The stoichiometry of kinetochore components is determined, suggesting conservation between multiple microtubule-binding vertebrate and single microtubule-binding yeast kinetochores.
Abstract
To define the molecular architecture of the kinetochore in vertebrate cells, we measured the copy number of eight kinetochore proteins that link kinetochore microtubules (MTs [kMTs]) to centromeric DNA. We used a fluorescence ratio method and chicken DT40 cell lines in which endogenous loci encoding the analyzed proteins were deleted and complemented using integrated green fluorescent protein fusion transgenes. For a mean of 4.3 kMTs at metaphase, the protein copy number per kMT is between seven and nine for members of the MT-binding KNL-1/Mis12 complex/Ndc80 complex network. It was between six and nine for four members of the constitutive centromere-associated network: centromere protein C (CENP-C), CENP-H, CENP-I, and CENP-T. The similarity in copy number per kMT for all of these proteins suggests that each MT end is linked to DNA by six to nine fibrous unit attachment modules in vertebrate cells, a conclusion that indicates architectural conservation between multiple MT-binding vertebrate and single MT-binding budding yeast kinetochores.
Introduction
The architecture of the proteins that link plus ends of kinetochore microtubules (MTs [kMTs]) to centromeric DNA is critical for understanding protein mechanisms for the four essential kinetochore functions: dynamic attachment to the plus ends of spindle MTs, force generation, attachment error correction, and the spindle assembly checkpoint (Skibbens et al., 1993; Inoué and Salmon, 1995; Pearson et al., 2001; Tirnauer et al., 2002; Musacchio and Salmon, 2007; Cimini, 2008). Particularly important for these essential functions are three highly conserved protein complexes (KNL-1/Mis12 complex/Ndc80 complex [KMN]) that assemble stably within the kinetochore to produce core attachment sites for kMTs: KNL-1 (hBlinkin)/ScSpc105, the Mis12 complex of four proteins (hMis12/ScMtw1, Dsn1, Nsl1, and Nnf1), and the four-subunit Ndc80 complex (Ndc80 (hHec1), Nuf2, Spc24, and Spc25), which, like KNL-1, binds MTs (Santaguida and Musacchio, 2009). The KMN network is linked to centromeric DNA at vertebrate kinetochores by members of the constitutive centromere-associated network (CCAN): centromere protein C (CENP-C), CENP-H, CENP-I, CENP-K-U, CENP-W, and CENP-X (Kline et al., 2006; Liu et al., 2006; Okada et al., 2006; Cheeseman et al., 2008; Amano et al., 2009; Santaguida and Musacchio, 2009). A CENP-T–CENP-W dimer and CENP-C are DNA-binding proteins that independently associate with histone H3 nucleosomes in the proximity of CENP-A nucleosomes (Hori et al., 2008). CENP-A is a modified histone H3 that specifies where kinetochores are assembled on centromeric DNA (Santaguida and Musacchio, 2009).
Important information about the architecture of the proteins linking kMTs to centromere DNA has been obtained using two-color fluorescence light microscopy methods to achieve nanometer scale measurements of the relative positions of these kinetochore proteins or their functional homologues along the axis of kMTs at metaphase in budding yeast (Joglekar et al., 2009) and human cells (Wan et al., 2009). Protein copy number per kMT is equally important for understanding kinetochore protein architecture and function (Musacchio and Salmon, 2007; Santaguida and Musacchio, 2009; Joglekar et al., 2010). We have made this measurement in budding and fission yeast with a fluorescence ratio method that used GFP fusion proteins expressed from endogenous promoters (Joglekar et al., 2006, 2008). For the KMN network, the numbers for both yeast species at metaphase are six to eight per kMT. In contrast, Emanuele et al. (2005) used biochemical methods to estimate ∼30 Ndc80 complexes per kMT for isolated Xenopus laevis chromosomes.
Protein homology and architecture within kinetochores of budding yeast and vertebrate cells predict a conserved MT attachment site (Joglekar et al., 2009; Wan et al., 2009). However, it is possible that protein copy number per kMT could be significantly different between budding yeast and vertebrates. There is significant divergence in their centromeres, in the primary sequences of homologous proteins, and in their kMT-binding capacity: kinetochores in budding yeast attach to one kMT, whereas kinetochores in vertebrates attach to multiple kMTs.
