Kinesin-2 motor KIF17 autoinhibition is visualized in vivo; in the absence of cargo, this homodimer’s C-terminal tail blocks microtubule binding, and a coiled-coil segment blocks motility.
Abstract
Long-distance transport in cells is driven by kinesin and dynein motors that move along microtubule tracks. These motors must be tightly regulated to ensure the spatial and temporal fidelity of their transport events. Transport motors of the kinesin-1 and kinesin-3 families are regulated by autoinhibition, but little is known about the mechanisms that regulate kinesin-2 motors. We show that the homodimeric kinesin-2 motor KIF17 is kept in an inactive state in the absence of cargo. Autoinhibition is caused by a folded conformation that enables nonmotor regions to directly contact and inhibit the enzymatic activity of the motor domain. We define two molecular mechanisms that contribute to autoinhibition of KIF17. First, the C-terminal tail interferes with microtubule binding; and second, a coiled-coil segment blocks processive motility. The latter is a new mechanism for regulation of kinesin motors. This work supports the model that autoinhibition is a general mechanism for regulation of kinesin motors involved in intracellular trafficking events.
Introduction
Long-distance transport in mammalian cells is driven by motor proteins that use the energy of ATP hydrolysis to carry vesicle, organelle, and multiprotein cargoes along microtubule tracks. In general, kinesin motors undergo directed motility toward the plus ends of microtubules in the cell periphery and thus drive anterograde transport, whereas cytoplasmic dynein moves toward the minus ends of microtubules in the cell center and thus drives retrograde transport. The activity of these motors must be tightly regulated in cells to ensure that ATP hydrolysis and microtubule-based motility are coupled to cargo transport. Regulation also involves attachment of each motor to the correct cargo and transport of that cargo to the proper subcellular destination at the relevant time.
A general model has emerged for kinesin motors that in the absence of cargo, the motors are kept in an inactive state by autoinhibition (Verhey and Hammond, 2009). The mechanism of autoinhibition has been best defined for kinesin-1 (formerly conventional kinesin or KIF5; for reviews see Adio et al., 2006; Verhey and Hammond, 2009). Work from a variety of laboratories has shown that inactive kinesin-1 motors exist in a folded conformation, which allows the tail segment to directly contact and inhibit the enzymatic activity of the motor domain. A similar mechanism has been proposed to regulate kinesin-3 family members. Indeed, the mammalian KIF1A and KIF13B/GAKIN motors are autoinhibited and exist in a folded conformation, although direct interactions between motor and nonmotor domains have not been demonstrated (Lee et al., 2004; Yamada et al., 2007; Hammond et al., 2009).
Two members of the kinesin-2 family have been described: the heterotrimeric KIF3A–KIF3B–KAP complex and the homodimeric KIF17 motor. Kinesin-2 motors participate in a variety of intracellular transport events, from axonal transport in neurons to intraflagellar transport in cilia and flagella (Hirokawa et al., 2009; Silverman and Leroux, 2009). An attractive possibility for regulation of kinesin-2 motors is that they are kept inactive in the absence of cargo and then activated for transport upon cargo binding. Strong support for this possibility was provided by Imanishi et al. (2006), who showed that full-length (FL) recombinant Caenorhabditis elegans OSM-3, the homologue of mammalian KIF17, shows little to no motility in vitro and undergoes a salt-dependent shift from a compact to an extended conformation. The heterotrimeric KIF3A/3B/KAP kinesin-2 motor also undergoes a salt-dependent shift to a more extended conformation, but the activity state of folded and extended motors has not been determined (Wedaman et al., 1996).
We set out to uncover the regulatory mechanisms that control the activity of the kinesin-2 motor KIF17 both in vitro and in live cells. We show that, in the absence of cargo, FL KIF17 exists in a folded conformation and is inactive for microtubule-based motility. We define two molecular mechanisms that contribute to autoinhibition of KIF17. First, inhibition of microtubule binding is caused by blockage of the motor domain by the C-terminal tail domain; and second, inhibition of processive motility is caused by a direct interaction between a coiled-coil (CC) segment and the motor domain. The latter is a new, as-yet undiscovered mechanism for regulation of kinesin motors. In addition, we suggest a simple model for KIF17 activation in which cargo binding relieves the autoinhibited state.
Results
KIF17 motors are in an inactive state when expressed in mammalian cells
To study the regulation of mammalian KIF17 in its native environment, tagged versions of human KIF17 were expressed in COS cells whose flat morphology is particularly amenable to live cell imaging. This approach has been used successfully to study kinesin-1 and kinesin-3 motors produced and labeled under native conditions, and avoids potential problems associated with in vitro purification and/or labeling (Cai et al., 2007a).
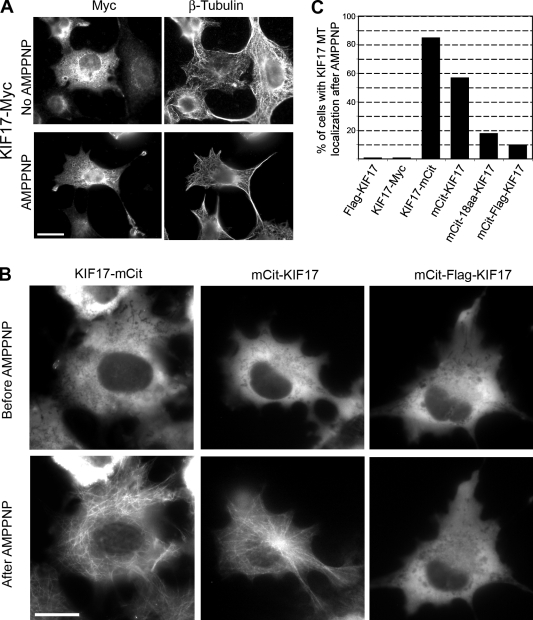
When expressed in cells, KIF17 motors tagged at the N or C termini with small epitope tags (Flag-KIF17 and KIF17-Myc) localized diffusely throughout the cytoplasm (Fig. 1 A and not depicted). As steady-state expression patterns do not reveal whether the motor is actively binding to and/or moving on microtubules, we took advantage of the fact that the nonhydrolyzable ATP analogue 5′-adenylylimididiphosphate (AMPPNP) locks kinesin motors in a microtubule-bound state. Although many mechanistic and thermodynamic factors contribute to microtubule binding by kinesin motors, the AMPPNP assay can be used to assess the inhibited state of kinesin motors in live cells (Cai et al., 2007a; Hammond et al., 2009). The cells were permeabilized with the bacterial toxin streptolysin O (SLO), then AMPPNP was added, and the cells were fixed and stained with antibodies to the epitope tags. Upon addition of AMPPNP, Flag-KIF17 and KIF17-Myc remained cytosolic and did not become locked in a microtubule-bound state (Fig. 1, A and C; and not depicted). This suggests that KIF17 motors exist in an inhibited state when expressed in mammalian cells.
Figure 1.
The kinesin-2 motor KIF17 is inactive for microtubule binding in mammalian cells. (A) Microtubule binding assay in fixed cells. COS cells expressing KIF17-Myc were untreated or treated with AMPPNP, then fixed and immunostained with anti-Myc and anti-tubulin antibodies. Bar, 20 µm. (B) Microtubule binding assay in live cells. COS cells expressing the indicated mCit-tagged versions of KIF17 were imaged, transiently permeabilized with SLO, and treated with AMPPNP. Shown are representative mCit images of the same cells before and after AMPPNP treatment. Bar, 20 µm. (C) Percentage of cells in which the indicated tagged versions of KIF17 localized to microtubules (MT) after AMPPNP treatment. At least 150 cells in three independent experiments were analyzed for each construct.
