Abstract
Objectives
Lysosome-associated membrane protein-1 (LAMP-1) has been suggested to be a cell surface receptor for a specific amelogenin isoform, leucine-rich amelogenin peptide or LRAP. However, it is unclear if LAMP-1 is an amelogenin receptor for dental mesenchymal cells. The goal of this study was to determine if LAMP-1 serves as a cell surface binding site for full length amelogenin on tooth root/periodontium associated mesenchymal cells.
Design
Murine dental follicle cells and cementoblasts (OCCM-30) were cultured for 2 days followed by addition of full length recombinant mouse amelogenin, rp(H)M180. Dose-response (0 to 100 μg/ml) and time course (0 to 120 minutes) assays were performed to determine the optimal conditions for live cell surface binding using immuno-fluorescent microscopy. A competitive binding assay was performed to determine binding specificity by adding Emdogain (1 mg/ml) to the media. An antibody against LAMP-1 was used to detect the location of LAMP-1 on the cell surface and the pattern was compared to cell surface bound amelogenin. Both amelogenin and cell surface LAMP-1 were immuno-co-localized to compare the amount and distribution pattern.
Results
Maximum surface binding was achieved with 50 μg/ml of rp(H)M180 for 120 minutes. This binding was specific as demonstrated by competitive inhibition (79% lower) with the addition of Emdogain. The binding pattern for rp(H)M180 was similar to the distribution of surface LAMP-1 on dental follicle cells and cementoblasts. The high co-localization coefficient (0.92) for rp(H)M180 and LAMP-1 supports rp(H)M180 binding to cell surface LAMP-1.
Conclusions
The data from this study suggest that LAMP-1 can serve as a cell surface binding site for amelogenin on dental follicle cells and cementoblasts.
Keywords: amelogenin, LAMP-1, tooth root, periodontium, dental follicle cells, cementoblasts
Introduction
Periodontal disease is marked by destruction of periodontal tissues, which can lead to tooth loss if left untreated. Recognition that periodontal regeneration can be achieved, i.e., formation of new bone, new cementum and supportive periodontal ligament (PDL), has resulted in increased attempts to understand the cellular and molecular mechanisms and factors that regulate formation of these tissues. It is well established that epithelial-mesenchymal (E-M) interactions are required for formation of the tooth crown (enamel and dentin).1-3 Recent data suggest tooth root development begins with Sonic Hedgehog signaling emanating from Hertwig's epithelial root sheath (HERS) cells (stimulated by a currently unknown source) inducing abutting dental papilla cells to express the transcription factor nuclear factor 1C (NF1C). Without NF1C expression, root odontoblast driven root development does not occur.4, 5 This confirms that E-M signaling initiates root dentin formation, a requirement for full root development including cementum formation and a functional periodontium.
Amelogenin expression has been detected by some investigators in HERS cells and root tissue during this developmental time point.6-9 Given the developmental history of these tissues, the use of an epithelial signaling protein to regenerate periodontal tissues is conceptually appealing and thus research directed at understanding the role of epithelium derived molecules in promoting tooth root formation should assist efforts to improve upon existing regenerative therapies. In fact, Emdogain® (EMD, Straumann, Switzerland) a predominately amelogenin containing medicament (derived from porcine enamel matrix) has been marketed for use in regenerating periodontal tissues based on the concept that E-M interactions are required for root formation.8, 10 While not completely predictable, meaningful regenerative results have been reported when EMD was used to treat bone and periodontal defects in both animal models and human patients.11-18 An early event in periodontium formation/regeneration is the development of acellular cementum and EMD is reported to alter the activity of dental follicle cells 19 and cementoblasts 20 in vitro. The mechanism of these effects remains to be defined, although amelogenin has been considered as the major factor responsible for these activities.21 However, as discussed below, additional factors may also be playing a role.
Amelogenins are the most abundant proteins of the enamel matrix and belong to a family of proteins formed as a result of alternative splicing of a single primary transcript.22, 23 One of these alternatively spliced products is known as LRAP, or M59/[A-4] to emphasize the absence of the polar hydrophilic amino acid sequence translated from amelogenin exon 4.24-26 In addition to their structural role in enamel formation, amelogenins have been shown to be involved in a range of activities, including mineral nodule formation and intercellular signaling.25, 27, 28 Amelogenin and associated peptides are secreted primarily by dental epithelial cells known as ameloblasts.29 Although there are conflicting reports,30 amelogenins were found to have low level expression in other cells, including odontoblasts31 and HERS cells that line the root during early phases of cementogenesis.32, 33 HERS cells are thought to either undergo apoptosis and/or transform into cementoblast-like cells, as well as remain as remnant epithelial rest cells within the mature PDL.34-36
Mice null for the amelogenin gene exhibited a defect in crown enamel formation37 and also expressed low levels of transcripts and proteins for bone sialoprotein (BSP) and osteocalcin (OCN),28 two markers of the mature cementoblast and osteoblast phenotype. A root phenotype (cementum defects) was also reported in these amelogenin null mice, although this defect seems to relate to changes in osteoclast behavior after the root is fully formed so it is not developmental in nature, per se.10 These data suggest that epithelial cells or products are required for proper development of periodontal tissues, including cementum and a functional PDL.36 A large body of work suggests a signaling function for amelogenins. Viswanathan and colleagues demonstrated that expression of both Ocn and Bsp were decreased when immortalized cementoblasts were treated with high dose of amelogenin.28 Interestingly, when the same cell type was treated with LRAP or TRAP (tyrosine rich amelogeninpeptide, a degradation product of full length amelogenin), similar effects were observed: Ocn was down-regulated while osteopontin (Opn) was up-regulated in a dose-response fashion.27, 38 These effects were seen as early as 6 hours post-treatment for Opn. Amelogenin spliced product LRAP/M59/[A-4] has been shown to enhance the expression of runt-related transcription factor 2 (Runx2), a master switch for defining the osteoblastic phenotype.25 LRAP treatment of either wild-type or amelogenin-null mouse embryonic stem cells induced a significant increase in mineral matrix formation, and significant increases in Bsp and osterix gene expression.39, 40 These in vitro data complement in vivo data in support of a role for amelogenins in modulating the expression of mesenchymal mineralized tissue-associated genes.