To test whether the protein copy numbers for vertebrate kinetochores are similar or different from the known numbers for budding yeast kinetochores, we used chicken DT40 cells because endogenous coding regions can be deleted and complemented by GFP fusions (Mikami et al., 2005; Hori et al., 2008). We applied the same ratio fluorescence method used previously for budding and fission yeast (Joglekar et al., 2006, 2008). The proteins analyzed included key members of the major protein complexes of the core kMT attachment site and its linkage to centromeric DNA (Santaguida and Musacchio, 2009).
Results and discussion
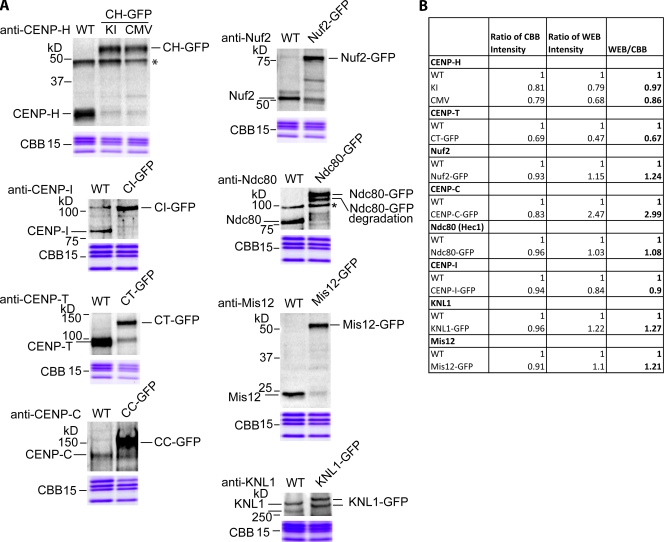
Measurements of protein copy number at metaphase kinetochores in DT40 cells were made for Nuf2, Ndc80 (Hec1), Mis12, KNL-1, CENP-H, CENP-I, CENP-T, and CENP-C using cell lines where the endogenous kinetochore protein was genetically knocked out and replaced by the protein fused to GFP (EGFP) expressed either from its endogenous promoter or a cytomegalovirus (CMV) promoter (Fukagawa et al., 2001, Mikami et al., 2005; Hori et al., 2008). In these cell lines, GFP fusion proteins were expressed on average near the level of the endogenous protein (except for CENP-C–GFP, which was about threefold higher), and unlabeled protein represented a very small fraction of the total protein (Fig. 1). Faint bands around the size of endogenous proteins were detected in all cells expressing GFP fusion proteins (Fig. 1 A). We confirmed that these were not endogenous proteins because endogenous proteins were not detected by Western blotting in parental knockout cell lines (Fukagawa et al., 2001; Hori et al., 2003, 2008). Under the conditions used in our experiments, all of the GFP fusion cell lines grew vigorously at a rate similar to wild-type cells.
Figure 1.
Measurements from Western blots of the mean level of GFP (EGFP) fusion protein expression relative to wild type. The nine DT40 cell lines used for measurement of protein copy number in the linkage between chromatin and kMTs are shown. The genomic region coding endogenous protein has been knocked out by homologous recombination for all cell lines. (A) Western blots for comparing level of GFP fusion protein in the cell lines compared with wild-type (WT) levels (top) and Coomassie brilliant blue (CBB) staining of a loading control protein (bottom). We prepared two gels and applied the same amount of protein for each. We used one gel for Western blots, and we used another one for Coomassie staining. Note that in CENP-H (CH)–GFP, the fusion protein was expressed from either the endogenous promoter (KI) or a CMV promoter. The rest of the cell lines have GFP fusion protein expression from a CMV promoter. Asterisks indicate nonspecific bands. CI, CENP-I; CT, CENP-T; CC, CENP-C. (B) Calculation of the mean level of GFP fusion protein relative to wild type (fourth column) from the ratio of Western blot (WEB) intensity and Coomassie intensity relative to wild type measured from Western blots and protein stains in A. Note that division of the Western blot intensity ratio to WT by the CBB intensity ratio to wild type corrects for variations in sample size. Also note that CENP-H–GFP (KI) has the same mean level of CENP-H–GFP protein expression relative to wild type (0.97) as CENP-H–GFP expressed from a CMV promoter (0.86).