To study KIF17 motors in live cells, the motor was tagged at the N or C termini with a monomeric version of Citrine (Cit, a variant of yellow fluorescent protein [FP]). The monomeric citrine (mCit)-KIF17 and KIF17-mCit motors localized diffusely throughout the cytoplasm at steady state but became locked in a microtubule-bound state upon addition of AMPPNP (Fig. 1, B and C). This indicates that the addition of larger, e.g., FP tags, to either end of the molecule interferes with the inhibitory mechanisms that prevent AMPPNP-induced microtubule localization. We reasoned that the FP tags may sterically hinder motor–tail interactions that contribute to autoinhibition. To test this possibility, we created additional tagged versions of KIF17 that contained spacer sequences (18-aa random sequence or a 20-aa sequence containing a Flag tag) between the mCit tag and the KIF17 polypeptide. Interestingly, the addition of spacer residues between the FP tag and KIF17 resulted in motors (mCit-18aa-KIF17 and mCit-Flag-KIF17) that remained largely cytosolic upon addition of AMPPNP (Fig. 1, B and C; and not depicted), which suggests that moving the FP further from the motor domain allows a return to the autoinhibited state. Our interpretation of these results is that KIF17 is regulated by an autoinhibition mechanism that prevents AMPPNP-induced microtubule binding in the absence of cargo. However, unlike other motors tested to date (Cai et al., 2007a; Hammond et al., 2009), attachment of a bulky epitope tag via short linkers to the N or C termini interferes with autoinhibition.
Truncated KIF17 motors retain a dimeric state
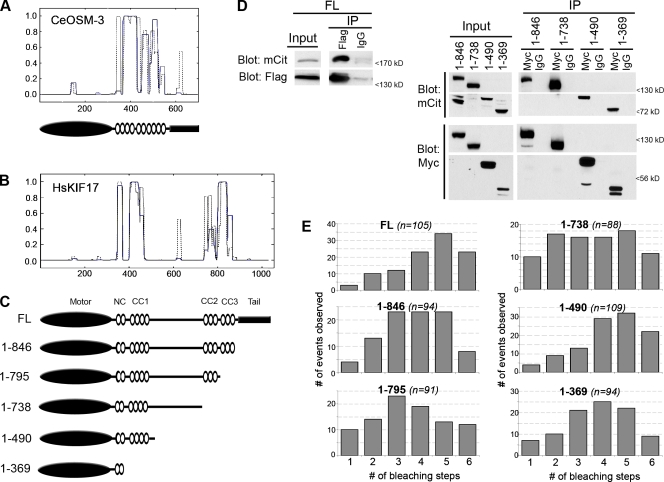
To identify the domains of KIF17 involved in autoinhibition, we created truncated versions by removing successive segments from the C-terminus (Fig. 2 C). Analysis of the human KIF17 sequence (Fig. 2 B) shows four regions of potential coil following the N-terminal motor domain—the neck coil (NC) and coiled-coil segments 1–3 (CC1-CC3)—followed by a C-terminal tail domain that can bind to cargo proteins (Setou et al., 2000; Takano et al., 2007). When compared with CC predictions of OSM-3 (Fig. 2 A), the overall domain organization appears similar, with the exception of the region between CC1 and CC2. In OSM-3, this region contains a short hinge segment that is required for the folded conformation of the autoinhibited motor (Imanishi et al., 2006). In KIF17, this central region contains an ∼300 residue segment of undefined structure, which suggests that the mechanisms of autoinhibition of OSM-3 and KIF17 may be different.
Figure 2.
FL and truncated KIF17 constructs exist in a dimeric state. (A, top) Coiled-coil prediction for C. elegans OSM-3 (GenBank/EMBL/DDBJ accession no. NP_001023308) using windows of 14 (gray dotted line) or 21 (blue line) amino acids (COILS; http://www.ch.embnet.org/software/COILS_form.html). (A, bottom) Schematic of the domain organization of OSM-3. Black oval, motor domain; white ovals, CC segments; black rectangle, tail domain. (B) Coiled-coil prediction for human KIF17 (GenBank/EMBL/DDBJ accession no. NP_065867) using windows of 14 (broken line) or 21 (solid line) amino acids. In A and B, the y axis gives the probability of CC conformation, and the x axis gives amino acid number. (C) Schematic of the domain organization of FL and truncated KIF17 proteins. (D) Coimmunoprecipitation of FL (left) or truncated (right) KIF17 constructs. COS cells were cotransfected with plasmids encoding Flag- and mCit-tagged FL KIF17 constructs (left) or Myc- and mCit-tagged truncated KIF17 constructs (right). Lysates were analyzed by Western blotting either before (input) or after immunoprecipitation with anti-Flag or control IgG antibodies (for FL; left), or with anti-Myc or control IgG antibodies (for truncated constructs; right). (E) Single-molecule photobleaching. Lysates of COS cells expressing the indicated FL or truncated KIF17 constructs tagged with 3xmCit were imaged by TIRF microscopy. The number of bleaching steps was counted for individual motors, and the values for the population were plotted as a histogram. n = total number of motors counted in at least two independent experiments. The mean number of bleaching steps for each construct was 4.4 for FL, 3.8 for 1–846, 3.5 for 1–795, 3.5 for 1–738, 4.3 for 1–490, and 3.8 for 1–369.
Because truncation of the OSM-3 tail domain destabilized the dimeric state of the molecule (Imanishi et al., 2006), we analyzed the dimeric state of our truncated KIF17 motors. We first used coimmunoprecipitation of Flag/Myc-KIF17 and mCit-KIF17 proteins coexpressed in COS cells. As expected, mCit-KIF17 coprecipitated with Flag-KIF17 in the presence of anti-Flag antibodies but not control (IgG) antibodies (Fig. 2 D, left). Likewise, when Myc- and mCit-tagged versions of the KIF17 truncations 1–846, 1–738, 1–490, and 1–369 were coexpressed, the mCit-tagged truncations coprecipitated with their Myc-tagged counterparts upon addition of anti-Myc but not control antibodies (Fig. 2 D, right). These results demonstrate that FL and truncated versions of KIF17 exist in an oligomeric state.
We then used the step-wise photobleaching of mCit to confirm the dimerization state of FL and truncated KIF17 motors at the single-molecule level. The KIF17 constructs were tagged with three tandem copies of mCit (3xmCit) and expressed in COS cells, then the lysates were analyzed by total internal reflection fluorescence (TIRF) microscopy. For each construct, the fluorescence intensity of ∼100 individual fluorescent molecules was recorded over time. The number of bleaching steps was determined for each molecule (Cai et al., 2007b; Hammond et al., 2009) and then plotted in a histogram to show the population distribution (Fig. 2 E). In control experiments, although we can detect at least 10 bleaching steps for 3xmCit-tagged proteins by this method, a maximum of six bleaching events were observed for dimeric 3xmCit-tagged kinesin-1 motors, whereas a maximum of three bleaching events were observed for monomeric 3xmCit-tagged kinesin-3 motors (Cai et al., 2007b; Hammond et al., 2009; unpublished data). For all FL and truncated KIF17 constructs, no more than six bleaching steps were observed (Fig. 2 E). These data indicate that FL and truncated KIF17 motors exist in a dimeric state and that the NC segment is sufficient for dimerization.