Dental follicle cells constitute the dental follicle region (a loose connective tissue) surrounding the developing tooth. Dental follicle cells play a critical role in the process of root development and tooth eruption.41 In addition, substantial evidence indicates that dental follicle cells are progenitors of periodontal mesenchymal cells including cementoblasts, PDL fibroblasts, and alveolar osteoblasts.36, 42, 43 Dental follicle cells and/or cementoblasts are the proposed target cells for amelogenin signaling in the periodontal region. Addition of EMD to immortalized murine dental follicle cells resulted in increased Opn mRNA level and decreased Ocn mRNA expression. EMD also blocked the induced mineralization by dental follicle cells in vitro.19 However, whether this was caused by amelogenin or other factors in EMD is not clear.21 Nevertheless, how amelogenins and other factors regulate tooth root/periodontium development needs to be further elucidated.
Tompkins et al. demonstrated that LRAP/M59/[A-4] regulates mesenchymal cells (mouse myoblast cell line C2C12) at least partly through a 95 kDa cell surface receptor (LAMP-1).44 They also showed another possible receptor appearing as a 75 kDa band in electrophoresis. In a simultaneous study, Wang et al. reported that enamel matrix proteins interacted with a number of secreted membrane proteins and integral proteins, including human CD63 antigen (LAMP-3),45 using the yeast two-hybrid assay. However, in a more recent study using a dental epithelial cell line (HAT-7), Xu et al. demonstrated that LAMP-1 is not a receptor for full length amelogenin (M180) on HAT-7 cells, while LAMP-3 is involved in amelogenin mRNA degradation.46 The reason for these discrepancies is unclear. Therefore, the critical first step to prove that amelogenins directly regulate the behavior of tooth root/periodontium formation associated mesenchymal cells is to identify a definitive receptor on tooth root/periodontium associated cells.
Materials and Methods
Recombinant protein
One hundred eighty amino acid recombinant mouse amelogenin, rp(H)M180, with a poly-Histidine (6-His) tag that can be used for immunofluorescent labeling, was provided by Dr. Malcolm Snead (University of Southern California). Previous studies have shown that the 6-His tag did not affect the biological effects of amelogenin on dental mesenchymal cells.28 A recombinant protein (Positope™, Invitrogen, Carlsbad, CA) containing 6-His tag was used as a control for the surface binding assay to exclude the possibility of non-specific binding caused by the 6-His tag.
Cell cultures
Murine dental follicle cells were obtained by removing the tissues surrounding the developing first molar of CD-1 mice, 22-24 days after the appearance of the vaginal plug.19 These primary cells were passaged once every 7 days, and dental follicle cells in passage two and three were used in these experiments. Immortalized murine cementoblast (OCCM-30) cells were isolated as previously described.47. Both cell types were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen) with 10% fetal bovine serum (FBS) containing 100 u/ml of penicillin, 100 μg/ml of streptomycin, and L-glutamine in a humidified atmosphere of 5% CO2 at 37°C. Each experimental group consisted of 6 culture dishes (n = 6).
rp(H)M180 binding assay
OCCM-30 cells were plated on glass cover slips at 104/cm2 and cultured as described above for 2 days. Before treating with rp(H)M180, cover slips were pre-cooled at 4°C for 10 minutes. Media were replaced by DMEM with 5% FBS and specific concentrations of rp(H)M180 (with 6-His tags) (0, 12.5, 25, 50, and 100 μg/ml) or control 6-His tagged protein (Positope™, Invitrogen, 50 μg/ml) for 2 hours at 4°C. After washing with phosphate buffered saline (PBS), 3 times, the cells were fixed with 8% paraformadehyde for 30 minutes at room temperature, and treated with 3% H2O2 for 30 minutes at room temperature to block endogenous peroxidase. After washing with PBS, cells were blocked with 10% normal goat serum (NGS) for 1 hour followed by blocking buffer (Blotto, Pierce, Rockford, IL) with 1% NGS for an additional 30 minutes. Primary antibody (mouse anti-6-His, 5 μg/ml in Blotto/1%NGS, Qiagen Inc., Valencia, CA) was applied to the cells for 2.5 hours at room temperature. After washing with PBS, the secondary antibody (HRP conjugated donkey anti-mouse, 2 μg/ml in Blotto/1% NGS, Jackson ImmunoResearch Laboratories Inc., West Grove, PA) was applied for 1.5 hours at room temperature followed by washing and the tyramide signal amplification (TSA) assay (Invitrogen). Briefly, labeling of HRP conjugated secondary antibody was amplified by Alexa Fluor 488 labeled tyramide. Cell nuclei were counter-stained with TO-PRO iodide (0.01 mM, Invitrogen) for one hour at room temperature. Cell surface bound rp(H)M180 was detected by immunofluorescent confocal microscopy (Carl Zeiss Laser Scanning Microscope, Carl Zeiss MicroImaging, Inc., Thornwood, NY). The maximum cell binding concentration was determined and used in the assays described below.