The standard for our fluorescence ratio method was the fluorescence from a cluster of anaphase kinetochores in budding yeast cells where GFP (GFP (S65T)) was fused to Ndc80 at the endogenous promoter (Fig. 2, budding yeast). A cluster contains 16 kinetochores each with one kMT. Previous measurements found seven Ndc80-GFP proteins on average per kMT, which is slightly less than the copy number of eight at metaphase (Joglekar et al., 2006). The total GFPs per cluster was 7 × 16 = 112. GFP (S65T) has the same fluorescence properties as the EGFP used in DT40 cells at the room temperature used to image both cell types (see Materials and methods; Yang et al., 1996; Patterson et al., 1997).
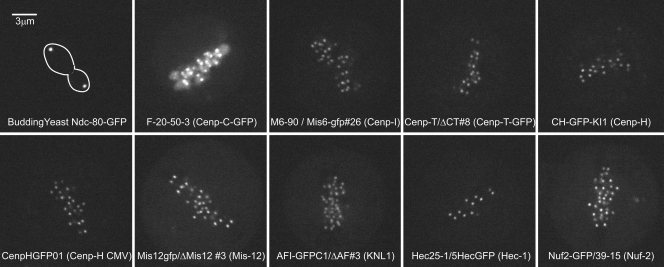
Figure 2.
Confocal fluorescence images of cells expressing different kinetochore proteins fused to GFP. The names of each cell line are given on its corresponding image. For visual comparison, all images were obtained under the same excitation light intensity, exposure, approximate depth from the coverslip surface, and microscope/camera instrumentation, and displayed at the same brightness and contrast.
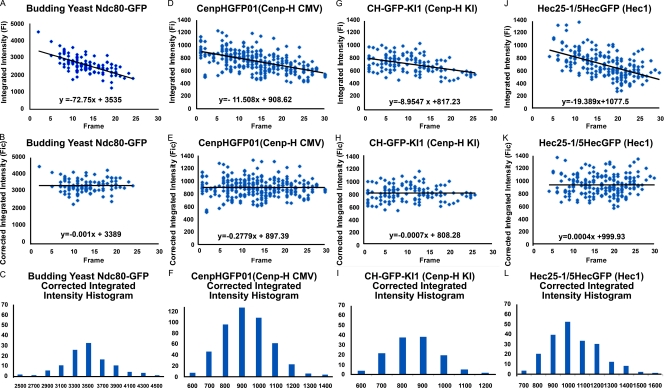
We measured Fi (integrated fluorescence intensity minus background) from images (Fig. 2) of anaphase kinetochore clusters in budding yeast or individual metaphase kinetochores in DT40 cells using the method previously described by Hoffman et al. (2001; Fig. S1 A). For each cell type, kinetochore Fi was highest near the coverslip inner surface and decreased with image plane number (Fig. 3, A, D, G, and J), where image plane number multiplied by 200 nm gives a measure of depth within the cell. Image plane number also represents the number of exposures. Repetitive exposures at one image plane showed that photobleaching did not make a significant contribution to the decrease in measured value with image plane number (Fig. S1 B); most of the decrease was caused by refraction at the glass–media interface that causes the central intensity of fluorescence to be reduced (Joglekar et al., 2006). To correct for this effect, we fitted a linear regression line to the primary data, and used the frame number at the first data measurement and the slope, to correct the Fi data at higher frame numbers (Fig. 3, B, E, H, and K). This produced corrected data, Fic, whose distribution was approximately normal (Fig. 3, C, F, I, and L) and yielded a mean value and SD for the population of measured kinetochores where n > 150. This correction method produced means close to the values measured near the coverslip while providing a measure of the SD for each dataset.
Figure 3.
