Microtubules with long polyglutamylated C-terminal tails are more prone to severing by spastin, establishing the importance of tubulin posttranslational modifications.
Abstract
Posttranslational glutamylation of tubulin is present on selected subsets of microtubules in cells. Although the modification is expected to contribute to the spatial and temporal organization of the cytoskeleton, hardly anything is known about its functional relevance. Here we demonstrate that glutamylation, and in particular the generation of long glutamate side chains, promotes the severing of microtubules. In human cells, the generation of long side chains induces spastin-dependent microtubule disassembly and, consistently, only microtubules modified by long glutamate side chains are efficiently severed by spastin in vitro. Our study reveals a novel control mechanism for microtubule mass and stability, which is of fundamental importance to cellular physiology and might have implications for diseases related to microtubule severing.
Introduction
When tubulin dimers polymerize into microtubules (MTs), the C-terminal tails of tubulin become exposed to the outer surface of the tubules (Nogales et al., 1998), where they provide binding sites for several MT-associated proteins (MAPs) and molecular motors (Wang and Sheetz, 2000; Lakämper and Meyhofer, 2005). The tubulin tails are subjected to various posttranslational modifications. One of them, polyglutamylation, adds variable numbers of glutamates to the C-terminal tails of tubulin (Eddé et al., 1990), and is specifically enriched on the MTs of neurons, centrioles, cilia, and of the mitotic spindle (Audebert et al., 1994; Mary et al., 1996; Bobinnec et al., 1998; Regnard et al., 1999; Fig. S1). As it increases the negative charge of MT tails, polyglutamylation could regulate the interactions of MAPs with MTs (for review see Verhey and Gaertig, 2007; Janke et al., 2008).
A group of proteins potentially regulated by polyglutamylation are the MT-severing enzymes katanin (McNally and Vale, 1993; Hartman et al., 1998) and spastin (Evans et al., 2005; Roll-Mecak and Vale, 2005), which belong to the family of AAA ATPases. One structural model of MT severing suggested that hexameric spastin rings seize the acidic tubulin tails and destabilize the MT lattice by pulling on the tails (Roll-Mecak and Vale, 2008). In the proposed structure, the sequence domain of spastin that binds the tubulin tails is positively charged, and could thus attract the negatively charged tubulin tails via electrostatic interactions. Polyglutamylation, which can further increase these charges by adding glutamate side chains, was therefore suggested as a potential regulator of MT severing (for review see Roll-Mecak and McNally, 2010).
A first link between tubulin modifications and MT severing was provided by the mutagenesis of the C-terminal tails of β-tubulin in the protist Tetrahymena thermophila. Although the mutations conserved the overall negative charge of the tubulin tails, they abolished glutamylation and also glycylation, another tubulin polymodification that uses the same modification sites as glutamylation (Redeker et al., 1994). Expression of the nonmodifiable tubulin resulted in defects in ciliary assembly and cytokinesis (Thazhath et al., 2002), a phenotype similar to that of a Tetrahymena strain deficient for the MT-severing enzyme katanin (Sharma et al., 2007). These experiments suggested a potential role for tubulin glutamylation or glycylation in katanin-mediated MT severing, but did not discriminate between the two modifications. Evidence favoring glutamylation as potential regulator of MT severing came from the observation that a mutation in a potential modification site on β-tubulin in Caenorhabditis elegans, an organism without glycylation, decreased the sensitivity of MTs to katanin-mediated severing (Lu et al., 2004).
However, direct evidence for tubulin polyglutamylation as a regulator of MT severing is lacking, and it is unclear if the different severing enzymes are regulated by the same mechanism. Here, we use a set of glutamylating enzymes with different enzymatic specificities (van Dijk et al., 2007) to study the role of distinct MT polyglutamylation types in MT severing by a combined cell biology and in vitro approach.
Results and discussion
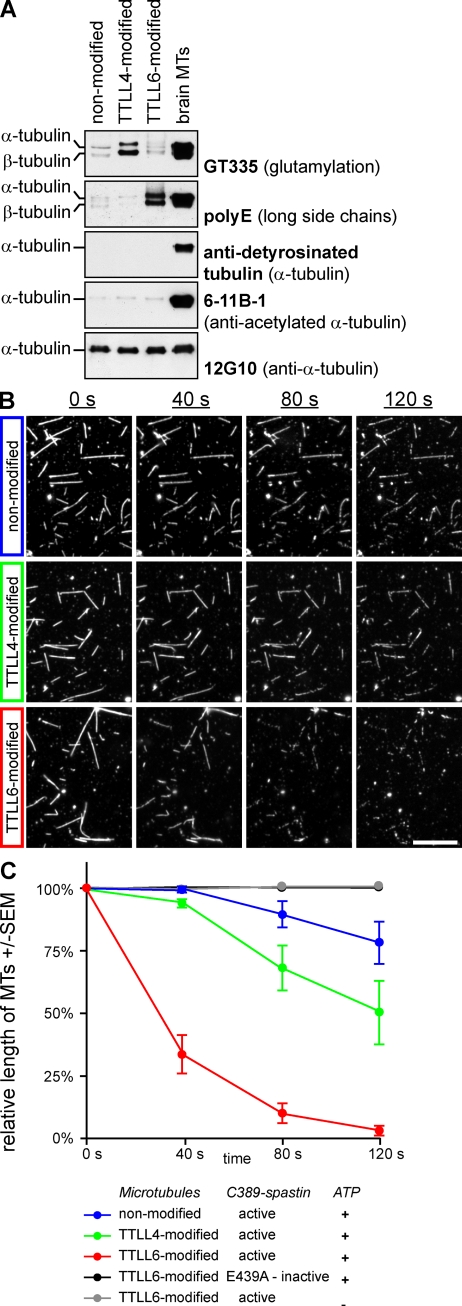
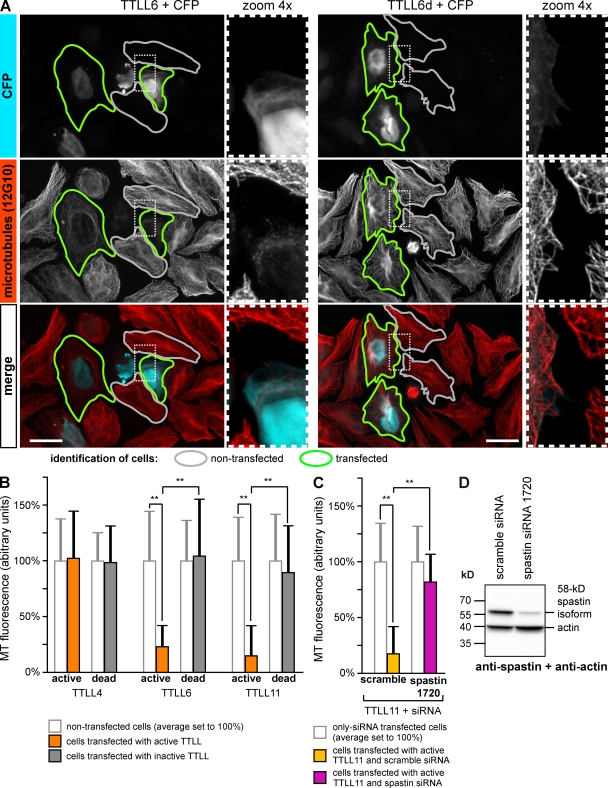
