assignments deposited in the Protein Data Bank and the BioMagResBank are 2KVM and 16778, respectively.
Supplemental Data include Experimental Procedures, 7 figures, and 2 tables that can be found with this article online at…
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Martinez-Leal J, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis C. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan E, Masui O, Gil J, Heard E, Allis C. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs S, Kim Y, Allis C, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hughes R, Wiggins K, Khorasanizadeh S, Waters M. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci USA. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose R, Erdjument-Bromage H, Tempst P, Jones R, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- Li D, Yea S, Dolios G, Martignetti JA, Narla G, Wang R, Walsh MJ, Friedman SL. Regulation of Kruppel-like factor 6 tumor suppressor activity by acetylation. Cancer Res. 2005;65:9216–9225. doi: 10.1158/0008-5472.CAN-05-1040. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu R. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, Manzur KL, Gurnon JR, Kang M, Van Etten JL, Zhou MM. Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol. 2008;10:1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nielsen P, Nietlispach D, Mott H, Callaghan J, Bannister A, Kouzarides T, Murzin A, Murzina N, Laue E. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rinn J, Kertesz M, Wang J, Squazzo S, Xu X, Brugmann S, Goodnough L, Helms J, Farnham P, Segal E, Chang H. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, Braund PS, Erdmann J, Viikari J, Moilanen L, et al. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1679–1683. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, Martinez D, Visakorpi T, Mu D, Cordon-Cardo C, Peters G, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Fraser P, Nagano T. The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol Chem. 2008;389:323–331. doi: 10.1515/BC.2008.047. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Walsh MJ, LeLeiko NS, Sterling KM., Jr Regulation of types I, III, and IV procollagen mRNA synthesis in glucocorticoid-mediated intestinal development. J Biol Chem. 1987;262:10814–10818. [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine D, Martienssen R. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.