Examples of Fi measurements. (A–L) Clusters of anaphase sister kinetochores in budding yeast expressing Ndc80-GFP from an endogenous promoter (A–C) and for kinetochores in metaphase DT40 cells depleted of the endogenous protein and expressing CENP-H–GFP from an endogenous (KI; D–F) or CMV promoter (G–I) or expressing Ndc80 (Hec1)–GFP from a CMV promoter (J–L). Fic (B, E, H, and K) and histograms of the corrected data (C, F, I, and L) were obtained as described in Results and discussion.
We first examined the CENP-H–GFP knock-in cells because CENP-H–GFP was expressed on the endogenous promoter. The mean level of CENP-H–GFP expression was nearly identical to the level of endogenous protein in wild-type cells (Fig. 1). The total cell fluorescence minus background varied between 6 × 104 and 9 × 104 counts for an image near the middle of the cell (Fig. S1 A). The sum of Fic measurements for kinetochores within the same plane revealed that kinetochores contributed <5–10% total cell fluorescence, and most was from fluorescence within the cytosol. As a result, for the other DT40 cell lines that expressed GFP fusion protein from a CMV promoter, we chose cells for analysis with total cell fluorescence near the aforementioned range (4–14 × 104) to minimize possible effects from protein overexpression, although cells outside this range were rare. The exception was the CENP-C–GFP cell line, which had on average significantly higher total cell fluorescence compared with the CENP-H–GFP knock-in cell line, as predicted by the Western blot analysis in Fig. 1. Most of the CENP-C–GFP cells showed various levels of fluorescence within the chromosome arms (Fig. 2), which was background subtracted from measurements of kinetochore fluorescence (Fig. S1 A). For the different cell lines, kinetochore Fic was not sensitive to different amounts of total cell fluorescence minus background (Fig. S1 C). This insensitivity is consistent with the stable association of the Ndc80 complex at kinetochores (Hori et al., 2003).
DT40 cells typically display a stable karyotype with a chromosome number of 80, which comprises 11 autosomal macrochromosomes, the heterogametic sex chromosomes, and 67 minichromosomes (Sonoda et al., 1998). The largest macrochromosomes have ∼20 times more DNA than the minichromosomes. In contrast to this wide range in chromosome size, fluorescent images of kinetochores in live cells (Fig. 2) did not show distinctly different populations of kinetochore fluorescence within the same cell, and measurement histograms typically showed a single peak with ∼20% or less SD (Fig. 3, C, F, I, and L; and Table I). As a further test for a distinct difference in kinetochore size between macrochromosomes and minichromosomes, we measured kinetochore fluorescence in metaphase chromosome spreads from hypotonically lysed cells (Fig. S2). The ratio of mean fluorescence obtained from kinetochores of macrochromosomes and minichromosomes was about one using immunofluorescent staining with either CENP-T or Ndc80 antibodies. Collectively, the aforementioned data indicate that kinetochores of macrochromosomes and minichromosomes contain similar amounts of proteins, although their DNA content can differ by more than an order of magnitude.
Table I.