The C-terminal tail domain of KIF17 is required for inhibition of microtubule binding
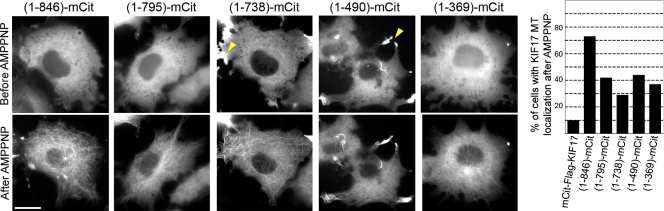
We then used the truncated KIF17 constructs to identify regions involved in autoinhibition of microtubule binding. Deletion of the KIF17 tail domain resulted in a motor, 1–846, that was diffuse and cytosolic at steady state but became trapped in a microtubule-bound state upon addition of AMPPNP, regardless of whether 1–846 was tagged with mCit (Fig. 3) or Myc tags (Fig. S1). These results indicate that removal of the C-terminal tail region results in a motor that is no longer inhibited for AMPPNP-induced microtubule binding.
Figure 3.
The C-terminal tail domain is required for inhibition of microtubule binding. Live cell microtubule binding assay. (left) COS cells expressing the indicated truncated KIF17 proteins (C-terminal mCit tag) were imaged, transiently permeabilized with SLO, and treated with AMPPNP. Shown are representative images of the same cells before and after AMPPNP treatment. Arrowheads indicate active truncated KIF17 motors that have accumulated at the plus ends of microtubules. Bar, 20 µm. (right) The percentage of cells in which the indicated KIF17-mCit proteins became locked in a MT-bound state after AMPPNP treatment was quantified. At least 150 cells in three independent experiments were analyzed for each construct.
Further truncations of KIF17 that removed the CC3 segment ([1–795]-mCit], the CC2 segment ([1–738]-mCit), the central region of unknown structure ([1–490]-mCit), or the CC1 segment ([1–369])-mCit) resulted in no change in microtubule binding behavior. That is, all truncations showed diffuse cytosolic localization before addition of AMPPNP and redistributed to a microtubule-trapped localization upon addition of AMPPNP (Fig. 3). The percentage of cells showing microtubule-bound motors in the presence of AMPPNP was lower for these truncations than for 1–846 motors (Fig. 3), which indicates that the CC3 segment may play a role in facilitating microtubule binding, either by interacting directly with the microtubule as has been reported for kinesin-1’s kinesin heavy chain (KHC) tail domain (Dietrich et al., 2008), or by promoting a conformation of the KIF17 motor that facilitates microtubule binding and/or the retention of AMPPNP in the nucleotide pocket.
It should also be noted that in the presence of AMPPNP, active KIF17 motors still maintain a pool of motors that is localized diffusely throughout the cytoplasm (Figs. 1 and 3; and unpublished data). This is in contrast to active versions of kinesin-1, where addition of AMPPNP traps virtually all expressed motors on microtubules, effectively clearing the cytoplasm (Cai et al., 2007a; Hammond et al., 2009). Whether this indicates a weaker binding of KIF17 than KHC to microtubules in the presence of AMPPNP, or whether these KIF17 motors are still partially inhibited for microtubule binding, is presently unclear.
The CC2 segment inhibits the processive motility of KIF17
At steady-state, expressed (1–738)-mCit and (1–490)-mCit motors showed an accumulation at the periphery of the cell, presumably at the plus ends of the microtubules, in addition to their diffuse cytosolic localization (Fig. 3, arrowheads). This suggests that, unlike the FL and other truncated motors, 1–738 and 1–490 may have the ability to move processively toward the plus ends of microtubules. To test this directly, we performed two assays that measure the ability of kinesin motors to undergo processive motility.
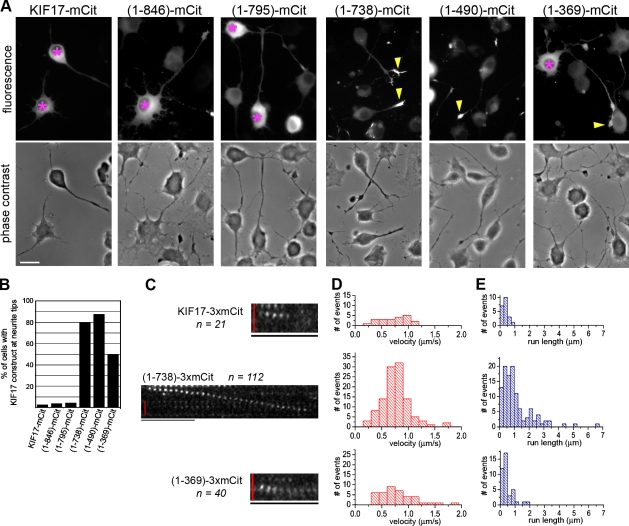
As a first test of the ability of FL and truncated KIF17 motors to move processively along microtubules, we used neuronal cells whose microtubule minus and plus ends are spatially segregated to the cell body and neurite tips, respectively. In this assay, active motors accumulate at neurite tips, and this localization generally correlates with their processive motility in vitro (Nakata and Hirokawa, 2003; Lee et al., 2004; Jacobson et al., 2006; Hammond et al., 2009). When expressed in differentiated CAD cells, mCit- and Myc-tagged versions of FL KIF17 were primarily localized in the cell body and did not concentrate in neurite tips (Fig. 4, A and B; and Fig. S2), which suggests that FL KIF17 motors are inactive for processive motility when expressed in mammalian cells. Similar results were obtained for the truncated constructs 1–846 and 1–795 (Fig. 4, A and B), even though these motors can bind to microtubules in the presence of AMPPNP (Fig. 3). These results indicate that microtubule binding and processive motility are two separate events that are regulated by different segments of the KIF17 protein.
Figure 4.
The CC2 segment is required for inhibition of processive motility. (A) Processive motility in vivo. CAD cells expressing FL or truncated KIF17 constructs (C-terminal mCit tag) were imaged. Shown are mCit fluorescence (top) and phase contrast (bottom) images of representative cells. Asterisks, cell bodies containing inactive KIF17 motors; arrowheads, neurite tips with accumulation of active KIF17 motors. Bar, 20 µm. (B) The percentage of cells with the indicated KIF17 construct accumulated at neurite tips. At least ten cells in two independent experiments were counted for each construct. (C–E) Processive motility in vitro. Single-molecule motility assays were performed using lysates of COS cells expressing the indicated FL or truncated KIF17 motors (C-terminal 3xmCit tag). (C) Representative time series showing the motility of individual FL KIF17-3xmCit, (1–738)-3xmCit, and (1–369)-3xmCit motors observed by TIRF microscopy. Bars: (vertical red bar) 2 µm; (horizontal black bar) 1 s. (D and E) The measured velocities (D) and run lengths (E) were plotted as histograms. n = number of motility events observed for each motor in three independent experiments.
Further truncation to remove the CC2 segment of KIF17 resulted in a motor, 1–738, that accumulated at the plus ends of microtubules in the neurite tips (Fig. 4, A and B). This suggests that the CC2 segment is critical for preventing the processive movement of KIF17 in mammalian cells. Similar results were obtained upon further truncation to remove the central region ([1–490]-mCit, Fig. 4 A; and [1–490]-Myc, Fig. S2). However, removal of the CC1 segment resulted in a decrease in processive motility, as (1–369)-mCit motors maintained a pool of motor in the cell body in addition to accumulation at neurite tips (Fig. 4 A).