To investigate the effects of glutamylation on the interphase MT network, which carries very low levels of glutamylation (Regnard et al., 1999; Fig. S1), we expressed various polyglutamylases in HeLa cells to generate specific subtypes of glutamylation (van Dijk et al., 2007). Cells expressing glutamylases were identified by coexpression of CFP, and they showed strongly increased glutamylation on the interphase MT network (Fig. S2, A and B). The length of the glutamylated side chains depends on the reaction specificities of the expressed enzymes (Fig. S2 C). To address whether different types of glutamylation on MTs have specific functions, we first investigated two enzymes with divergent enzymatic characteristics, tubulin tyrosine ligase-like proteins 4 and 6 (TTLL4 and TTLL6). TTLL4 generates short side chains that can be detected specifically with the GT335 antibody, whereas TTLL6 adds long side chains detected with polyE antibody (Fig. S2 C). At the overexpression conditions used here, both enzymes modify both α- and β-tubulin (example in Fig. 4 A; van Dijk et al., 2007). Strikingly, 24 h after transfection, TTLL6-expressing cells had lost more than 70% of their MT mass, whereas no changes were found after TTLL4 expression (Fig. 1, A and B). This effect was specific to the polyglutamylation activity and not to the presence of the TTLL6 protein because expression of an enzymatically dead version of TTLL6 had no effect (Fig. 1, A and B). TTLL11, another long side chain–generating enzyme (van Dijk et al., 2007), also reduced MT mass by more than 70% (Fig. 1 B), which further strengthens the conclusion that long glutamate side chains on tubulin provide a signal for reduction of MT mass.
Figure 4.
Microtubule polyglutamylation activates spastin-mediated severing in vitro. (A) Immunodetection of modification levels on purified tubulin that was copolymerized with 6.5% rhodamine-labeled brain tubulin. GT335 detects glutamylation irrespective of the side chain length, whereas polyE is specific to long chains. Tubulin purified from HeLa cells differs only in polyglutamylation levels, while acetylation and detyrosination are not altered. (B) In vitro severing of differentially modified MTs. MTs purified from HeLa cells were copolymerized with 6.5% rhodamine-labeled brain tubulin and imaged over time. C389-spastin and ATP were added to the imaging chamber at t = 0 s. Bar, 10 µm. (C) Quantification of the relative length of MTs in time-lapse movies as shown in B. The total MT length was determined for each time frame and normalized to the first time frame (100% at 0 s). MT fragments <0.85 µm were omitted. Each time point represents mean ± SEM from five experiments (except for negative controls, which were not included in replica experiments).
Figure 1.
Polyglutamylation induces microtubule severing in HeLa cells. (A) MT morphology in HeLa cells expressing either active TTLL6 or an inactive mutant (TTLL6d). Transfected cells (green contours) were identified by CFP, which was coexpressed from the same plasmid (Fig. S2). Cells were stained with the general tubulin antibody 12G10, which detects α-tubulin in a glutamylation-independent manner (not depicted). Gray contours indicate examples of untransfected cells. Bar, 20 µm. (B) Quantification of MT mass in cells expressing active and inactive TTLL4, 6, and 11. Mean 12G10 fluorescence intensity was measured as indicated by contours in A for >30 cells per experimental condition and plotted as normalized to the mean of nontransfected cells (100%). Error bars indicate SD. Statistical significance was determined by two-tailed Student’s t test. All P values (**) are below 10−10. (C) Quantification as in B after expression of TTLL11 in cells transfected with nonsilencing control (scramble) or spastin-specific siRNA 1720 (transfection scheme of siRNA; see Materials and methods). (D) Validation of spastin siRNA. HeLa cells were transfected with scramble and spastin siRNA, and actin and spastin levels were detected on the same blot with specific antibodies. The siRNA 1720 reduces the levels of spastin to 13.2%.
The polyglutamylation-induced loss of interphase MTs was likely to be mediated by the MT-severing enzyme spastin, which is known to be a regulator of MT mass in metazoan cells (Sherwood et al., 2004). To test this, we expressed TTLL11 in the presence of two different siRNAs specific to spastin. While expression of TTLL11 alone led to the disassembly of more than 70% of the MTs in the cells, only ∼20% of the MT mass was lost after RNAi depletion of spastin (Fig. 1, C and D; unpublished data). This indicates that the modification of tubulin with long glutamate side chains induces spastin-dependent disassembly of MTs in HeLa cells.