Kinetochore Fic and protein number per kinetochore and kMT for GFP fusion cell lines
| GFP fusion cell lines | Fic | Ratio to yeast Ndc80 | Protein No./kinetochore | Protein No./kMT |
| Budding yeast Ndc80 | 3,390 ± 373 | 1.00 ± 0.11 | NA | NA |
| CENP-C | 1,192 ± 312 | 0.35 ± 0.09 | 39 ± 10 | 9.2 ± 2.4 |
| CENP-I | 757 ± 113 | 0.22 ± 0.03 | 25 ± 4 | 5.8 ± 0.9 |
| CENP-T | 851 ± 119 | 0.25 ± 0.04 | 28 ± 4 | 6.5 ± 0.9 |
| CENP-H CMV | 879 ± 147 | 0.26 ± 0.04 | 29 ± 5 | 6.8 ± 1.1 |
| CENP-H (KI) | 808 ± 121 | 0.24 ± 0.04 | 27 ± 4 | 6.2 ± 0.9 |
| Mis12 | 1,172 ± 271 | 0.35 ± 0.08 | 39 ± 9 | 9.0 ± 2.1 |
| KNL-1 | 932 ± 170 | 0.28 ± 0.05 | 31 ± 6 | 7.2 ± 1.3 |
| Ndc80 (Hec1) | 1,000 ± 178 | 0.29 ± 0.05 | 33 ± 6 | 7.7 ± 1.4 |
| Nuf2 | 1,128 ± 251 | 0.33 ± 0.07 | 37 ± 8 | 8.7 ± 1.9 |
NA, not applicable. Mean values for Fic were measured from kinetochores for each GFP fusion cell line, and calculations of protein copy number per kinetochore and kMT were obtained by the ratio fluorescence method. Ratio values to yeast Ndc80-GFP were obtained by dividing the mean Fic by the value for yeast Ndc80-GFP. The protein number per kinetochore in DT40 cells was obtained by multiplying the ratio to yeast Ndc80 by the number of Ndc80-GFPs in an anaphase kinetochore cluster (112; Joglekar et al., 2006). The protein number per kMT was obtained by dividing the ratio to yeast Ndc80 by the mean number of kMTs in metaphase DT40 cells (4.3; Ribeiro et al., 2009).
Table I and Fig. 4 show the results from our analysis of metaphase kinetochore fluorescence for the different GFP fusion proteins. For the anaphase budding yeast standard, the mean integrated intensity of a kinetochore cluster was 3,390 ± 373, corresponding to 112 ± 13 Ndc80-GFP molecules per cluster and 7 ± 0.8 molecules per kinetochore. For kinetochores in the DT40 cell lines, the protein copy number per kinetochore was obtained by 112 × Fic/ 3,390.
Figure 4.
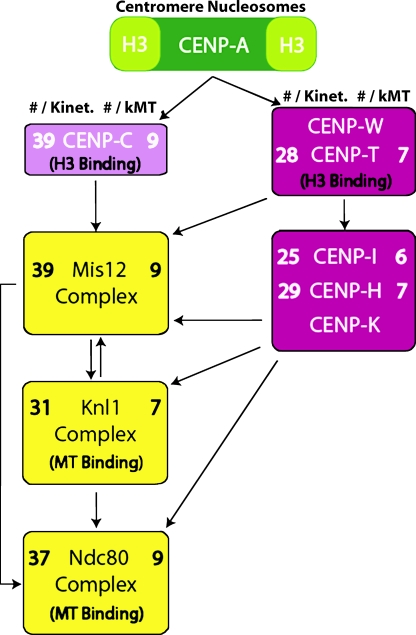
Protein numbers at kinetochores of DT40 cells indicate a fibrous unit attachment module linking chromatin to kMTs near their plus ends. Measured values from Table I for protein copy number per kinetochore (left) and kMT (right) are listed for each protein measured in the linkage between chromatin and MTs. The SD for all mean values listed is ∼20% (Table I). The diagram is the relevant part of a recent epistatic map (Santaguida and Musacchio, 2009). Such maps are generated in part by protein depletion (mRNA degradation) in cells by siRNA and immunofluorescence microscopy of kinetochore protein localization and by biochemical assays of protein–protein interactions. The DNA-binding protein, CENP-C, is needed for normal levels of Mis12 at kinetochores (Liu et al., 2006; Kwon et al., 2007), whereas normal levels of CENP-H, CENP-I, and CENP-K require the DNA-binding complex CENP-T–CENP-W (Hori et al., 2008). These proteins are all needed for normal amounts of the KMN network at kinetochores.
Kinetochore Fic for CENP-H–GFP expressed from the endogenous promoter or from a CMV promoter was 879 ± 47 compared with 808 ± 121, respectively. These values are very similar, as expected from comparing mean expressed protein levels in the cells (Fig. 1), thereby validating cell lines using CMV promoters for GFP fusion protein expression. Previously, Ribeiro et al. (2009) used a ratio measurement method similar to ours to compare the copy numbers in DT40 cells for CENP-H–GFP at kinetochores of knock-in cells (29/kinetochore) and knock-in cells with condensin knocked out (31/kinetochore). For both cell lines, their measurements for CENP-H–GFP per kinetochore were nearly identical to ours (27–29/kinetochore; Table I). This result demonstrates the reproducibility of the ratio method.