Time course of rp(H)M180 binding assay
OCCM-30 cells were cultured and treated in the same manner as in the binding assay. The optimal concentration of rp(H)M180 based on maximal surface staining was applied in the media for designated lengths of time (0, 15, 30, 60, 120 minutes) at 4°C. Immunofluorescent microscopy was performed as described above. The time point with the maximum labeling and least background was used in the following assays.
Homologous competitive binding assay
OCCM-30 cells and dental follicle cells were plated and treated in the same fashion as described above. Cells were pretreated with 20-fold excess of EMD for 1 hour followed by rp(H)M180 (50 μg/ml) or no ligand control for additional 2 hours at 4°C. The reduction of the cell surface fluorescence labeling was evaluated by the TSA assay and immunofluorescent microscopy as described above. LSM5 PASCAL (Carl Zeiss) software was used to compare the fluorescent pixels per μm2 of membrane surface distribution of bound rp(H)M180 on the cell surfaces of dental follicle cells and OCCM-30 cells with and without pre-treatment of EMD. EMD was used rather than recombinant amelogenin due to EMD availability and the knowledge that EMD is primarily composed of amelogenin.
Immuno-co-localization of surface bound rp(H)M180 and LAMP-1 on cell surfaces
OCCM-30 cells were cultured as described above. The optimal concentration of rp(H)M180 (50 μg/ml) and the antibody against LAMP-1 (rat anti-LAMP-1, 10μg/ml, Abcam, Cambridge, MA) were added to media (with 5% FBS) simultaneously in order to detect rp(H)M180 binding sites on cell surface and localization of cell surface LAMP-1. Cells were incubated at 4°C for 2 hours (as determined above) followed by the TSA assay and immunofluorescent microscopy. The images were evaluated as described above (with LSM5 software) to compare the distribution of bound rp(H)M180 and LAMP-1 on the cell surfaces of dental follicle cells and OCCM-30 cells. The co-localization coefficient was calculated based on the relative number of co-localizing pixels for rp(H)M180 as compared to the number of pixels above a threshold that was set by the software.
Statistical Analysis
All experiments were repeated at least three times with consistent results. Data from homologous competitive binding assay were analyzed by Student's t-test to determine the difference between the two groups. P < 0.05 was used to indicate significance. Sigmastat 3.1 (Systat Software, Inc., Point Richmond, CA) was used for statistical analysis.
Results
Cell Surface Binding Saturation Assay
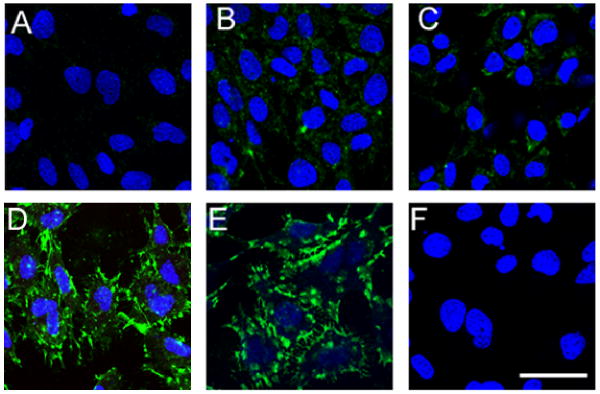
We first strove to determine the concentration of rp(H)M180 which would result in maximum cell surface binding (receptor saturation) using immunofluorescence microscopy. Cementoblasts (OCCM-30) were brought to 4°C and incubated with concentrations of rp(H)M180-6His of 0, 12.5, 25, 50, and 100 μg/ml. After a 2 hour incubation, the cells were washed, fixed, and cell bound rp(H)M180 was detected with an anti-6His antibody. In the absence of rp(H)M180, OCCM-30 cells showed low levels of background staining (Fig. 1A). OCCM-30 cells treated with either 12.5 or 25 μg/ml rp(H)M180 showed a dose-dependent increase in the amount of cell surface bound protein (Fig. 1B, C). Cells treated with 50 μg/ml of rp(H)M180 exhibited greater cell binding of protein compared to the lower concentrations. Cell surface bound rp(H)M180 was readily detected as large “patch” domains which were abundant at the cell margins (Fig. 1D). A further increase in rp(H)M180 concentration to 100 μg/ml failed to show an increase in cell binding, indicating that the rp(H)M180 receptors on OCCM-30 were saturated at 50 μg/ml of rp(H)M180 (Fig. 1E).
Figure 1.
Dose response: rp(H)M180 binding to cementoblast cell surfaces. OCCM-30 cells were incubated with rp(H)M180 (0-100 μg/ml) or control peptide (Positope, 50 μg/ml) for 2 hours at 4°C before anti-6 His antibody was applied, followed by immunofluorescent (confocal) microscopy using a TSA assay and TO-PRO staining (for nuclei).
Panels A-E: rp(H)M180 doses at: 0, 12.5, 25, 50, and 100 μg/ml, respectively.
Panel F: Positope (50 μg/ml).
Blue -- nuclei. Green -- locations of cell surface bound rp(H)M180. Scale bar = 50 μm.
To exclude the possibility that the 6-His tag on the recombinant protein was responsible for cell binding, a control peptide containing a 6-His tag (Positope) was used in the cell surface binding assays in lieu of rp(H)M180. OCCM-30 cells treated with Positope (50 μg/ml) exhibited a level of fluorescence equal to the untreated control (compare Fig 1A to 1F).