To verify the processive motility of these KIF17 motors and to obtain quantitative information about their motile properties, we performed in vitro single-molecule motility assays. For this assay, KIF17 motors were tagged with three tandem copies of mCit for increased signal and decreased photoblinking and photobleaching (Cai et al., 2007b). Lysates from COS cells expressing KIF17-3xmCit motors were added to a flow chamber containing polymerized microtubules and assayed by time-lapse imaging on a TIRF microscope. Only motility events that lasted at least six frames (300 ms) were included in the data analysis. To compare motility properties between KIF17 constructs, the amount of expressed protein was normalized by Western blotting with an antibody to mCit (unpublished data), and each construct was analyzed for the same total amount of time.
Few in vitro motility events were detected for FL KIF17-3xmCit motors (n = 21; Fig. 4, C–E; and Table I), which is consistent with their inability to accumulate at the plus ends of microtubules in neuronal cells (Fig. 4 A). Analysis of these motility events showed that FL KIF17-3xmCit motors moved for only short distances (mean run length 0.36 ± 0.04 µm; Fig. 4 E and Table I). A limited ability to undergo processive motility in vitro was also seen upon removal of the tail domain ([1–846]-3xmCit) and the CC3 segment ([1–795]-3xmCit; not depicted), which is consistent with their in vivo behavior (Fig. 4 A). However, further truncations that removed the CC2 segment resulted in a dramatic increase in the number of motility events observed for (1–738)-3xmCit motors (n = 112; Fig. 4, C–E; and Table I) and (1–490)-3xmCit motors (see Fig. 8 C and Table III). (1–738)-3xmCit motors moved with a mean speed of 0.77 ± 0.02 µm/s (Fig. 4 D and Table I), and moved in runs that were as long as 6–7 µm but averaged 1.13 ± 0.10 µm (Fig. 4 E and Table I). Truncations that remove the CC1 segment ([1–369]-3xmCit) resulted in a reduction in the number of motility events observed (n = 40) and the mean run length (0.45 ± 0.06 µm; Fig. 4 E and Table I), which is consistent with the in vivo analysis (Fig. 4 A), which suggests that the CC1 segment may facilitate processive motility. We conclude that the CC2 segment plays an important role in autoinhibition of KIF17 by preventing processive motility.
Table I.
Motile properties of truncated KIF17 constructs
| KIF17 | No. of events observed | Speed | Run length |
| µm/s | µm | ||
| FL KIF17-3xmCit | 21 | 0.71 ± 0.06 | 0.36 ± 0.04 |
| (1-738)-3xmCit | 112 | 0.77 ± 0.02 | 1.13 ± 0.01 |
| (1-369)-3xmCit | 40 | 0.79 ± 0.06 | 0.45 ± 0.06 |
Figure 8.
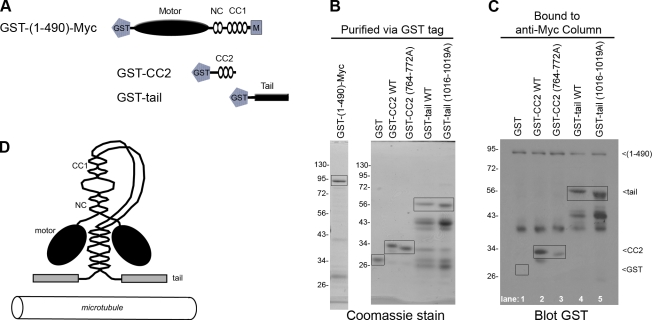
The CC2 and tail domains interact directly with the KIF17 motor. (A) Schematic of GST-tagged constructs. (B) Coomassie-stained gel showing recombinant, purified GST-(1–490)-Myc motor, WT and mutant GST-CC2, and WT and mutant GST-tail proteins. (C) GST-(1–490)-Myc protein was bound to an anti-Myc column, then GST-tagged versions of WT or mutant (764–772A) CC2 domains as well as WT or mutant (1016–1019A) tail domains were added to the column. GST was used as a control. The column-bound proteins were identified by immunoblotting with an anti-GST antibody. Boxed regions indicate proteins of interest. The numbers to the left of the gel blots indicate protein size markers in kD.
Table III.
Effects of the CC2 and tail domains on the motility of KIF17 motors
| KIF17 | No. of events observed | Speed | Run length |
| µm/s | µm | ||
| (1-490)-3xmCit | 122 | 0.90 ± 0.03 | 0.87 ± 0.04 |
| (1-490)-3xmCit + GST | 125 | 0.93 ± 0.02 | 1.13 ± 0.05 |
| (1-490)-3xmCit + GST-CC2 | 0 | NA | NA |
| (1-490)-3xmCit + GST-tail | 23 | 0.80 ± 0.04 | 0.76 ± 0.08 |
NA, not applicable.
Mutation of the CC2 segment relieves autoinhibition of processive motility
The results presented thus far suggest that two regions of KIF17 are involved in autoinhibition of motor activity: the tail domain interferes with microtubule binding (Fig. 3), whereas the CC2 segment blocks processive motility (Fig. 4). We hypothesized that KIF17 exists in a folded state that allows these segments to inhibit motor activity. If so, then KIF17 should undergo conformational changes that correlate with its activation, as observed for OSM-3 (Imanishi et al., 2006), and the inhibitory CC2 and tail segments should directly interact with and inhibit the activity of the motor domain.
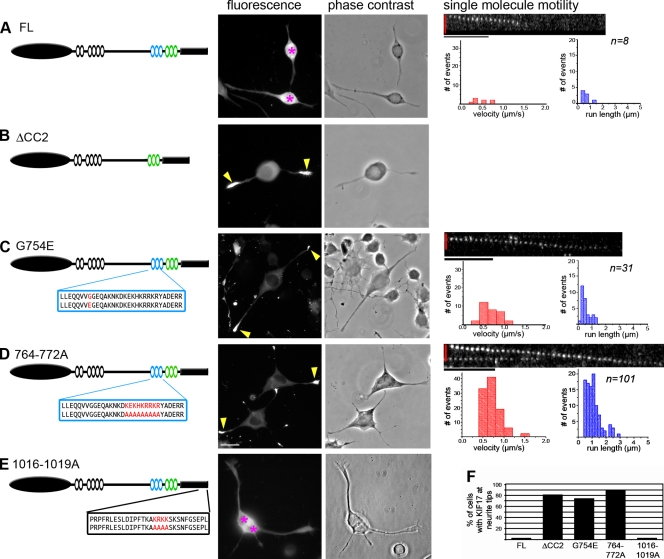
To test the relationship between KIF17 conformation and activity, we performed sucrose gradient sedimentation assays. However, we first had to generate mutations in FL KIF17 that result in activation of processive motility. Deletion of the entire CC2 segment in a manner that preserves the predicted CC architecture of KIF17 (not depicted) resulted in an active motor, ΔCC2, that accumulated at neurite tips in differentiated CAD cells (Fig. 5, B and F), which confirms that the CC2 segment plays a critical role in autoinhibition of KIF17 motility. To generate more subtle activating mutations, we created a point mutation based on the OSM-3 allele sa125 in which mutation of Gly444 to Glu (G444E) results in activation of OSM-3 ATPase activity, processive motility in vitro, and chemosensory defects in worms (Imanishi et al., 2006). Because Gly444 and surrounding residues in OSM-3 align best with Gly754 and adjacent residues of KIF17 (unpublished data), we mutated Gly754 to Glu (G754E) in FL KIF17. This mutation resulted in a dramatic accumulation of motors at neurite tips in differentiated CAD cells (Fig. 5, C and F), which demonstrates that a single point mutation in CC2 is sufficient to relieve autoinhibition of FL KIF17.