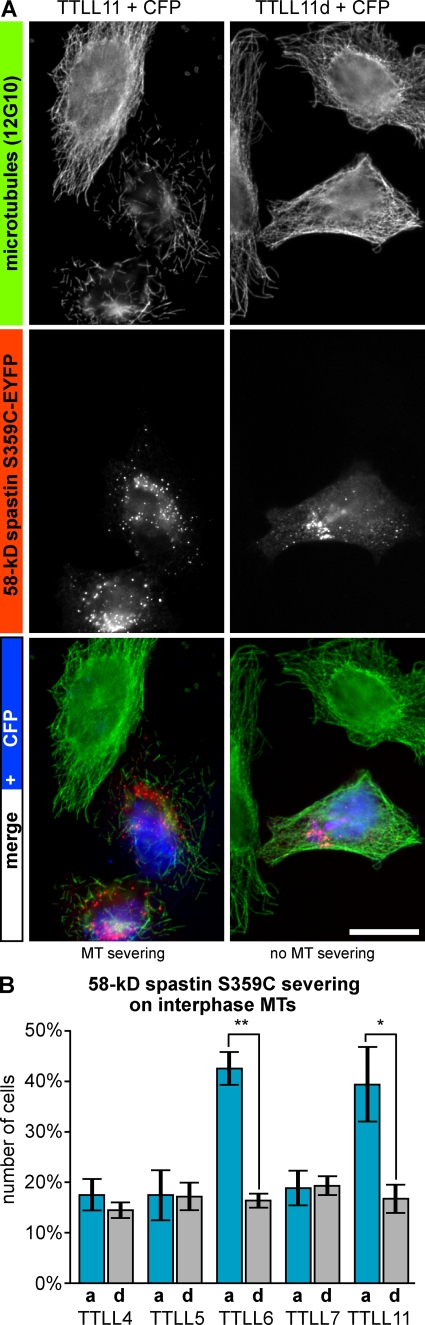
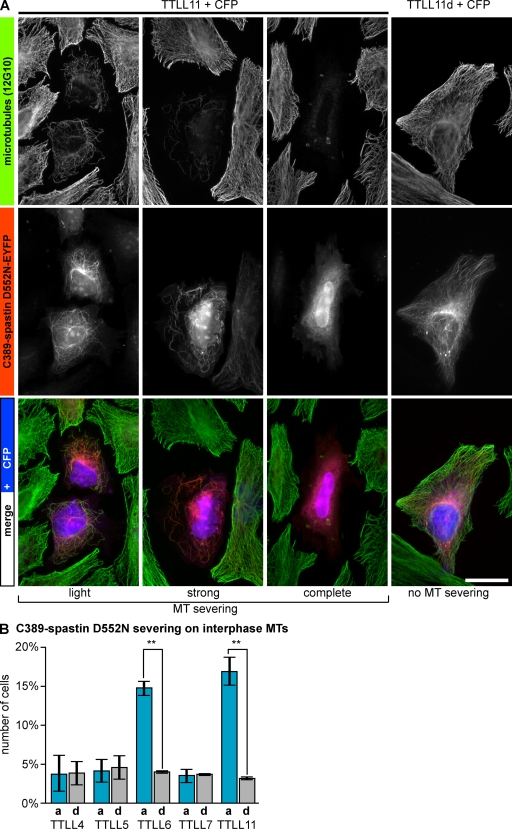
We next investigated whether exogenously expressed spastin could be activated by MT polyglutamylation by using the most widely expressed 58-kD isoform (Fig. 1 D). To further test if the regulation of MT severing by polyglutamylation is due to a regulation on the catalytic domain of spastin, we also investigated a truncated version of the enzyme that only contains domains essential for severing activity (C389-spastin; Fig. S3; White et al., 2007). Upon expression in HeLa cells, both the 58-kD and the C389-spastin completely disassembled MTs, which made it impossible to monitor their activation by polyglutamylation. To circumvent this problem, we tested different mutations that have previously been shown to decrease the severing activity of spastin. After introduction of the S359C mutation (61% activity; Roll-Mecak and Vale, 2005; Fig. S3) into the 58-kD spastin isoform, and of the D552N mutation (19% activity; Roll-Mecak and Vale, 2005; Fig. S3) into C389-spastin, both proteins induced MT severing only in a low percentage of the transfected HeLa cells. Co-expression of TTLL6 or TTLL11 together with either the mutated 58-kD spastin (S359C) for 16 h (Fig. 2) or with the mutated C389-spastin (D552N) for 7 h (Fig. 3) significantly increased the number of cells with MT severing. In contrast, coexpression of TTLL4, TTLL5, or TTLL7 did not change the frequency of MT severing in the transfected cells (Figs. 2 and 3). The observed severing must be due to the activation of the exogenously expressed spastin because endogenous spastin was not activated at these early time points after transfection of polyglutamylating enzymes. Furthermore, coexpression of active TTLL6 or TTLL11 with enzymatically dead spastin did not induce MT disassembly (unpublished data). Together, these results indicate that the severing activity of spastin can be stimulated by the addition of long glutamate side chains on MTs, and that the mechanism of this activation resides in a domain of the spastin protein that is directly involved in the catalysis of MT severing.
Figure 2.
Polyglutamylation activates 58-kD spastin in vivo. (A) HeLa cells were cotransfected with the 58-kD isoform of spastin-EYFP (mutated S359C; Fig. S3) and an active (TTLL11) or inactive (TTLL11d) polyglutamylase. Cells were grown for 16 h before fixation and immunofluorescence staining with 12G10. CFP allowed identification of TTLL11-expressing cells. Bar, 20 µm. (B) Fraction cells expressing spastin S359C-EYFP and CFP that show MT severing as shown in A. A minimum of 100 cells was analyzed in three independent experiments. Error bars indicate SD. Significance was determined by two-tailed Student’s t test. P values are below 0.001 (**), or below 0.01 (*).
Figure 3.