As saturated cell surface binding was achieved at 50 μg/ml rp(H)M180, this concentration of rp(H)M180 protein was used in the time course binding assays, including time points of 0, 15, 30, 60, 120, and 240 min. Maximal cell surface binding for rp(H)M180 was achieved after 120 min of incubation (data not shown). Therefore, 120 min was chosen as the standard incubation time for all the experiments in this study.
rp(H)M180 Binding Specificity Binding Assay
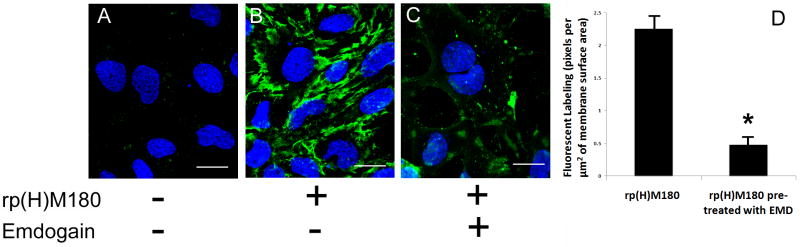
Next, the specificity of cell surface binding for rp(H)M180 protein was determined using EMD as a competitive binding antagonist. OCCM-30 cells were pretreated with either media or a 20 fold excess EMD prior to incubation with 50 μg/ml rp(H)M180. OCCM-30 cells treated with rp(H)M180 alone showed clear cell surface binding with focal areas of enhanced rp(H)M180 binding (Fig. 2B) as compared to the untreated control (Fig. 2A). Cells pretreated with EMD showed a significant decrease in rp(H)M180 detection (Fig. 2C). To quantitate the inhibition of rp(H)M180 binding by EMD the number of anti-6His fluorescent pixels per μm2 of membrane surface area was determined using the confocal LSM software. In the absence of the EMD inhibitor (Fig. 2B), OCCM-30 cells had approximately 2.26 fluorescent pixels per μm2 of membrane surface. When the cells were treated with EMD prior to rp(H)M180, the fluorescent pixels dropped to 0.48 pixels per μm2 of membrane surface (Fig. 2D). Therefore there was a 79% inhibition of rp(H)M180 binding by EMD, suggesting that they utilize a common cell surface receptor.
Figure 2.
Binding specificity: EMD blocked rp(H)M180 binding on cementoblasts. OCCM-30 cells were pretreated with 20 fold of EMD (C) for 1 hour followed by rp(H)M180 (B, C) or no ligand control (A) for additional 2 hours at 4°C. The anti-6-His antibody was applied and immunofluorescent (confocal) microscopy was performed as described in Fig. 1. The fluorescent pixels per μm2 of membrane surface decreased significantly (79% lower, P < 0.05) with the cells pre-treated with 20 fold of EMD (D). Representative fields are shown. Blue – nuclei. Green -- locations of surface bound amelogenins. Scale bar = 20 μm. Bars on the bar graph are standard deviations (SD).
rp(H)M180 and LAMP-1 Cell Surface Binding Pattern Comparison
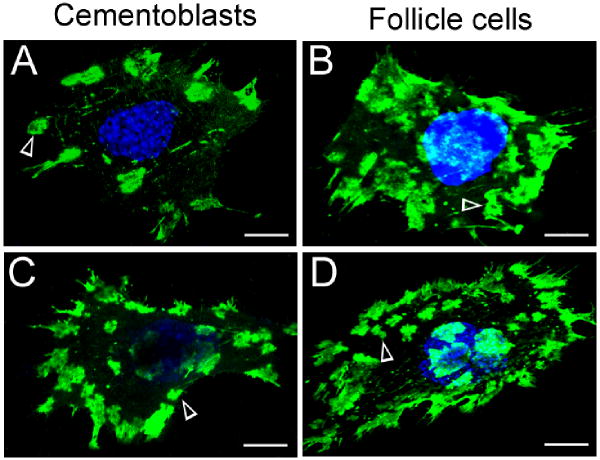
After demonstrating the specificity of rp(H)M180 binding, we determined whether rp(H)M180 and LAMP-1 had similar cell surface binding patterns using both OCCM-30 and dental follicle cells. Similar to above, OCCM-30 cells treated with 50 μg/ml rp(H)M180 for 2 hours had punctate areas of rp(H)M180 binding, which were mostly located at the periphery of the cell (Fig. 3A). Dental follicle cells treated under the same conditions exhibited some differences in rp(H)M180 cell surface binding patterns. The extent of cell surface protein binding was greater, with punctate areas seen more widely distributed across the cell surface (Fig. 3B). We then examined the distribution of LAMP-1 protein on the cell surface of OCCM-30 and dental follicle cells. Confocal microscopic examination of individual OCCM-30 and dental follicle cells demonstrated similar LAMP-1 antibody binding patterns to that observed for rp(H)M180. LAMP-1 binding was observed on the surface of OCCM-30 cells in focal areas located primarily along the cell periphery (Fig. 3C) and appears similar to the binding pattern of rp(H)M180 (Fig. 3A). Like rp(H)M180 (Fig. 3B), LAMP-1 on dental follicle cells was more widely distributed (Fig. 3D) than on cementoblasts (Fig. 3C). Thus, we observed that rp(H)M180 and LAMP-1 bound the cell surface of OCCM cells and dental follicle cells in a similar pattern, but this pattern was unique to the cell type.
Figure 3.
Immunofluorescent localization of amelogenin binding sites and LAMP-1 on the surface of OCCM-30 and dental follicle cells.
Panels A and B: Cells were incubated with rp(H)M180 (50 μg/ml) at 4°C for 2 hours followed by immunofluorescent (anti-6-His) microscopy with the TSA assay and TO-PRO staining.
Panels C and D: Cells were incubated with anti-LAMP-1 at 4°C for 2 hours followed by immunofluorescent microscopy with the TSA assay and TO-PRO staining.