Figure 5.
Deletion or mutation of CC2 is sufficient to relieve autoinhibition of processive motility. (A–E, left) Schematics of FL KIF17 (A), constructs in which the CC2 domain was deleted (B, ΔCC2), or constructs in which mutations were introduced into the CC2 (C, G754E; and D, 764–772A) or tail (E, 1016–1019A) domains. For the mutants, the amino acid sequences of the WT and mutated CC2 (C and D, blue box) or tail (E, black box) segments are indicated below the schematic. (A–E, middle) Processive motility in vivo. Differentiated CAD cells expressing the indicated KIF17 constructs (C-terminal mCit tag) were imaged. Shown are mCit (fluorescence) and phase contrast images of representative cells. Pink asterisks, cell bodies containing inactive KIF17 motors; yellow arrows, neurite tips with accumulation of active KIF17 motors. Bar, 20 µm. (F) The percentage of cells with the indicated KIF17 construct accumulated at neurite tips. At least 50 cells in two independent experiments were counted for each construct. (A–D, right) Processive motility in vitro. Single-molecule motility assays were performed using lysates of COS cells expressing 3xmCit-tagged WT KIF17 (A), G754E mutant (C), or 764-772A mutant (D). The measured velocities (red graphs) and run lengths (blue graphs) were plotted as histograms. n values indicate the number of motility events observed for each construct in two independent experiments. Bars: (vertical red bar) 2 µm; (horizontal black bar) 1 s.
Because ionic interactions are critical for inhibition of the kinesin-1 motor by its tail domain (Stock et al., 1999), we looked for charged regions in the tail and CC2 segments of KIF17 that could be involved in inhibition of microtubule binding and processive motility, respectively. In both the tail domain and the CC2 segment, a stretch of basic residues was identified and mutated to alanines (1016–1019A and 764–772A, respectively; Fig. 5, D and E). When expressed in CAD cells, 764-772A motors but not 1016-1019A motors showed a dramatic accumulation in neurite tips (Fig. 5, D–F), which indicates that charged residues in CC2 may be critical for interaction with the motor domain in the autoinhibited state.
To confirm that mutation of residues G754 and 764–772 in the CC2 domain results in activation of processive motility, we performed in vitro single-molecule motility assays. The motility of individual motors in lysates of COS cells expressing 3xmCit-tagged versions of FL KIF17, G754E, or 764–772A motors was determined by time-lapse TIRF microscopy. Again, few motility events were observed for FL KIF17 (n = 8; Fig. 5 A and Table II). However, mutation of either G754E or 764–772A resulted in a dramatic increase in the number of motility events observed as well as the overall speed and processivity of the motor (Fig. 5, C and D; and Table II). Notably, mutation of the basic residues 764–772 resulted in a larger increase in KIF17 activation than mutation of the single glycine residue. Collectively, these results demonstrate that the CC2 segment plays a critical role in autoinhibition of processive motility and that the ionic interactions between CC2 and the KIF17 motor domain may be critical for this regulation.
Table II.
Motile properties of mutant FL KIF17 constructs
| KIF17 | No. of events observed | Speed | Run length |
| µm/s | µm | ||
| FL KIF17-3xmCit | 8 | 0.48 ± 0.07 | 0.64 ± 0.11 |
| FL(G754E)-3xmCit | 31 | 0.64 ± 0.04 | 0.58 ± 0.06 |
| FL(764-772A)-3xmCit | 101 | 0.72 ± 0.02 | 1.07 ± 0.05 |
Release of inhibition correlates with a shift to an extended conformation
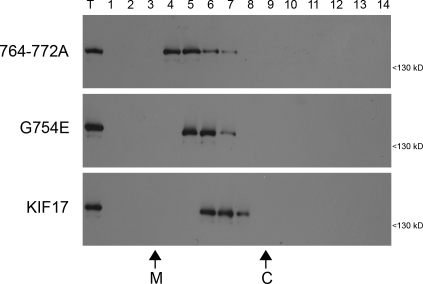
We then used sucrose gradient sedimentation analysis to test the relationship between KIF17 activity and conformation. Lysates of COS cells expressing Flag-tagged versions of inactive (FL) and active (G754E and 764–772A) KIF17 motors were loaded on the top of linear sucrose gradients and separated by sedimentation. The majority of FL KIF17 motor was found in fractions 6 and 7 (Fig. 6). Mutation of G754E or 764–772A resulted in a slower migration in the sucrose gradients (Fig. 6), which is consistent with the possibility that the active motors are in an extended conformation. A similar shift in sedimentation behavior was observed for the activated OSM-3(G444E) motor (Imanishi et al., 2006). The change in sedimentation is consistent with the hypothesis that KIF17 undergoes a conformational transition between an inactive folded state and an active extended state.
Figure 6.
Mutations in CC2 that relieve autoinhibition convert the motor to an extended conformation. Lysates of COS cells expressing Flag-tagged FL KIF17, or the G754E or 764-772A mutants were subjected to sucrose gradient sedimentation. Fractions were removed from the top (lane 1), run on SDS-PAGE, and immunoblotted with an antibody to the Flag tag. T, total lysate. The sedimentation of the size standards catalase (C; 11.3 S) and malate dehydrogenase (M; 4.3 S) is indicated on the bottom.
The CC2 and tail segments inhibit KIF17 motility via direct interactions with the motor domain
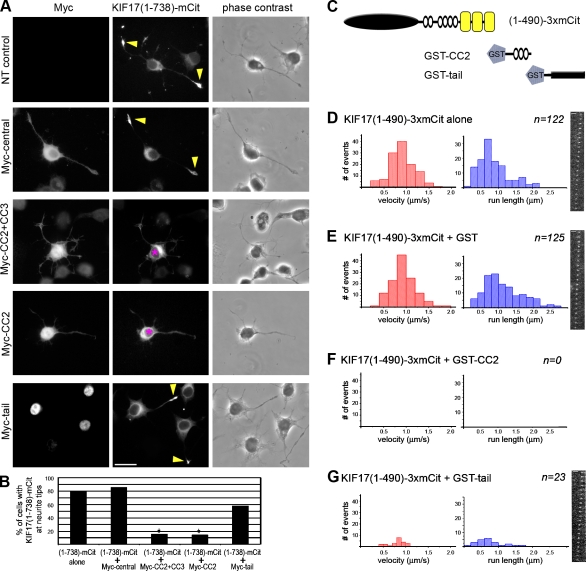
We tested whether the CC2 and tail constructs could act in trans to inhibit the processive motility of active KIF17 motors. In differentiated CAD cells, the ability of active (1–738)-mCit motors to accumulate in neurite tips (Fig. 7, A and B) was not affected by expression of a Myc-tagged version of KIF17’s central region (aa 466–738; Fig. 7, A and B), but was abolished upon coexpression of Myc-tagged CC2 + CC3 or CC2 segments (Fig. 7, A and B). Although expression of the Myc-tail domain reduced the accumulation of (1–738)-mCit motors in neurite tips, the result was not statistically significant, perhaps because of nuclear accumulation of the tail construct (Fig. 7, A and B).