Polyglutamylation activates a short version of spastin in vivo. (A) HeLa cells transfected with the N-terminally truncated form of spastin, C389-spastin-EYFP (mutated D552N; Fig. S3), were grown for 15 h and subsequently transfected with either active TTLL11 or inactive mutant (TTLL11d) 7 h before fixation and immunofluorescence staining by 12G10. CFP coexpressed from the TTLL11 plasmids allowed identification of transfected cells. Bar, 20 µm. (B) Fraction of cells expressing C389-spastin D552N-EYFP and CFP showing MT severing (all severing phenotypes as shown in A were compiled). A minimum of 100 cells was analyzed in three independent experiments. Error bars SD. Significance was determined by two-tailed Student’s t test. All P values (**) are below 0.001.
To test whether spastin is directly activated by polyglutamylation, we set up an in vitro MT severing assay using the truncated version of spastin (C389-spastin; Fig. S3). MTs used in most published in vitro studies are highly polyglutamylated, as they are prepared from brain tubulin (Eddé et al., 1990). Because we wanted to test the role of polyglutamylation on spastin-mediated MT severing, MTs with very low glutamylation levels were purified from HeLa cells. With this as a reference, we also prepared differentially glutamylated tubulin by transfecting HeLa cells 24 h before the tubulin purification with either TTLL4 or TTLL6. To visualize the MTs, we copolymerized the differently modified HeLa tubulin with 6.5% of rhodamine-labeled brain tubulin and analyzed the modification state of the resulting MTs by immunoblot. TTLL4 induced high levels of glutamylation with short side chains detected with GT335, while TTLL6 mostly generated long glutamate side chains as shown with polyE antibody (Fig. 4 A; Fig. S2 C). Other tubulin modifications, such as detyrosination and acetylation, were not altered in the differentially glutamylated HeLa MTs (Fig. 4 A).
To measure the efficiency of spastin-mediated severing of the differentially modified MTs in vitro, MTs were attached to mini-chambers, which were assembled as described in Materials and methods, and placed under the microscope. The severing reaction was started by flowing spastin and ATP into the chambers while imaging. Within a period of 120 s, the addition of spastin induced disassembly of nonmodified MTs to 78.2 ± 8.5% (mean ± SEM) of their original lengths. In the same time frame, TTLL4-modified MTs were disassembled to 50.0 ± 12.7% of their original lengths, whereas TTLL6-modified MTs were completely severed (2.2 ± 1.7%; Fig. 4, B and C). Neither the addition of spastin in the absence of ATP nor the addition of inactive spastin (E439A; Fig. S3) induced severing of TTLL6-modified MTs, indicating the specificity of the assay (Fig. 4 C). Together with the observed effects of MT glutamylation in cells (Figs. 1–3), these in vitro data demonstrate that glutamylation of tubulin directly stimulates spastin-mediated severing of MTs, and that the length of the glutamate side chain determines the efficiency of severing. Short side chains, such as those added by TTLL4, have less effect than long side chains added by TTLL6. In agreement with these observations, brain MTs, which are highly modified with long glutamate side chains (Fig. 4 A), were disassembled within less than 20 s when assayed under similar conditions (unpublished data).
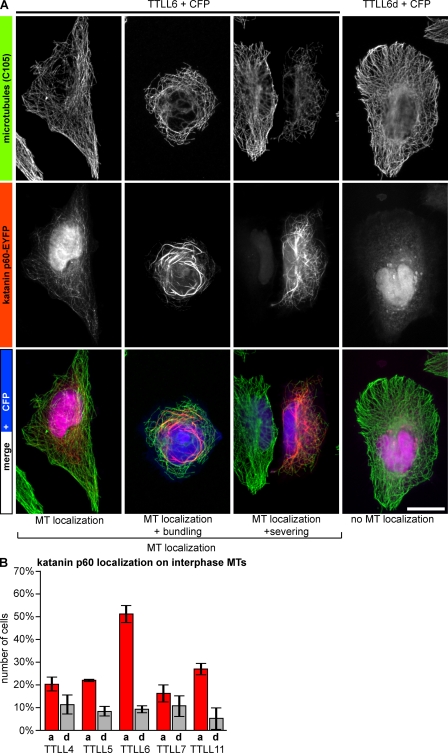
To test whether tubulin polyglutamylation could be a general activator for MT-severing enzymes, we investigated another severing protein, katanin p60 (Hartman et al., 1998; Fig. S3). In contrast to spastin, katanin p60 does not localize to MTs and, more strikingly, it does not induce detectable MT severing upon expression in HeLa or U2OS cells (Fig. 5 A, panel TTLL6d + CFP). This made it difficult to quantify katanin activation by cellular MT morphology as it was done for spastin (Figs. 2 and 3). However, expression of YFP-tagged katanin p60 together with different TTLL enzymes in U2OS cells induced localization of katanin p60 to MTs, which was accompanied by MT bundling and occasional severing (Fig. 5 A). Localization of katanin p60 to MTs was previously proposed as a possible readout for its activation (Hartman and Vale, 1999; McNally et al., 2000). Accordingly, two inactive mutants of katanin p60 (E309A and E332Q; Fig. S3) were not targeted to MTs upon glutamylation (unpublished data). To determine the activation of katanin by polyglutamylation, we quantified MT localization, bundling, and severing after coexpression of katanin p60 with different TTLL enzymes. Similar to spastin-mediated severing, the long chain–generating glutamylase TTLL6 induced a more pronounced katanin p60 activation than the short side chain–generating enzymes TTLL4, TTLL5, and TTLL7 (Fig. 5 B). However, TTLL11 showed a weaker impact on katanin p60 activation as compared with its strong effect on spastin-mediated severing (Figs. 1 B and 2 B). Likely explanations for these results could be that TTLL6 and TTLL11 either use different modification sites within the C-terminal tails of tubulin, or they generate glutamate side chains of different lengths. Thus, although spastin is insensitive to these differences, katanin p60 is more efficiently activated by glutamylation generated with TTLL6.
Figure 5.
Effects of differential microtubule glutamylation on katanin p60. (A) U2OS cells were cotransfected with katanin p60-EYFP and either active TTLL6 or an inactive mutant (TTLL6d), with CFP as transfection control. Activation of katanin leads to MT localization of katanin p60, and sometimes to MT bundling and/or severing. Bar, 20 µm. (B) Fraction of cells showing colocalization of katanin p60-EYFP with MTs (all phenotypes as shown in A were compiled). A minimum of 200 cells was analyzed in three independent experiments. Error bars indicate SD.