For the KMN network, the fluorescence ratios between DT40 kinetochores and anaphase clusters in yeast yielded mean protein copy numbers per DT40 kinetochore of 31 ± 6 for KNL-1–GFP, 39 ± 9 for Mis12-GFP, 37 ± 8 for Nuf2-GFP, and 32 ± 6 for Ndc80 (Hec1)–GFP (Table I and Fig. 4). Because Ndc80 (Hec1) and Nuf2 form a heterodimer within the Ndc80 complex, it is not a surprise that their measured mean values are very similar. The values for Mis12 and KNL-1 are also very similar to those of the Ndc80 complex, suggesting equal copy numbers for all members of the KMN network.
For the proteins that link the KMN network to chromatin at the inner kinetochore, we measured per kinetochore 39 ± 10 of CENP-C, 28 ± 4 of CENP-T–GFP, 25 ± 4 of CENP-I–GFP, and 27–29 of CENP-H–GFP (Table I and Fig. 4). We have not obtained GFP-expressing cells for CENP-K, but CENP-K is expected to have the same copy number as CENP-H because both CENP-H and CENP-K have extensive α-helical coiled-coil domains, and they have been shown biochemically to form an extended heterodimer (Qiu et al., 2009). Therefore, the kinetochore protein copy numbers for the linkage between the KMN network and chromatin are similar (25–39).
To show that this similarity is not an artifact of our ratio measurement method, we obtained protein copy numbers for proteins not expected to have similar values. We measured copy numbers per kinetochore of 62 for CENP-A–GFP, 12 for Bub1-GFP, and 72 for Mad2-GFP in nocodazole-treated metaphase cells (Fig. S3). All of these DT40 cell lines expressed GFP fusion protein from a CMV promoter in the presence of unlabeled endogenous protein. Because of the presence of endogenous protein, no other conclusion can be reached from these data except that the ratio measurement method gives distinctly different values for each in comparison with the similar values obtained for the proteins within the linkage between kMTs and chromatin.
The number of kMTs per kinetochore in DT40 cells at metaphase is reported to be 4.3 ± 0.3 from assays using serial section electron microscopy (Ribeiro et al., 2009). To obtain an estimate of the mean copy number per kMT and its SD, we divided the mean copy number and its SD measured per kinetochore by 4.3 (Table I and Fig. 4). This yielded about eight Ndc80 (Hec1) complexes and a similar number for the Mis12 (nine) complex and KNL-1 (seven). These numbers are almost identical to the copy numbers for the homologous proteins per kMT for both budding and fission yeast (Joglekar et al., 2006, 2008). Thus, an important finding of our study is that the highly conserved KMN network of core MT attachment proteins is also highly conserved in protein copy number per kMT between yeast and vertebrates.
The copy number for proteins that link the KMN network to chromatin is very different between vertebrates and yeast cells. Our measured values for mean copy number per kMT for CENP-H, CENP-I, and CENP-C in DT40 cells at metaphase are six, seven, and nine, whereas the corresponding values for the homologues in budding and fission yeast are two to three, two to three, and one, respectively. In addition, the proteins in vertebrates that bind DNA, CENP-C, and CENP-T–CENP-W are very similar in number per kMT to the KMN network of proteins (Table I and Fig. 4), whereas this is not the case in budding yeast.
We hypothesize that the protein linkage between chromatin and kMTs in vertebrates is produced on average by about eight “unit attachment modules” that are anchored to chromatin at one end by CENP-C and CENP-T–CENP-W independently binding DNA associated with H3 histone in the proximity of CENP-A nucleosomes (Hori et al., 2008). At the other end, the attachment modules can become anchored to kMTs near their plus ends by the Ndc80 (Hec1) complex and KNL-1. The Mis12 complex may link the two different functional ends of this unit attachment module together (Fig. 4). The unit attachment module is likely fibrous because many of the proteins have extensive α-helical coiled-coil domains (e.g., the Ndc80 complex, CENP-K, and CENP-H). The end to end length of the module is likely 62–72-nm long within vertebrate kinetochores based on measurements obtained by two-color super-resolution fluorescence microscopy (Wan et al., 2009).