Figure 7.
The CC2 and tail domains inhibit KIF17 motility in trans. (A) Processive motility in vivo. Differentiated CAD cells expressing (1–738)-mCit motor alone (NT, nontransfected control) or together with the indicated Myc-tagged constructs were fixed and stained with an antibody to the Myc tag. Asterisks, cell bodies containing inactive KIF17 motors; arrowheads, neurite tips with accumulation of active KIF17 motors. (B) Quantification of the percentage of cells with (1–738)-mCit tip accumulation from three independent experiments. *, P < 0.05. (C–F) Processive motility in vitro. Single-molecule motility assays were performed for (1–490)-3xmCit motors alone (D) or in the presence of recombinant purified GST (E), GST-CC2 (F), or GST-tail (G) proteins. The speed (red graphs) and run lengths (blue graphs) of the observed events were plotted as histograms. n = number of motility events observed in two independent experiments. Each frame is 50 ms. Bars: (A) 20 µm; (D, E, and G) 2 µm.
To obtain quantitative information about the ability of the CC2 and tail segments to inhibit the activity of the KIF17 motor, we used single-molecule motility assays. Lysates of COS cells expressing active (1–490)-3xmCit motors were incubated alone or in the presence of recombinant purified GST-tagged CC2 or tail domains (Fig. 7 C), and the motility of individual motors was analyzed by TIRF microscopy. A large number of motility events were observed for (1–490)-3xmCit motors alone (Fig. 7 D and Table III) and in the presence of GST (Fig. 7 E and Table III). When added in trans, the CC2 segment completely blocked motor activity, as no motility events were observed for (1–490)-3xmCit motors in the presence of GST-CC2 (Fig. 7 F and Table III). Addition of the GST-tail domain did not completely block motor activity but did result in a dramatic reduction in the number of motility events observed (Fig. 7 G and Table III).
To test whether inhibition of KIF17 motility is caused by direct interactions between the motor domain and the CC2 or tail segments, we performed binding assays using recombinant GST-tagged motor, CC2, and tail constructs (Fig. 8 A). For these experiments, we define the KIF17 motor domain as a dimer of heads, including the NC and CC1 segments. We generated both wild-type (WT) and mutant (764–772A and 1016–1019A) versions of the CC2 and tail domains, and in both cases, mutation of the basic residues resulted in a faster mobility in SDS-PAGE gels (Fig. 8 B). The WT and mutant GST-tail constructs were highly susceptible to proteolysis (Fig. 8 B).
Direct binding assays were performed by loading GST-KIF17(1–490)-Myc motors in a column via anti-Myc antibodies. Purified GST-tagged WT and mutant (764–772A) versions of the CC2 segment, and WT and mutant (1016–1019A) versions of the tail domain were allowed to bind to the column. After extensive washing, the bound fraction was analyzed by Western blotting with an anti-GST antibody to detect all proteins. GST alone was used as a control and did not bind to the motor (Fig. 8 C, lane 1). The CC2 segment was able to interact directly with the motor as GST-CC2 protein bound to the column (Fig. 8 C, lane 2), but this interaction was dramatically reduced by mutation of the basic residues 764–772 (Fig. 8 C, lane 3). In contrast, both WT and mutant 1016-1019A forms of the tail domain bound to the motor (Fig. 8 C, lanes 4 and 5). These results indicate that both CC2 and tail segments interact directly with the KIF17 motor domain and that residues 764–772 are critical for the interaction of CC2 with the motor. We conclude that autoinhibition of KIF17 results from intramolecular interactions between the KIF17 motor domain and the CC2 and tail segments in the folded inactive state.
Discussion
In the absence of cargo, kinesin activity must be tightly controlled to prevent futile ATP hydrolysis and congestion of microtubule tracks. Recent work has demonstrated that autoinhibition may be a general mechanism for the regulation of kinesin motors that drive intracellular trafficking events (Verhey and Hammond, 2009). We provide the first demonstration that mammalian kinesin-2 motors are regulated by autoinhibition and determine two intramolecular mechanisms that contribute to regulation of KIF17 in vitro and in live cells.
Autoinhibition is caused by a folded conformation that enables direct interactions between KIF17 motor and nonmotor regions
We demonstrate that FL KIF17 is in an inactive state when expressed under native conditions, as the motor can neither be locked on microtubules by AMPPNP nor undergo processive motility to the plus ends of microtubules. We also show that activation of KIF17 correlates with a shift to a more extended conformation. In this way, the mechanism of KIF17 regulation is similar to that of kinesin-1 (Verhey and Hammond, 2009).
The folded conformation enables two regions of KIF17—the CC2 segment and the tail domain—to interact directly with and inhibit the enzymatic activity of dimeric KIF17 motors. In the case of the inhibitory tail domain, we have not been able to identify residues that contribute to the direct interaction with the motor domain and/or autoinhibition of motor activity. In the case of the inhibitory CC2 segment, we identified two mutations, G754E and 762-772A, that result in activation of processive motility and a corresponding conversion to a more extended conformation. We propose that G754 contributes to a hinge region that enables the folded conformation, similar to the role of G444 in OSM-3 (Imanishi et al., 2006). We propose that residues 764–772 are involved in ionic interactions with the motor domain in the folded conformation, although we cannot exclude the possibility that residues 764–772 are critical for an overall structural conformation of the CC2 segment required for interaction with the motor domain.
The fact that ionic interactions play a critical role in mediating the interaction of inhibitory segments with the KIF17 motor domain in the autoinhibited state is similar to the regulatory mechanism of kinesin-1 (Verhey and Hammond, 2009). These intramolecular interactions appear to be quite weak in trans, as direct interactions between motor and nonmotor segments could not be detected in conventional pull-down experiments (unpublished data) but only when present at high concentrations on a column. Within the FL molecule, the folded conformation enables weak interactions to become biochemically strong and mechanistically important.
Two molecular mechanisms contribute to KIF17 autoinhibition
We define two molecular mechanisms (Fig. 8 D) that contribute to autoinhibition of folded KIF17 motors both in vitro and in live cells. In the first mechanism, the KIF17 tail domain interacts directly with the dimeric motor and interferes with microtubule binding. Whether the tail domain blocks the initial interaction of the KIF17 motor with the microtubule or accelerates the release of ADP-bound KIF17 requires further mechanistic analysis. In the second mechanism, the CC2 segment interacts directly with the dimeric motor and blocks processive motility. This is the first direct demonstration that kinesin motors can be regulated by an autoinhibitory mechanism that directly interferes with the ability of the motor to undergo processive movement.
Dual regulatory mechanisms have recently been shown to contribute to the autoinhibitory mechanisms of mammalian kinesin-1 and kinesin-3 motors (Cai et al., 2007a; Hammond et al., 2009). In the case of kinesin-1, the KHC tail domain interacts with the motor domain and interferes with the initial microtubule interaction, whereas the kinesin light chain (KLC) subunit blocks processive motility (Coy et al., 1999; Hackney and Stock, 2000; Cai et al., 2007a). The fact that the exact sequences and secondary structural elements required for autoinhibition differ between kinesin motors suggests that the evolutionary development of the kinesin superfamily involved the adoption of unique cargo-binding regions together with mechanisms for regulation of the common kinesin catalytic domain.