The demonstration that long glutamate side chains are much more efficient at activating the MT-severing proteins spastin and katanin than short side chains supports the idea that tubulin modification with long glutamate side chains may be a general activator of MT severing. This would imply that another AAA MT-severing protein, fidgetin (Zhang et al., 2007), could also be activated by polyglutamylation. The observed differences in the sensitivity of spastin and katanin to TTLL11-generated polyglutamylation raises the exciting possibility that fine-tuning of these side chains could be implicated in a differential activation of these proteins within a single cell.
By showing that tubulin glutamylation, and in particular long glutamate side chains, stimulates MT severing in vivo and in vitro, we provide evidence that tubulin polyglutamylation can act as a direct regulator of MT functions. This novel regulatory mechanism could play a role in the local confinement of severing events to specific subcellular regions, or even along single MTs. For example, the mitotic spindle shows distinct glutamylation patterns on subsets of MTs, with a highly glutamylated midspindle, but not astral MTs (Fig. S1 A), and MTs with long glutamate chains restricted to the spindle poles and the midbody (Fig. S1 B). Polyglutamylation might thereby regulate localized severing for spindle dynamics and chromosome movement (McNally et al., 2006; Zhang et al., 2007), as well as for cytokinetic abscission (Connell et al., 2009). The relevance of polyglutamylation for ciliogenesis and cytoskeleton dynamics during cell division is also supported by the ciliary defects and the cytokinesis arrests induced by mutated glutamylation/glycylation sites on tubulin (Thazhath et al., 2002) and katanin knockout in Tetrahymena cells (Sharma et al., 2007). Another process that requires local MT severing and might also be regulated by the polyglutamylation status of MTs is neurite outgrowth (Ahmad et al., 1999; Wood et al., 2006).
In conclusion, MT polyglutamylation might provide a permissive signal within complex regulatory networks that control where and when MTs are severed within a single cell. How opposing activities of modifying (van Dijk et al., 2007) and yet undiscovered de-modifying enzymes control the spatial and temporal distribution of MT glutamylation is an important question to be addressed in the future. Considering that mutations affecting spastin activity have been linked to neurodegeneration in hereditary spastic paraplegia (Evans et al., 2005; Roll-Mecak and Vale, 2005), the discovery of polyglutamylation as a novel regulator of spastin activity opens the exciting possibility that changes in MT glutamylation levels could play a role in the pathogenesis of neurodegenerative disorders.
Materials and methods
Cloning, mutagenesis, and siRNA
The cloning of mouse TTLL4, 5, 6, 7, and 11 has been described previously (van Dijk et al., 2007). Untagged TTLL proteins were expressed under the control of the cytomegalovirus promoter, together with CFP, which was under the control of a second cytomegalovirus promoter on the same expression plasmid (see Fig. S2 for functionality tests of these vectors). Katanin p60 was cloned from mouse cDNA into an EYFP-tagged vector. For spastin, we cloned the most widely expressed 58-kD isoform of spastin (starting at position 85; Fig. S3), and the truncated version C389-spastin (C-terminal 389 amino acids; minimal version necessary for severing activity; White et al., 2007; Fig. S3) from mouse cDNA into an EYFP-tagged vector as well as into a bacterial expression vector harboring an N-terminal GST tag. Functionality tests showed that both versions of spastin are active. A truncated version of mouse KIF5B (N-terminal 555 amino acids) was cloned with a 6-histidine (6His) tag into a bacterial expression vector.
Point mutations were introduced by a PCR-based quick-change method. The expression plasmids containing the cDNAs of the genes of interest were amplified with two mutagenic primers and Pfu polymerase (Promega). Template DNA was removed by digesting with DpnI (New England Biolabs, Inc.) for 4 h at 37°C; the DNA was then transformed into Escherichia coli cells and single clones were isolated. The presence of the mutation was verified by DNA sequencing. Using this method, we generated two previously described inactive versions for each of katanin p60 and spastin (see Fig. S3 for details). Additionally, we generated spastin versions with reduced activity, carrying the S359C (C1076G) or the D552N (G1654A) mutation (Fonknechten et al., 2000; Roll-Mecak and Vale, 2005; Fig. S3). The Walker motif of KIF5B was mutated T92N to gain a nonmotile kinesin (C275A; Nakata and Hirokawa, 1995), which was used to fix MTs onto glass slides in the in vitro experiments.
Two siRNAs for spastin, siRNA 1720 (GCCAGUGAGAUGAGAAAUA; Fig. 1 D) and siRNA 623 (AAACGGACGUCUAUAAUGA; Connell et al., 2009; not depicted) were used in our assays.
Purification of recombinant proteins
Recombinant proteins produced in E. coli were fused to either 6His or GST. The cells were grown at 37°C into the exponential growth phase, then cooled down to 20°C, and protein production was induced with 0.5 mM IPTG. Cells were harvested after 4 h.
For purification of GST-tagged C389-spastin, bacteria were resuspended in lysis buffer (20 mM Tris/HCl, pH 7.5, 300 mM NaCl, 2 mM MgCl2, 1 mM ATP, and protease inhibitors) and sonicated. The lysate was centrifuged at 80,000 g for 10 min at 4°C. The supernatant was incubated with glutathione sepharose (GE Healthcare) for 30 min at 4°C. The flow-through was discarded and the resin washed with wash buffer (20 mM Tris/HCl, pH 7.5, and 500 mM NaCl) until the flow-through was clear of protein. GST fusion proteins were eluted with elution buffer (50 mM Tris/HCl, pH 8.0, 300 mM NaCl, 10 mM MgCl2, 5 mM DTT, and 20 mM reduced glutathione). After purification, GST-C389-spastin was dialyzed against Hepes buffer (20 mM Hepes/NaOH, pH 7.5, 100 mM NaCl, 3 mM MgCl2, and 5 mM DTT).