Blue -- nuclei. Green -- locations of rp(H)M180 (A, B) or LAMP-1 (C, D). Arrow heads -- cell surface patch domains. Scale bar = 8 μm.
rp(H)M180 and LAMP-1 Immuno-co-localization Assay
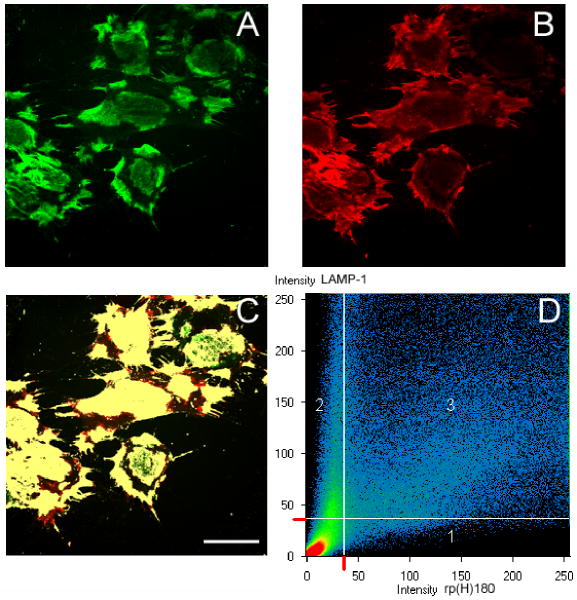
The above assay demonstrated similarities in cell surface binding patterns shared by rp(H)M180 and LAMP-1. In additional investigations, we determined whether these two proteins co-localized on the cell surface. Since both cell types exhibited a similar pattern of binding, only OCCM-30 cells were used for these studies. OCCM-30 cells were incubated simultaneously with both 50 μg/ml rp(H)M180 and LAMP-1 antibody for 2 hours and observed by confocal microscopy. The binding patterns for rp(H)M180, stained in green (Fig. 4A, green) and LAMP-1, seen in red (Fig. 4B, red) were noted as previously, i.e. rp(H)M180 exhibited punctuate areas while LAMP-1 stain exhibited areas of more focal staining. By superimposing the rp(H)M180 and LAMP-1 images, the areas that were labeled both were identified by the software and given a different color (yellow, Fig. 4C), indicating co-localization of rp(H)M180 and LAMP-1 protein on the OCCM cell surface (Fig. 4C). A scattergram showing the distribution of rp(H)180 only (region 1) and LAMP-1 only (region 2) compared to co-localized pixels, i.e. pixels that are both red and green (region 3), illustrates the high degree of co-localization of LAMP-1 and rp(H)M180. Furthermore, a co-localization coefficient for rp(H)M180 and LAMP-1 was calculated using the Zeiss LSM software. The co-localization coefficient is the relative number of co-localizing pixels for rp(H)M180 as compared to the number of pixels above the threshold which was set at 37. A value of 0 is indicative of no co-localization while a value of 1 means all pixels co-localize. In our study, the co-localization coefficient for rp(H)M180 with LAMP-1 was 0.92, showing that cell bound rp(H)M180 is found at sites that contain LAMP-1. Therefore, dental mesenchyme cells used in this study, like myoblasts used previously,44 may use LAMP-1 for amelogenin binding.
Figure 4.
Immuno-co-localization of surface bound amelogenin and LAMP-1 on the cell surface (OCCM-30). rp(H)M180 (50 μg/ml) and anti-LAMP-1 were incubated with OCCM-30 cells for 2 hours at 4°C. Anti-6-His was applied followed by fluorescent microscopy with the TSA assay and TO-PRO staining. All panels are showing the same field. Scale bar = 40 μm.
A: locations of surface bound rp(H)M180 (green).
B: locations of cell surface LAMP-1 (red).
C: Co-localization of both surface bound rp(H)M180 and LAMP-1 (yellow).
D: Co-localization scattergram demonstrating overlapping of the labeling. Area 1: labeling of rp(H)M180 alone. Area 2: labeling of LAMP-1 alone. Area 3: co-localization of both rp(H)M180 and LAMP-1.
Discussion
Our strategy was to determine if a previously identified receptor for amelogenins on mouse myoblasts and ameloblast-like cells,44, 48 LAMP-1, was associated with tooth root/periodontium associated cells (cementoblasts and dental follicle cells). We found that rp(H)M180 bound to both OCCM-30 and dental follicle cells in a saturable manner, and that this binding was specific as the rp(H)M180 binding could be competitively antagonized by EMD at 20-fold excess concentration. We also determined that the cell surface localization patterns of rp(H)M180 and LAMP-1 protein were similar in OCCM and dental follicle cells, and that these proteins were co-localized in the OCCM double labeling experiments. These results shed insights into a possible pathways by which amelogenins may mediate biological responses during development and regeneration of periodontal tissues.33 In in vitro experiments using murine ameloblast-like LS8 cells LRAP was able to regulate the activity of inducible and endothelial nitric oxide synthase, thus engaging the nitric oxide signaling pathway.49 Full length recombinant amelogenin has been demonstrated to stimulate the WNT signaling pathway in murine osteoblasts and human PDL cells, while LRAP is reported to induce canonical WNT signaling, with subsequent expression of WNT antagonists and inhibitors in mouse embryonic stem cells.40, 50 The connection linking amelogenin binding with LAMP1 membrane protein to these observed signaling pathways is an avenue for future research.