Role of the NC and subsequent hinge segments in dimerization and processive motility
We demonstrate that human KIF17 exists in a dimeric state, which is consistent with hydrodynamic analysis of KIF17/OSM-3 motors from other species (Signor et al., 1999; Setou et al., 2000), and that the NC segment is sufficient for dimerization of truncated KIF17(1–369) motors. Yet surprisingly, (1–369)-mCit motors showed reduced processivity in vivo and in vitro as compared with (1–490)-mCit motors. We propose that the hinge segment between the NC and CC1 segments facilitates processive motility, perhaps by stabilizing the coiled structure of the NC and/or by facilitating the motor–microtubule interaction. Similarly positioned hinge segments in kinesin-1 and kinesin-3 motors have been noted to positively affect processive motility via stabilizing effects on dimer formation and the motor–microtubule interaction (Thormählen et al., 1998; Thorn et al., 2000; Kallipolitou et al., 2001). Thus, it seems likely that processive motility of dimeric kinesin motors is influenced by sequences in the motor–proximal hinge segment.
Cargo-mediated activation of KIF17
Autoinhibition of motor activity leads to a simple model where cargo binding relieves the inhibitory mechanism, enabling subsequent microtubule-based motility. In this work, we demonstrate that KIF17 can be activated for microtubule binding but not processive activity by attachment of an FP to the N or C termini of KIF17 via short linkers (i.e., mCit-KIF17 and KIF17-mCit). Similar FP fusions do not result in activation of kinesin-1 or kinesin-3 motors (Cai et al., 2007a; Hammond et al., 2009). These results suggest that the presence of the FP interferes with inhibitory interactions between motor and tail regions of KIF17 in the folded state.
For both kinesin-1 and the kinesin-2 motor OSM-3, release of autoinhibition by cargo binding can be mimicked in vitro by attachment of beads to the tail of the motor (Coy et al., 1999; Imanishi et al., 2006). How motors are activated in vivo is largely unknown. Recent work has shown that activation of kinesin-1 in cells requires binding partners of both inhibitory segments (tail of the KHC and KLC subunits) to relieve autoinhibition (Blasius et al., 2007). We propose that KIF17 is activated in vivo by cargo proteins, e.g., intraflagellar transport components that enable targeting to and processive motility in cilia. As two mechanisms contribute to autoinhibition of KIF17, it seems likely that at least two binding partners are needed for full activation of motor activity. Further elucidation of the mechanisms of KIF17 activation will require the identification of binding partners of the inhibitory CC2 and tail segments.
Materials and methods
Plasmids and antibodies
Full-length or truncated human KIF17 constructs were generated by PCR amplification from HsKIF17 cDNA (GenBank/EMBL/DDBJ accession no. NP_065867). KIF17 internal deletions and mutations were made by overlapping PCR or QuikChange (Agilent Technologies). PCR products were cloned into the mCit-N1 or C1 vectors (modified from Takara Bio Inc.’s EYFP-N1/C1 vectors), the 3xmCit-N1 vector (Cai et al., 2007b), pcDNA3-Flag/Myc vectors, or pGEX-5X (GE Healthcare) using convenient restriction sites. Linker sequences for KIF17 constructs with N-terminal epitope tags were: SGAG for mCit-KIF17, AGGILSGGAVSGGASGAG for mCit-18aa-KIF17, SGLRSMDYKDDDDKSHGGGA for mCit-Flag-KIF17, and GGGA for Flag-KIG17. Linker sequences for KIF17 constructs with C-terminal epitope tags were: TV for KIF17-Myc and TVPRARDPPVAT for KIF17-mCit. All plasmids were verified by DNA sequencing. KHC-mCit and mCFP-KLC have been described previously (Cai et al., 2007a). The following antibodies were used: Myc (clone 9E10; Sigma-Aldrich), Flag (clone M2; Sigma-Aldrich), GFP (used to recognize mCit; Invitrogen), GST (Santa Cruz Biotechnology, Inc.), and anti–β-tubulin (clone E7; Developmental Studies Hybridoma Bank).
Cell culture and immunofluorescence
COS and CAD cells were cultured, transfected, and processed for immunofluorescence as described previously (Blasius et al., 2007; Cai et al., 2007a). COS cells were grown in DME with 10% Fetal Clone III (Thermo Fisher Scientific), and CAD cells were grown in 1:1 F12:DME (Lonza) with 10% FBS (HyClone) at 37°C and 5% CO2. The cells were transfected with Trans-It LT1 (Mirus Bio LLC) according to the manufacturer’s instructions. Cells were fixed with 3.7% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 0.2% fish skin gelatin, all in PBS. After staining, the coverslips were mounted in ProLong Gold (Invitrogen). Images were collected on a microscope (TE2000; Nikon) using Plan-Fluor 40×, NA 0.75 or Plan-Apochromat 60× NA 1.4 objectives, a camera (CS ES2; Photometrics), and MetaMorph software (MDS Analytical Technologies).
Coimmunoprecipitation and sucrose gradient sedimentation
18–24 h after transfection, COS cells were resuspended in lysis buffer 1 (25 mM Hepes/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, 1% Triton X-100, and protease inhibitors; pH 7.4) or lysis buffer 2 (40 mM Hepes, pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 50 mM NaF, 0.5% NP-40, 0.1% Brij-35, and protease inhibitors), and insoluble material was removed by centrifugation at 20,000 g at 4°C for 10 min. For immunoprecipitation, lysates were incubated with the specified antibodies for 2.5–18 h at 4°C. Protein A agarose beads were then added and the mixture was incubated for an additional 30 min at 4°C. Beads were pelleted, washed two times with lysis buffer, and analyzed by SDS-PAGE and Western blotting. For sucrose density gradient centrifugation, lysates were loaded on the top of linear 9–17% sucrose density gradients and separated by centrifugation. Fractions were removed from the top and analyzed by SDS-PAGE and Western blotting. Catalase (11.3 S, 256 kD; EMD) and malate dehydrogenase (4.3 S, 66 kD; Sigma-Aldrich) were used as standards.
In vivo microtubule binding assay
COS cells were plated onto glass-bottomed dishes (MatTek Corporation) and transfected with plasmids encoding the proteins of interest. 24 h later, cells were viewed on an inverted microscope (TE2000; Nikon). Cells were treated with 0.1 µg/ml SLO in permeabilization buffer (PB; 25 mM Hepes/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 10 mg/ml BSA; pH 7.4) for 1 min. After washing three times with PB, cells were incubated with PB containing 2 mM AMPPNP. Cells were monitored every minute for an additional 15 min. Alternatively, cells were treated with SLO and AMPPNP in the presence of taxol for 10 min. Then, cells were fixed and stained for immunofluorescence using Flag, HA, or Myc antibodies.
In vitro single molecule motility and photobleaching assays
Motility assays were performed in flow chambers as described previously (Cai et al., 2007b), except that P25 buffer (25 mM Pipes/KOH, 1 mM EGTA, and 2 mM MgCl2, pH 6.8) was used in place of P12. In brief, motor proteins were prepared by lysing transfected COS cells in lysis buffer 2 with 1 mM ATP. 5–10 µl of cell lysate was added to flow chambers containing taxol-stabilized microtubules and 40–45 µl of oxygen scavenger buffer (1 mM DTT, 1 mM MgCl2, 2 mM ATP, 10 mM glucose, 0.1 mg/ml glucose oxidase, 0.08 mg/ml catalase, 10 mg/ml BSA, and 10 µM taxol in P25). Lysates were incubated with ATP for motility assays or with AMPPNP for photobleaching analysis. The 488-nm line of a tunable, single-mode, fiber-coupled argon ion laser (CVI Melles Griot) was used for excitation. An objective-type TIRF microscope and a back-illuminated EM charge-coupled device camera (Cascade 512B; Roper Industries) were used to capture images every 100 ms. Measurements for each construct come from at least two independent protein preparations. Velocity and track length measurements were obtained using homemade plug-ins written for ImageJ (National Institutes of Health) and include motile events lasting at least 0.3 s. All data are presented as mean ± standard error.