To isolate 6His-tagged KIF5B, bacteria were resuspended in lysis buffer (20 mM potassium phosphate, pH 7.0, 150 mM NaCl, 1 mM MgCl2, 0.1 mM ATP, 5 mM β-mercaptoethanol, 10% [wt/vol] glycerol, and protease inhibitors) and sonicated. The lysate was centrifuged at 80,000 g for 10 min at 4°C. In parallel, chelating sepharose (GE Healthcare) resin was incubated for 5 min with 4 volumes of 200 mM NiSO4 and subsequently equilibrated with equilibration buffer (50 mM potassium phosphate, pH 7.0, and 100 mM NaCl). The clarified supernatant was then incubated with the resin for 30 min at 4°C. The flow-through was discarded and the resin washed with wash buffer (50 mM potassium phosphate, pH 7.0, 1 M NaCl, 2 mM β-mercaptoethanol, 25 mM imidazole, and 10% [wt/vol] glycerol) until the flow-through was clear of protein. The 6His-tagged protein was eluted with elution buffer (150 mM imidazole, 50 mM KCl, 10 mM β-mercaptoethanol, and 10% [wt/vol] glycerol). After purification, KIF5B was concentrated and dialyzed with motor buffer (12 mM Pipes, pH 7.0, 10 mM NaCl, 1 mM EGTA, and 1 mM MgCl2).
Tubulin purification (detailed protocol: Lacroix and Janke, 2010)
Tubulin was purified from HeLa cells grown in adherent culture. The purification protocol was adapted from a method for yeast tubulin purification (Davis et al., 1993). For the induction of tubulin glutamylation, cells were transfected with either TTLL4_C639-EYFP for the generation of short glutamate side chains, or TTLL6_N513-EYFP for long chains. (The truncated versions of TTLLs have been shown to be expressed at higher levels and to be more active. Both enzymes modify α- as well as β-tubulin under the experimental conditions used here; van Dijk et al., 2007).
Purification experiments were performed with ∼1 g of cultured cells. Cells were detached from the culture dishes with PBS containing 0.5 mM of EDTA, collected and spun down (200 g for 5 min at 22°C). Cells were lysed (2 ml buffer for 1 g of cells) on ice in MEM buffer (50 mM MES/NaOH, pH 6.8, 2 mM EGTA, 1 mM MgCl2, and 0.2% [vol/vol] NP-40) containing 200 mM NaCl. The soluble fraction was obtained by centrifugation at 120,000 g for 10 min at 4°C, and directly loaded onto a DEAE fast-flow sepharose (GE Healthcare) column (2 ml) at 1 ml/min. The column was washed with MEM buffer with 200 mM NaCl at the same flow rate until no protein was detected in the flow-through. Tubulin was eluted with an NaCl gradient (200–520 mM NaCl in MEM buffer, gradient volume: 40 ml). The eluted fractions were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining of the proteins, and the fractions with the purest and most concentrated tubulin were pooled and precipitated with 60% (wt/vol) ammonium sulfate at 4°C for 20 min. The pellet was dissolved in 500 µl BRB80 buffer (80 mM Pipes/NaOH, pH 6.9, 1 mM EGTA, and 1 mM MgCl2) containing 0.1 mM GTP, and cleaned up on a NAP-5 gel filtration column (GE Healthcare) with BRB80 buffer containing 0.1 mM GTP. The tubulin purified in this way was ready for polymerization, and could also be frozen in liquid nitrogen and stored at −80°C. From 1 g of cells, ∼400 µg of protein was obtained. Although the purified HeLa tubulin was of high purity, we cannot exclude the possibility that trace amounts of other proteins that were copurified (for details see Lacroix and Janke, 2010) had an influence on the severing activity measured in our assays.
In vitro MT-severing assay (Fig. 4 , B and C)
100 µl of 1 mg/ml HeLa tubulin were polymerized into MTs for 30 min at 37°C in BRB80 buffer, containing 4 µM Paclitaxel, 1 µl of 7 mg/ml rhodamine-labeled tubulin (Hyman et al., 1991), and 5 µl of 25 mM GTP. Finally, the MTs were stabilized with 20 µM Paclitaxel, and the MT solution was kept in the dark. To avoid the possibility of the incorporated rhodamine-labeled tubulin biasing the experimental results, the same amounts were copolymerized with each batch of HeLa tubulin.
Mini-chambers were prepared on glass slides that were previously treated with hot ethanol steam. The chambers were kept on ice during preparation. After two washes with motor buffer (12 mM Pipes/NaOH, pH 7.0, 10 mM NaCl, 1 mM EGTA, and 1 mM MgCl2), a 0.25-mg/ml solution of purified KIF5B 555 T92N motor was perfused twice and incubated for 8 min on ice. The kinesin-coated chambers were transferred to room temperature and incubated for 2 min before washing three times with Hepes buffer (20 mM Hepes/NaOH, pH 7.5, 100 mM NaCl, and 3 mM MgCl2). The MT solution at 15 µg/ml was then twice perfused into the chambers, which were then incubated for 5 min at room temperature in the dark to allow the MTs to attach to the kinesin.
The chambers were washed four times with Hepes buffer and then transferred to an upright fluorescence microscope (DMRA, Leica) with a 100× immersion objective at 23°C. The focus and light intensity were adjusted to the rhodamine-labeled MTs. The chambers were perfused twice with severing buffer (20 mM Hepes/NaOH, pH 7.5, 100 mM NaCl, 3 mM MgCl2, 10 mM DTT, 1 mM ATP, 20 mM glucose, 0.4 mg/ml glucose oxidase, and 0.2 mg/ml catalase) containing 1 µM purified GST-C389-spastin, and the image acquisition was started immediately with a time lapse of 40 s for a total duration of 2 min (Fig. 4 B).
The total MT length of each movie frame was measured manually by drawing a straight line over each MT using ImageJ software (open source http://rsbweb.nih.gov/ij). Fluorescent particles smaller than 0.85 µm were excluded from quantification. The MT length of each movie frame was normalized to t = 0 s.