In previous studies, we showed that treatment of OCCM-30 cells with rp(H)M180 resulted in a decrease in Bsp transcripts, an increase in Opn transcripts and an inhibition of mineralized nodule formation.28 Amelogenin null mice were analyzed for Bsp expression at both the mRNA and protein levels with reductions in cementoblasts and surrounding osteoblasts noted in both cases.28 OCCM-30 cells cultured with LRAP in vitro had a decrease in Ocn mRNA levels, with increases in both Opn and osteoprotegerin (Opg) gene mRNA expression.38 These changes in gene expression appeared to be modulated via the MAPK signaling pathway.38 LRAP/M59/[A-4] was used in experiments with mouse C2C12 myoblasts to identify LAMP-1 as an amelogenin cell surface receptor.44 This finding was supported by evidence from experiments utilizing an ameloblast yeast 2 hybrid system indicating that amelogenin bound to LAMP-1 as well as LAMP-3 (CD63).51 The specific binding of rp(H)M180 to both OCCM-30 and dental follicle cells and co-localization with LAMP-1 at the cell surface presented here, indicate that rp(H)M180 could exert its signaling effect, at least in part, through the cell surface receptor LAMP-1. LAMP-1 was initially identified as a transmembrane protein playing a structural role in endosomal and lysosomal membranes.52 LAMP-1 has also been detected on the plasma membrane.53 Interactions between amelogenins and LAMP-1 were demonstrated in separate reports using confocal microscopy in which amelogenin isoforms rp(H)M180 or M59 were colocalized with LAMP-1 at the cell surface of LS8 ameloblast-like cells.48, 49 Our results confirm this interaction more precisely as shown in Fig. 4E and extend our understanding to include mesenchymal cells involved in tooth root and cementum formation.
To understand the nature of the interaction of amelogenin and LAMP-1, the structure of amelogenin needs to be considered. Efforts to determine the exact structure of the amelogenin protein via crystallographic methods have been unsuccessful.54 However, investigation into the native structure of amelogenin has shown that the high content of the amino acid proline contributes to its previously observed unstructured character, and has led to the suggestion that amelogenin belongs to a class of proteins known as intrinsically disordered proteins (IDPs),55-57 which do not have a folded structure under physiological conditions, and are enriched in prolines.58, 59 Another feature of IDPs is they can have multiple binding partners or functions and can be involved in signaling pathways.60 While highly speculative, it is intriguing to propose that the lack of innate global secondary structure of amelogenin may account for its observed roles in both enamel biomineralization and cell signaling. Interestingly, several of the SIBLING proteins involved in biomineralization of dentin and bone, bone sialoprotein, osteopontin and dentin matrix protein-1 fall into the flexible IDP class.59, 61, 62 It is known that IDPs often fold upon binding to their targets and that this binding can develop in a stepwise fashion referred to as fly-casting based on their flexible nature.60, 63, 64 Therefore, it is possible that LAMP-1 functions by binding and maintaining amelogenin in a more structured form in which amelogenin could then bind correctly to a receptor, yet to be identified, which initiates an intracellular signaling cascade.
The amelogenin primary transcript is alternatively spliced, with both full length and M59 amelogenin isoforms sharing exon 6D.26 Using the yeast 2-hyrbid system, Zou et al have identified the exon 6D amino acid region as that which binds to LAMP-1.65 This common cell binding epitope may explain why rp(H)M180 and M59 had similar biological effects on OCCM and dental follicle cells in culture, including an increase in Opn gene expression, and a decrease in Ocn gene expression.28, 38
Here we report that 180 amino acid recombinant mouse amelogenin was able to bind OCCM-30 cells and dental follicle cells in a saturable manner, and that this binding was specific. We also demonstrated that the cell surface localization pattern of rp(H)M180 was similar to that of the reported amelogenin receptor LAMP-1, and that these two proteins co-localize. This extends the understanding of how amelogenin proteins bind to target cells to include two mesenchymal cell types directly involved in tooth root and cementum formation, and further provides avenues for future studies into the mechanism whereby EMD is able to stimulate periodontal regeneration in the clinical setting.
Acknowledgments
This work is supported by NIH/NIDCR Grant DE09532 (to MJS), DE13045 (to MLS), and DE011089 (to CWG).
Abbreviations
- rp(H)M180
180 amino acid recombinant mouse amelogenin
- LAMP-1
lysosome-associated membrane protein-1
- LRAP
leucine-rich amelogenin peptide
- TRAP
tyrosine-rich amelogenin peptide
- PDL
periodontal ligament
- E-M
epithelial-mesenchymal
- HERS
Hertwig's epithelial root sheath
- NF1C
Nuclear Factor 1C
- BSP(Bsp)
bone sialoprotein
- OCN(Ocn)
osteocalcin
- OPG(Opg)
osteoprotegerin
- Runx2
runt-related transcription factor 2
- OPN(Opn)
osteopontin
- NGS
normal goat serum
- FBS
fetal bovine serum
- HRP
horseradish peroxidase
- TSA
tyramide signal amplification
- OCCM-30
murine immortalized cementoblasts
- DMEM
Dulbecco's Modified Eagle's Medium
- PBS
phosphate buffered saline
- IDP
intrinsically disordered proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bleicher F, Couble ML, Farges JC, Couble P, Magloire H. Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 1999;18:133–43. doi: 10.1016/s0945-053x(99)00007-4. [DOI] [PubMed] [Google Scholar]
- 2.Maas R, Bei M. The genetic control of early tooth development. Crit Rev Oral Biol Med. 1997;8:4–39. doi: 10.1177/10454411970080010101. [DOI] [PubMed] [Google Scholar]