It should also be noted that these attachment modules are assembled stepwise at the kinetochore because CCAN proteins, including CENP-C, CENP-H, CENP-I, and CENP-T, are present at the kinetochore throughout the cell cycle. The KMN network assembles at kinetochores in late G2 or prophase and leaves in telophase. Although our measured copy numbers are nearly stoichiometric for the CCAN and KMN network proteins, it is possible that a substoichiometric number of CCAN proteins is needed within the attachment modules to link the eight or so KMN network proteins to centromere chromatin, as has been measured for budding and fission yeast. More biochemical details of the linkage between the KMN proteins and the CCAN proteins are needed to resolve this important structural issue.
In summary, our protein copy number measurements suggest for vertebrate kinetochores a fibrous unit attachment module, linking chromatin to kMTs with about eight attachment modules on average per kMT at metaphase. This number is similar between yeast and vertebrates for the KMN network of proteins that form the core kMT attachment site. The fibrous unit attachment module between centromeric chromatin and the end of a kMT is likely important for essential protein functions at the kinetochore–MT interface, including MT plus end attachment, force generation, attachment error correction, and recruitment of spindle assembly checkpoint proteins to the kinetochore (DeLuca et al., 2005, 2006; Cheeseman et al., 2006; Kiyomitsu et al., 2007; Santaguida and Musacchio, 2009).
Materials and methods
Cell culture
The DT40 cell lines were constructed as described previously Fukagawa et al. (2001) and stored in liquid nitrogen. After thawing, cells were cultured in DME (Invitrogen) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 1% chicken serum (Invitrogen), 1% antibiotic antimycotic (Invitrogen), and 100 µM β-mercaptoethanol (EMD). CENP-H, CENP-I, Ndc80, Mis-12, CENP-C, KNL-1, and CENP-T knockout cell lines expressing each EGFP fusion protein under control of CMV promoter were also supplemented with 2 µg/ml tetracycline (Fukagawa et al., 2001; Nishihashi et al., 2002; Hori et al., 2003; Kline et al., 2006; Kwon et al., 2007; Cheeseman et al., 2008; Hori et al., 2008). Cells were maintained at 38°C in a 5% CO2 atmosphere. Yeast cells expressing GFP (S65T) fusion proteins were cultured and prepared as described previously in Joglekar et al. (2006). We expected equivalent fluorescence intensity from GFP (S65T) within kinetochore clusters in budding yeast and of EGFP within individual kinetochores in metaphase DT40 cells for the same excitation light intensity. This is because both GFP (S65T) and EGFP (S65T and F64L) (Yang et al., 1996) exhibit nearly identical excitation and emission spectra, extinction coefficient, quantum yield, fluorescence stability to pH changes >7.0, and photobleaching stability (Patterson et al., 1997). The additional mutation in EGFP prevents protein folding defects at the high temperatures (38°C) needed to culture DT40 cells (Yang et al., 1996); at 37°C, EFGP has the same fluorescence properties as GFP (S65T) at 28°C (Patterson et al., 1997).
Microscopy
The DT40 cells are not much larger in diameter than budding yeast cells, and both were adhered to the inner surface of coverslips pretreated with 0.5 mg/ml−1 concanavalin A. DT40 cells were seeded on coverslips 1 h before experimentation. Before imaging, they were mounted in rose chambers (Skibbens et al., 1993) filled with warm L-15 (Sigma-Aldrich) media lacking pH dye. Budding yeast cells were suspended and immobilized within a standard glass-slide coverslip preparation as described previously in Joglekar et al. (2006). All images were obtained at room temperature using a spinning-disk confocal (CSU-10; Yokogawa), 100× 1.4 NA objective (Nikon), and a cooled charge-coupled device camera (ORA AG; Hamamatsu Photonics) with the same 488-nm laser intensity and exposure time (Maddox et al., 2003). This gave a high signal to noise for each kinetochore fluorescent image and pixel sizes at the specimen scale of 65 nm. We obtained images of optical sections along the z axis through the cell at 200-nm intervals beginning near or at the coverslip–cellular interface.