Measurement of MT disassembly in cells (Fig. 1, B and C)
HeLa cells transfected with TTLL-encoding plasmids were identified by coexpressed CFP. Cells were fixed and tubulin was visualized using the 12G10 antibody. Images were acquired using the same settings for all slides, and analyzed in ImageJ software. On each slide, at least 30 CFP-positive cells, as well as 30 control cells without CFP expression, were analyzed by manual definition of cell contours and measurement of mean tubulin fluorescence. Mean and standard deviations were then determined for the cell population of each condition, and then normalized to nontransfected cells. Significant deviations were tested by two-tailed Student’s t test at a significance level of P ≤ 0.001.
Protein electrophoresis and immunoblot
SDS-PAGE was performed using standard protocols, or in the case of mammalian α- and β-tubulin by a special protocol, as in Eddé et al., 1987. Proteins were transferred to nitrocellulose or PVDF membranes (GE Healthcare) and detected with antibodies. Membranes were incubated with rabbit polyE (anti-polyglutamylation; 1:1,000) and anti-detyrosinated tubulin (1:1,000; Millipore) antibodies, or mouse GT335 (anti-glutamylation, 1:1,000; Wolff et al., 1992), 6-11B-1 (anti-acetylated tubulin; 1:2,000; Sigma-Aldrich), anti-actin clone C4 (1:200,000; Millipore), Sp 3G11/1 (anti-spastin; 1:200; Santa Cruz Biotechnology, Inc.), and 12G10 (anti-α-tubulin; 1:500). Protein bands were visualized with HRP-labeled donkey anti–rabbit or anti–mouse IgG (1:10,000; GE Healthcare) followed by detection with chemiluminescence (ECL Western blot detection kit; GE Healthcare). Protein bands were quantified (Fig. 1 D) using the ChemiGlow West ECL kit and the sFluorchem chemiluminescence imager (Alpha Innotech).
Cell culture, immunofluorescence, and microscopy
HeLa or U2OS cells were cultured on plastic dishes or glass coverslips under standard conditions. Expression plasmids were transfected using JetPEI (Polyplus transfection), and siRNA with Oligofectamine (Invitrogen). For siRNA treatment, cells were transfected with siRNA and grown for 24 h before plasmids were transfected. A second siRNA transfection was performed 2 h after the transfection of the DNA plasmid, and cells were analyzed 22 h later (Fig. 1, C and D).
Cells were fixed using a protocol for the preservation of cytoskeletal structures (Bell and Safiejko-Mroczka, 1995) and then incubated with polyE (1:1,500), GT335 (1:5,000), 12G10 (1:500), the rabbit anti-tubulin C105 (1:1,000; gift of M. Andreu, Centro de Investigaciones Biológicas, Madrid, Spain) antibodies for 1 h, followed by 30 min with anti–mouse or anti–rabbit Alexa 568, or anti–mouse or anti–rabbit Alexa 488 antibody (1:1,000; Invitrogen). DNA was visualized by DAPI staining (0.02 µg/ml). Coverslips were mounted with Mowiol polyvinyl alcohol 4-88 (Fluka). We used DMRA microscopes (Leica) at 23°C with the oil objectives 40× (NA 1.25), 63× (NA 1.32), and 100× (NA 1.4). For the imaging of the MT cytoskeleton, the mechanical aperture of the objectives was closed in order to have an in-focus representation of the whole MT network. The fluorochromes imaged were DAPI (filter: excitation 340–380 nm, emission 425 nm; Leica), ECFP (excitation 421–450 nm, emission 466–500 nm; Chroma Technology Corp.) Alexa 488 (excitation 450–490 nm, emission 500–550 nm; Leica), EYFP (excitation 482–512 nm, emission 532–568 nm; Chroma Technology Corp.), and Alexa 555 and 568 (excitation 530–560 nm, emission 572.5–647.5 nm; Leica). Images were acquired using a CoolSnap HQ camera (Photometrics) and MetaMorph 7 software (MDS Analytical Technologies). Images were analyzed using ImageJ software and assembled in Adobe Photoshop (Adobe Systems, Inc.).
Online supplemental material
Fig. S1 shows the distribution of glutamylated and polyglutamylated MTs in HeLa cells during the cell cycle. Fig. S2 shows a functionality and specificity test for expression vectors that express TTLL proteins and CFP separately. Fig. S3 shows annotated alignment of the protein sequences of mouse spastin and katanin p60. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001024/DC1.
Acknowledgments
We are grateful to M.M. Magiera (CRBM, Montpellier, France), M. Steinmetz (Paul-Scherrer-Institute, Villigen, Switzerland), and T. Surrey (EMBL, Heidelberg, Germany) for instructive discussions on the manuscript. The authors want to thank J.M. Andreu for the gift of the polyclonal antibody C105. The monoclonal antibody 12G10 developed by J. Frankel and M. Nelson was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. The authors declare no conflict of interest.
This work was supported by the Centre National de la Recherche Scientifique, the Universities of Montpellier 2 and 1, the Association pour la Recherche sur le Cancer (ARC) awards CR504/7817 and 3140, the French National Research Agency (ANR) awards 05-JCJC-0035 and 08-JCJC-0007, the HFSP program grant RGP 23/2008, and the EMBO Young Investigator Program grant to C. Janke. B. Lacroix was supported by a fellowship from the Ligue contre le Cancer and the EMBO short-term fellowship ASTF 157-2007. K. Rogowski received two postdoctoral fellowships from the Ligue contre le Cancer and the EMBO long-term fellowship ALTF 546-2006. D.W. Gerlich was supported by SNF research grant 3100A0-114120, a European Young Investigator (EURYI) award of the European Science Foundation, and by ETH research grant ETH-03 08-1. We thank J.-M. Donnay, M.M. Magiera, J.-C. Mazur (CRBM); and V. Bäcker, J. Cau, V. Georget, S. DeRossi, and P. Travo (RIO Imaging facility at the CRBM) for technical assistance.