- 3.Thesleff I, Nieminen P. Tooth morphogenesis and cell differentiation. Curr Opin Cell Biol. 1996;8:844–50. doi: 10.1016/s0955-0674(96)80086-x. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Xu X, Bringas P, Hung YP, Chai Y. Smad4-shh-nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2009 doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, et al. Essential role for nfi-c/ctf transcription-replication factor in tooth root development. Mol Cell Biol. 2003;23:1075–84. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong CD, Hammarstrom L. Expression of amelin and amelogenin in epithelial root sheath remnants of fully formed rat molars. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;90:218–23. doi: 10.1067/moe.2000.107052. [DOI] [PubMed] [Google Scholar]
- 7.Fukae M, Tanabe T, Yamakoshi Y, Yamada M, Ujiie Y, Oida S. Immunoblot detection and expression of enamel proteins at the apical portion of the forming root in porcine permanent incisor tooth germs. J Bone Miner Metab. 2001;19:236–43. doi: 10.1007/s007740170026. [DOI] [PubMed] [Google Scholar]
- 8.Hammarstrom L. Enamel matrix, cementum development and regeneration. J Clin Periodontol. 1997;24:658–68. doi: 10.1111/j.1600-051x.1997.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 9.Janones DS, Massa LF, Arana-Chavez VE. Immunocytochemical examination of the presence of amelogenin during the root development of rat molars. Arch Oral Biol. 2005;50:527–32. doi: 10.1016/j.archoralbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama J, Sreenath T, Hatakeyama Y, Thyagarajan T, Shum L, Gibson CW, et al. The receptor activator of nuclear factor-kappa b ligand-mediated osteoclastogenic pathway is elevated in amelogenin-null mice. J Biol Chem. 2003;278:35743–8. doi: 10.1074/jbc.M306284200. [DOI] [PubMed] [Google Scholar]
- 11.Boyan BD, Weesner TC, Lohmann CH, Andreacchio D, Carnes DL, Dean DD, et al. Porcine fetal enamel matrix derivative enhances bone formation induced by demineralized freeze dried bone allograft in vivo. J Periodontol. 2000;71:1278–86. doi: 10.1902/jop.2000.71.8.1278. [DOI] [PubMed] [Google Scholar]
- 12.Hammarstrom L, Heijl L, Gestrelius S. Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J Clin Periodontol. 1997;24:669–77. doi: 10.1111/j.1600-051x.1997.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 13.Heden G, Wennstrom J, Lindhe J. Periodontal tissue alterations following emdogain treatment of periodontal sites with angular bone defects. A series of case reports. J Clin Periodontol. 1999;26:855–60. doi: 10.1034/j.1600-051x.1997.00855.x. [DOI] [PubMed] [Google Scholar]
- 14.Heijl L. Periodontal regeneration with enamel matrix derivative in one human experimental defect. A case report. J Clin Periodontol. 1997;24:693–6. doi: 10.1034/j.1600-051x.1997.00693.x. [DOI] [PubMed] [Google Scholar]
- 15.Heijl L, Heden G, Svardstrom G, Ostgren A. Enamel matrix derivative (emdogain) in the treatment of intrabony periodontal defects. J Clin Periodontol. 1997;24:705–14. doi: 10.1111/j.1600-051x.1997.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 16.Lekovic V, Camargo PM, Weinlaender M, Nedic M, Aleksic Z, Kenney EB. A comparison between enamel matrix proteins used alone or in combination with bovine porous bone mineral in the treatment of intrabony periodontal defects in humans. J Periodontol. 2000;71:1110–6. doi: 10.1902/jop.2000.71.7.1110. [DOI] [PubMed] [Google Scholar]
- 17.Pontoriero R, Wennstrom J, Lindhe J. The use of barrier membranes and enamel matrix proteins in the treatment of angular bone defects. A prospective controlled clinical study. J Clin Periodontol. 1999;26:833–40. doi: 10.1034/j.1600-051x.1997.00833.x. [DOI] [PubMed] [Google Scholar]
- 18.Sculean A, Reich E, Chiantella GC, Brecx M. Treatment of intrabony periodontal defects with an enamel matrix protein derivative (emdogain): A report of 32 cases. Int J Periodontics Restorative Dent. 1999;19:157–63. [PubMed] [Google Scholar]
- 19.Hakki SS, Berry JE, Somerman MJ. The effect of enamel matrix protein derivative on follicle cells in vitro. J Periodontol. 2001;72:679–87. doi: 10.1902/jop.2001.72.5.679. [DOI] [PubMed] [Google Scholar]
- 20.Tokiyasu Y, Takata T, Saygin E, Somerman M. Enamel factors regulate expression of genes associated with cementoblasts. J Periodontol. 2000;71:1829–39. doi: 10.1902/jop.2000.71.12.1829. [DOI] [PubMed] [Google Scholar]
- 21.Foster BL, Popowics TE, Fong HK, Somerman MJ. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol. 2007;78:47–126. doi: 10.1016/S0070-2153(06)78003-6. [DOI] [PubMed] [Google Scholar]
- 22.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–8. [PubMed] [Google Scholar]
- 23.Simmer JP. Alternative splicing of amelogenins. Connect Tissue Res. 1995;32:131–6. doi: 10.3109/03008209509013715. [DOI] [PubMed] [Google Scholar]
- 24.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix: Sequences of two amelogenin polypeptides. Biosci Rep. 1981;1:771–8. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 25.Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, et al. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–72. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 26.Veis A. Amelogenin gene splice products: Potential signaling molecules. Cell Mol Life Sci. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanson EC, Fong HK, Foster BL, Paine ML, Gibson CW, Snead ML, et al. Amelogenins regulate expression of genes associated with cementoblasts in vitro. Eur J Oral Sci. 2006;114 1:239–43. doi: 10.1111/j.1600-0722.2006.00321.x. discussion 54-6, 381-2. [DOI] [PubMed] [Google Scholar]
- 28.Viswanathan HL, Berry JE, Foster BL, Gibson CW, Li Y, Kulkarni AB, et al. Amelogenin: A potential regulator of cementum-associated genes. J Periodontol. 2003;74:1423–31. doi: 10.1902/jop.2003.74.10.1423. [DOI] [PubMed] [Google Scholar]
- 29.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Amelogenins. Sequence homologies in enamel-matrix proteins from three mammalian species. Biochem J. 1983;211:149–54. doi: 10.1042/bj2110149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, Slavkin HC, Snead ML. Cells from hertwig's epithelial root sheath do not transcribe amelogenin. J Periodontal Res. 1991;26:42–7. doi: 10.1111/j.1600-0765.1991.tb01624.x. [DOI] [PubMed] [Google Scholar]