GST protein purification
The expression of GST fusion proteins was induced in Rosetta 2 Escherichia coli by addition of 1 mM IPTG for 2–4 h. Cell pellets were resuspended in 50 mM Tris, pH 7.5, 0.5 mM EDTA, and 0.3 M NaCl plus 1 mg/ml lysozyme, 4 mM DTT, proteosome inhibitors, and 0.2% NP-40, then stored overnight at −80°C. Samples were thawed on ice, NaCl was added to 750 mM, and MgCl2 was added to 6 mM, then the samples were incubated with 0.1 mg Dnase1 for 2 h at 4°C. Samples were then sonicated at 60% amplitude and centrifuged at 15,000 g for 20 min at 4°C. Soluble protein was incubated with glutathione sepharose beads (GE Healthcare) for 2 h at 4°C. The beads were washed with 1% Triton X-100 in PBS and again with PBS alone. Protein was eluted with 10 mM of reduced L-glutathione (Sigma-Aldrich), 50 mM Tris, pH 8.0, and 0.2 mM EDTA. Protein-containing fractions were pooled and dialyzed overnight against 10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 5% glycerol, and 1 mM DTT. For GST-KIF17(1–490)-Myc motors, 100 µM ATP was present throughout purification.
In vitro binding assay
Purified GST-KIF17(1–490)-Myc was incubated with anti-Myc agarose beads (Sigma-Aldrich) for 20 min at 4°C, loaded into columns, and washed with P12/ATP (12 mM Pipes/KOH, pH 6.8, 1 mM EGTA, 2 mM MgOAc, and 100 µM ATP). Purified GST, GST-CC2, or GST-tail proteins in P12/ATP buffer were incubated with the motor-bound beads for 20 min at 4°C. Unbound protein was removed by extensive washing with P12/ATP buffer. The beads were then removed from the column, resuspended in sample buffer, and analyzed by SDS-PAGE and Western blotting.
Online supplemental material
Fig. S1 shows that Myc-tagged 1–846 behaves similarly to mCit-tagged 1–846 in its localization to microtubules in the presence of AMPPNP. Fig. S2 shows that Myc-tagged FL and truncated motors behave similarly to mCit-tagged motors in their localization to neurite tips. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001057/DC1.
Supplementary Material
Acknowledgments
We thank Edgar Meyhofer for help with TIRF microscopy and members of the Verhey laboratory for helpful discussions.
This work was supported by National Institutes of Health grants to K.J. Verhey (GM070862 and GM083254) and a predoctoral fellowship to J.W. Hammond (National Science Foundation F017154).
Footnotes
Abbreviations used in this paper:
- CC
- coiled-coil
- FL
- full length
- FP
- fluorescent protein
- KHC
- kinesin heavy chain
- KLC
- kinesin light chain
- mCit
- monomeric citrine
- NC
- neck coil
- SLO
- streptolysin O
- TIRF
- total internal reflection fluorescence
- WT
- wild-type
References
- Adio S., Reth J., Bathe F., Woehlke G.. 2006. Review: regulation mechanisms of Kinesin-1. J. Muscle Res. Cell Motil. 27:153–160. 10.1007/s10974-005-9054-1 [DOI] [PubMed] [Google Scholar]
- Blasius T.L., Cai D., Jih G.T., Toret C.P., Verhey K.J.. 2007. Two binding partners cooperate to activate the molecular motor Kinesin-1. J. Cell Biol. 176:11–17. 10.1083/jcb.200605099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Hoppe A.D., Swanson J.A., Verhey K.J.. 2007a. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J. Cell Biol. 176:51–63. 10.1083/jcb.200605097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Verhey K.J., Meyhöfer E.. 2007b. Tracking single Kinesin molecules in the cytoplasm of mammalian cells. Biophys. J. 92:4137–4144. 10.1529/biophysj.106.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy D.L., Hancock W.O., Wagenbach M., Howard J.. 1999. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1:288–292. 10.1038/13001 [DOI] [PubMed] [Google Scholar]
- Dietrich K.A., Sindelar C.V., Brewer P.D., Downing K.H., Cremo C.R., Rice S.E.. 2008. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc. Natl. Acad. Sci. USA. 105:8938–8943. 10.1073/pnas.0803575105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D., Stock M.F.. 2000. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2:257–260. 10.1038/35010525 [DOI] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Blasius T.L., Li Z., Jiang Y., Jih G.T., Meyhofer E., Verhey K.J.. 2009. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7:e72. 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., Niwa S.. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Imanishi M., Endres N.F., Gennerich A., Vale R.D.. 2006. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J. Cell Biol. 174:931–937. 10.1083/jcb.200605179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C., Schnapp B., Banker G.A.. 2006. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 49:797–804. 10.1016/j.neuron.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Kallipolitou A., Deluca D., Majdic U., Lakämper S., Cross R., Meyhöfer E., Moroder L., Schliwa M., Woehlke G.. 2001. Unusual properties of the fungal conventional kinesin neck domain from Neurospora crassa. EMBO J. 20:6226–6235. 10.1093/emboj/20.22.6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.R., Shin H., Choi J., Ko J., Kim S., Lee H.W., Kim K., Rho S.H., Lee J.H., Song H.E., et al. 2004. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 23:1506–1515. 10.1038/sj.emboj.7600164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N.. 2003. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J. Cell Biol. 162:1045–1055. 10.1083/jcb.200302175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M., Nakagawa T., Seog D.H., Hirokawa N.. 2000. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 288:1796–1802. 10.1126/science.288.5472.1796 [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K.P., Rose L.S., Scholey J.M.. 1999. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol. Biol. Cell. 10:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M.A., Leroux M.R.. 2009. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 19:306–316. 10.1016/j.tcb.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Stock M.F., Guerrero J., Cobb B., Eggers C.T., Huang T.G., Li X., Hackney D.D.. 1999. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J. Biol. Chem. 274:14617–14623. 10.1074/jbc.274.21.14617 [DOI] [PubMed] [Google Scholar]
- Takano K., Miki T., Katahira J., Yoneda Y.. 2007. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res. 35:2513–2521. 10.1093/nar/gkm125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen M., Marx A., Sack S., Mandelkow E.. 1998. The coiled-coil helix in the neck of kinesin. J. Struct. Biol. 122:30–41. 10.1006/jsbi.1998.3986 [DOI] [PubMed] [Google Scholar]
- Thorn K.S., Ubersax J.A., Vale R.D.. 2000. Engineering the processive run length of the kinesin motor. J. Cell Biol. 151:1093–1100. 10.1083/jcb.151.5.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W.. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wedaman K.P., Meyer D.W., Rashid D.J., Cole D.G., Scholey J.M.. 1996. Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132:371–380. 10.1083/jcb.132.3.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K.H., Hanada T., Chishti A.H.. 2007. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 46:10039–10045. 10.1021/bi701169w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.