Footnotes
Abbreviations used in this paper:
- MT
- microtubule
- TTLL
- tubulin tyrosine ligase like
References
- Ahmad F.J., Yu W., McNally F.J., Baas P.W. 1999. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145:305–315 10.1083/jcb.145.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S., Koulakoff A., Berwald-Netter Y., Gros F., Denoulet P., Eddé B. 1994. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J. Cell Sci. 107:2313–2322 [DOI] [PubMed] [Google Scholar]
- Bell P.B., Jr., Safiejko-Mroczka B. 1995. Improved methods for preserving macromolecular structures and visualizing them by fluorescence and scanning electron microscopy. Scanning Microsc. 9:843–857, discussion :858–860 [PubMed] [Google Scholar]
- Bobinnec Y., Moudjou M., Fouquet J.P., Desbruyères E., Eddé B., Bornens M. 1998. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton. 39:223–232 [DOI] [PubMed] [Google Scholar]
- Connell J.W., Lindon C., Luzio J.P., Reid E. 2009. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 10:42–56 10.1111/j.1600-0854.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A., Sage C.R., Wilson L., Farrell K.W. 1993. Purification and biochemical characterization of tubulin from the budding yeast Saccharomyces cerevisiae. Biochemistry. 32:8823–8835 10.1021/bi00085a013 [DOI] [PubMed] [Google Scholar]
- Eddé B., de Nechaud B., Denoulet P., Gros F. 1987. Control of isotubulin expression during neuronal differentiation of mouse neuroblastoma and teratocarcinoma cell lines. Dev. Biol. 123:549–558 10.1016/0012-1606(87)90413-1 [DOI] [PubMed] [Google Scholar]
- Eddé B., Rossier J., Le Caer J.P., Desbruyères E., Gros F., Denoulet P. 1990. Posttranslational glutamylation of alpha-tubulin. Science. 247:83–85 10.1126/science.1967194 [DOI] [PubMed] [Google Scholar]
- Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., Lauring B.P. 2005. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 168:599–606 10.1083/jcb.200409058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonknechten N., Mavel D., Byrne P., Davoine C.S., Cruaud C., Bönsch D., Boentsch D., Samson D., Coutinho P., Hutchinson M., et al. 2000. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 9:637–644 10.1093/hmg/9.4.637 [DOI] [PubMed] [Google Scholar]
- Hartman J.J., Vale R.D. 1999. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 286:782–785 10.1126/science.286.5440.782 [DOI] [PubMed] [Google Scholar]
- Hartman J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., McNally F.J. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287 10.1016/S0092-8674(00)81578-0 [DOI] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., Mitchison T. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Janke C., Rogowski K., van Dijk J. 2008. Polyglutamylation: a fine-regulator of protein function? ‘Protein Modifications: beyond the usual suspects’ review series. EMBO Rep. 9:636–641 10.1038/embor.2008.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., Janke C. 2010. Generation of differentially polyglutamylated microtubules. Methods Mol. Biol. In press [DOI] [PubMed] [Google Scholar]
- Lakämper S., Meyhöfer E. 2005. The E-hook of tubulin interacts with kinesin’s head to increase processivity and speed. Biophys. J. 89:3223–3234 10.1529/biophysj.104.057505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Srayko M., Mains P.E. 2004. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol. Biol. Cell. 15:142–150 10.1091/mbc.E03-06-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary J., Redeker V., Le Caer J.P., Rossier J., Schmitter J.M. 1996. Posttranslational modifications in the C-terminal tail of axonemal tubulin from sea urchin sperm. J. Biol. Chem. 271:9928–9933 10.1074/jbc.271.17.9928 [DOI] [PubMed] [Google Scholar]
- McNally F.J., Vale R.D. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429 10.1016/0092-8674(93)90377-3 [DOI] [PubMed] [Google Scholar]
- McNally K.P., Bazirgan O.A., McNally F.J. 2000. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113:1623–1633 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F.J. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N. 1995. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 131:1039–1053 10.1083/jcb.131.4.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 391:199–203 10.1038/34465 [DOI] [PubMed] [Google Scholar]
- Redeker V., Levilliers N., Schmitter J.M., Le Caer J.P., Rossier J., Adoutte A., Bré M.H. 1994. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 266:1688–1691 10.1126/science.7992051 [DOI] [PubMed] [Google Scholar]
- Regnard C., Desbruyères E., Denoulet P., Eddé B. 1999. Tubulin polyglutamylase: isozymic variants and regulation during the cell cycle in HeLa cells. J. Cell Sci. 112:4281–4289 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., McNally F.J. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., Vale R.D. 2005. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15:650–655 10.1016/j.cub.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Vale R.D. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 451:363–367 10.1038/nature06482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Bryant J., Wloga D., Donaldson R., Davis R.C., Jerka-Dziadosz M., Gaertig J. 2007. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J. Cell Biol. 178:1065–1079 10.1083/jcb.200704021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood N.T., Sun Q., Xue M., Zhang B., Zinn K. 2004. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2:e429 10.1371/journal.pbio.0020429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhath R., Liu C., Gaertig J. 2002. Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 4:256–259 10.1038/ncb764 [DOI] [PubMed] [Google Scholar]
- van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 26:437–448 10.1016/j.molcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Gaertig J. 2007. The tubulin code. Cell Cycle. 6:2152–2160 [DOI] [PubMed] [Google Scholar]
- Wang Z., Sheetz M.P. 2000. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 78:1955–1964 10.1016/S0006-3495(00)76743-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.R., Evans K.J., Lary J., Cole J.L., Lauring B. 2007. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 176:995–1005 10.1083/jcb.200610072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., Denoulet P. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432 [PubMed] [Google Scholar]
- Wood J.D., Landers J.A., Bingley M., McDermott C.J., Thomas-McArthur V., Gleadall L.J., Shaw P.J., Cunliffe V.T. 2006. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum. Mol. Genet. 15:2763–2771 10.1093/hmg/ddl212 [DOI] [PubMed] [Google Scholar]
- Zhang D., Rogers G.C., Buster D.W., Sharp D.J. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]