Integrated intensity measurements
Fi measurements were obtained with image analysis software (MetaMorph; MDS Analytical Technologies) using the method of Hoffman et al. (2001) as described in Fig. S1 A.
Data analysis and correction of errors
For each cell type, the magnitude of kinetochore Fi was highest near the coverslip inner surface and decreased with image plane number (Fig. 3, A, D, G, and J) as a result of refraction at the coverslip surface. To correct for this effect, we fitted the primary data with a linear regression line and used the intercept value of frame number (FN) at the first data measurement (FNb) and the slope, m, to correct the Fi data at higher frame numbers: Fic(FN) = Fi(FN) + Fi(FNb) − m × (FN−FNb) (Fig. 3, B, E, H, and K). This correction produced a linear regression line with zero slope though the corrected integrated intensity data and approximately normal distributions of values about the mean (Fig. 3, C, F, I, and L). For each cell type, we obtained a mean value and SD for Fic from the n > 150 kinetochore clusters (budding yeast) or n > 200 individual kinetochores (DT40 cells).
Online supplemental material
Fig. S1 shows an illustration of the imaging methods used to obtain kinetochore Fi and cellular Ficell, a graph of the small effect of photobleaching during image acquisition, and a graph of kinetochore Fic as a function of Ficell. Fig. S2 illustrates the similar kinetochore Fi distributions for macrochromosomes and minichromosomes. Fig. S3 shows data for GFP-Mad2, GFP–CENP-A, and GFP-Bub1 in cells expressing endogenous protein. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912022/DC1.
Acknowledgments
We appreciate the stimulating discussions about kinetochore protein architecture by the members of the Salmon, Fukagawa, and Bloom laboratories.
This study was funded by the National Institutes of Health (grant GM 24364). Experiments in the Fukagawa laboratory were supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T. Fukagawa. A. Joglekar holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- CENP
- centromere protein
- CMV
- cytomegalovirus
- KMN
- KNL-1/Mis12 complex/Ndc80 complex
- kMT
- kinetochore MT
- MT
- microtubule
References
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta. 1786:32–40 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Emanuele M.J., McCleland M.L., Satinover D.L., Stukenberg P.T. 2005. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 16:4882–4892 10.1091/mbc.E05-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617 10.1093/emboj/20.16.4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Inoué S., Salmon E.D. 1995. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 6:1619–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., Salmon E.D. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K.S., Salmon E.D. 2010. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol. 22:57–67 10.1016/j.ceb.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kline S.L., Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17 10.1083/jcb.200509158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168 10.1091/mbc.E07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Moree B., Canman J.C., Salmon E.D. 2003. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 360:597–617 10.1016/S0076-6879(03)60130-8 [DOI] [PubMed] [Google Scholar]
- Mikami Y., Hori T., Kimura H., Fukagawa T. 2005. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25:1958–1970 10.1128/MCB.25.5.1958-1970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W.C., Fukagawa T. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2:463–476 10.1016/S1534-5807(02)00144-2 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Knobel S.M., Sharif W.D., Kain S.R., Piston D.W. 1997. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73:2782–2790 10.1016/S0006-3495(97)78307-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266 10.1083/jcb.152.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Wang J., Yu C., He D. 2009. CENP-K and CENP-H may form coiled-coils in the kinetochores. Sci. China C Life Sci. 52:352–359 10.1007/s11427-009-0050-3 [DOI] [PubMed] [Google Scholar]
- Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P. 2009. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 20:2371–2380 10.1091/mbc.E08-11-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki M.S., Buerstedde J.M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598–608 10.1093/emboj/17.2.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Canman J.C., Salmon E.D., Mitchison T.J. 2002. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 13:4308–4316 10.1091/mbc.E02-04-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Cheng L., Kain S.R. 1996. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 24:4592–4593 10.1093/nar/24.22.4592 [DOI] [PMC free article] [PubMed] [Google Scholar]