- 31.Ye L, Le TQ, Zhu L, Butcher K, Schneider RA, Li W, et al. Amelogenins in human developing and mature dental pulp. J Dent Res. 2006;85:814–8. doi: 10.1177/154405910608500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeichner-David M, Oishi K, Su Z, Zakartchenko V, Chen LS, Arzate H, et al. Role of hertwig's epithelial root sheath cells in tooth root development. Dev Dyn. 2003;228:651–63. doi: 10.1002/dvdy.10404. [DOI] [PubMed] [Google Scholar]
- 33.Haze A, Taylor AL, Haegewald S, Leiser Y, Shay B, Rosenfeld E, et al. Regeneration of bone and periodontal ligament induced by recombinant amelogenin after periodontitis. J Cell Mol Med. 2009;13:1110–24. doi: 10.1111/j.1582-4934.2009.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosshardt DD, Nanci A. Hertwig's epithelial root sheath, enamel matrix proteins, and initiation of cementogenesis in porcine teeth. J Clin Periodontol. 2004;31:184–92. doi: 10.1111/j.0303-6979.2004.00473.x. [DOI] [PubMed] [Google Scholar]
- 35.Foster BL, Somerman MJ. Regenerating the periodontium: Is there a magic formula? Orthod Craniofac Res. 2005;8:285–91. doi: 10.1111/j.1601-6343.2005.00351.x. [DOI] [PubMed] [Google Scholar]
- 36.Kemoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, et al. Human dental follicle cells acquire cementoblast features under stimulation by bmp-2/-7 and enamel matrix derivatives (emd) in vitro. Cell Tissue Res. 2007;329:283–94. doi: 10.1007/s00441-007-0397-3. [DOI] [PubMed] [Google Scholar]
- 37.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–5. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 38.Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FH, Jr, et al. Leucine-rich amelogenin peptide: A candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–36. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- 39.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warotayanont R, Frenkel B, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis by activation of the wnt pathway. Biochem Biophys Res Commun. 2009;387:558–63. doi: 10.1016/j.bbrc.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise GE, Frazier-Bowers S, D'Souza RN. Cellular, molecular, and genetic determinants of tooth eruption. Crit Rev Oral Biol Med. 2002;13:323–34. doi: 10.1177/154411130201300403. [DOI] [PubMed] [Google Scholar]
- 42.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (dpscs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625–30. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–55. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 44.Tompkins K, George A, Veis A. Characterization of a mouse amelogenin [a-4]/m59 cell surface receptor. Bone. 2006;38:172–80. doi: 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Wang HJ, Tannukit S, Shapiro JL, Snead M, Paine ML. Using the yeast two-hybrid assay to discover protein partners for the leucine-rich amelogenin peptide (lrap) and for tuftelin-interacting protein 11 (tfip11) Eur J Oral Sci. 2006;114 1:276–9. doi: 10.1111/j.1600-0722.2006.00289.x. discussion 85-6, 382. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Harada H, Taniguchi A. The effects of lamp1 and lamp3 on m180 amelogenin uptake, localization and amelogenin mrna induction by amelogenin protein. J Biochem. 2008;144:531–7. doi: 10.1093/jb/mvn096. [DOI] [PubMed] [Google Scholar]
- 47.D'Errico JA, Berry JE, Ouyang H, Strayhorn CL, Windle JJ, Somerman MJ. Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol. 2000;71:63–72. doi: 10.1902/jop.2000.71.1.63. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro JL, Wen X, Okamoto CT, Wang HJ, Lyngstadaas SP, Goldberg M, et al. Cellular uptake of amelogenin, and its localization to cd63, and lamp1-positive vesicles. Cell Mol Life Sci. 2007;64:244–56. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iacob S, Veis A. Identification of the functional activity of the [a-4] amelogenin gene splice product in newborn mouse ameloblasts. Bone. 2008;42:1072–9. doi: 10.1016/j.bone.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuzawa M, Sheu TJ, Lee YJ, Chen M, Li TF, Huang CT, et al. Putative signaling action of amelogenin utilizes the wnt/beta-catenin pathway. J Periodontal Res. 2009;44:289–96. doi: 10.1111/j.1600-0765.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 51.Bartlett JD, Ganss B, Goldberg M, Moradian-Oldak J, Paine ML, Snead ML, et al. 3. Protein-protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- 52.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: Emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–45. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 53.Deng YP, Storrie B. Animal cell lysosomes rapidly exchange membrane proteins. Proc Natl Acad Sci U S A. 1988;85:3860–4. doi: 10.1073/pnas.85.11.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85:775–93. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 55.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, et al. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form (dagger) Biochemistry. 2009;48:2272–81. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93:3664–74. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lakshminarayanan R, Yoon I, Hegde BG, Fan D, Du C, Moradian-Oldak J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins. 2009;76:560–9. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–64. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–33. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 60.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of sibling proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun. 2001;280:460–5. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 62.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–56. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 65.Zou Y, Wang H, Shapiro JL, Okamoto CT, Brookes SJ, Lyngstadaas SP, et al. Determination of protein regions responsible for interactions of amelogenin with cd63 and lamp1. Biochem J. 2007;408:347–54. doi: 10.1042/BJ20070881. [DOI] [PMC free article] [PubMed] [Google Scholar]