Abstract
We identify a distinctive circular dichroism (CD) signature for self-assembled 14-helical β-peptides. Our data show that self-assembly leads to a mimimum at 205 nm, which is distinct from the well-known minimum at 214 nm for a monomeric 14-helix. The onset of assembly is indicated by [θ]205/[θ]214 > 0.7. Our results will facilitate rapid screening for self-assembling β-peptides and raise the possibility that far-UV CD will be useful for detecting higher-order structure for other well-folded oligoamide backbones.
The development of unnatural oligomers that can adopt specific folding patterns (“foldamers”)1 represents a profound challenge in molecular design. Considerable success has been achieved at the level of secondary structures, especially for helices, and attention has begun to turn toward tertiary structures.2 A helix-bundle tertiary structure, in which two or more helices associate with their long axes approximately aligned, is common among globular proteins and constitutes an attractive goal for foldamer-based efforts. Folding in water can be driven by burial of stripes of hydrophobic side chains that are projected from one side of each helix.3 The design of α-peptide sequences that adopt globally amphiphilic α-helical conformations and assemble to form helix-bundle quaternary structure has proven to be a useful step toward de novo protein (i.e. tertiary structure) design,4 and several groups have sought to recapitulate this approach with foldamers.2 We reported the first example of β-peptide helix-bundle quaternary structure formation in aqueous solution.2a Subsequent studies by the groups of DeGrado2b and Schepartz2c–g have advanced this field. Each of these efforts has involved the β-peptide 14-helix, which is defined by backbone i,i-2 C=O–H–N H-bonds (14-atom ring). Progress toward helix-bundle formation has been achieved with other foldamer backbones as well.2g–m
Here we report that formation of a helix-bundle quaternary structure by 14-helical β-peptides is accompanied by a characteristic change in the circular dichroism (CD) signature relative to a monomeric 14-helix. Discovery and optimization of α-peptides or foldamers that adopt a specific tertiary or quaternary structure are facilitated if formation of the target structure can be detected spectroscopically. CD provides a convenient method for monitoring helix-bundle quaternary assembly among α-peptides, which are typically unfolded as monomers in aqueous solution. Self-assembly leads to a dramatic concomitant increase in α-helicity that can be detected by CD and provides an indirect indication of assembly.5 In our initial studies of β-peptide quaternary structure formation, we concluded that CD was not useful for detecting self-assembly of 14-helices because the β-peptides we employed were already 14-helical in the monomeric state.2a This high level of secondary structure formation was achieved by using sequences rich in trans-2-aminocyclohexanecarboxylic acid (ACHC) residues, which strongly promote 14-helicity.6
The isolated 14-helix gives rise to a strong minimum around 214 nm, as established in many studies.1,7 Here, we show that a specific 14-helix quaternary structure is signaled by a distinctive minimum around 205 nm. This characteristic CD signature is observed whether or not the self-assembling β-peptides contain ACHC. Our findings rationalize previously unexplained features in reported CD data of β-peptides that form helix bundles.2c–g
We examined three β-peptides (1–3), which are designed to self-assemble in aqueous solution. These ACHC-rich β-peptides should adopt the 14-helical conformation, which has approximately three residues per turn. The lipophilic-lipophilic-hydrophilic triads common to 1–3 lead to segregation of the lipophilic ACHC residues on one side of the 14-helix and the hydrophilic β3-hLys residues on the other side (Figure 1). The global amphiphilicity of these 14-helices favors the assembly of 14-helices with lipophilic ACHC residues at the core. Previous work showed that 10-mer 1 begins to form discrete assemblies, in equilibrium with the monomeric state, at concentrations above ~1 mM in aqueous solution; analytical ultracentrifugation (AU) indicated a preference for either hexameric or tetrameric assembly depending upon pH.2a We expected 13-mer 2 and 16-mer 3 to have a higher propensity to self-associate than does 10-mer 1 because α-peptide self-association is known to increase as the hydrophobic surface area increases.8 Indeed, AU of 1 mM solutions of either 2 or 3 in aqueous buffer indicated that these β-peptides are both highly self-associated, while 1 mM 1 is mostly monomeric under similar conditions. β-Peptides 2 and 3 form much larger assemblies than does 1: for 1 mM solutions of 2 or 3, we estimate the average assembly to contain ~20 or ~40 molecules, respectively.9 1H NMR data support the conclusion that lengthening the globally amphiphilic 14-helix increases the degree of self-association. For 10-mer 1, as previously reported, amide proton resonances are sharp below 1 mM (100 mM acetate buffer, pH 3.8), but the resonances broaden at or above 1.4 mM which is consistent with the onset of self-association.9 In contrast, for 0.1 mM 13-mer 2 in aqueous solution, amide proton resonances are broadened beyond the point of detectability.9
Figure 1.
Sequence and helical wheel diagrams of self-assembling β-peptides 1–3 and nonassociating sequence isomer iso-1.
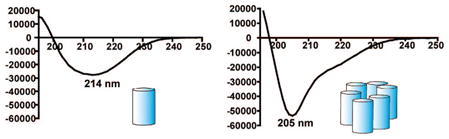
The far-UV CD spectra of 0.1 mM aqueous solutions of 2 or 310 differ considerably from the canonical 14-helical signature (Figure 2A), and these differences can be shown to arise from a high degree of self-association. Both samples display an intense minimum at 205 nm, along with a shoulder at 214 nm (isolated 14-helices give rise to a minimum at 214 nm). Comparison of CD and AU data suggests that the [θ]205/[θ]214 ratio is a useful indicator for self-association of 14-helices (Figure 2, Table 1). [θ]205/[θ]214 ≈ 0.7 for 10-mer 1 under conditions that do not support self-association according to AU (0.1 mM in aqueous Tris buffer, pH 7). A similar [θ]205/[θ]214 value is observed for iso-1, an isomeric 10-mer that has been shown via AU not to self-associate.2a Therefore, we conclude that [θ]205/[θ]214 ≈ 0.7 is the signature for a β-peptide that is 14-helical but does not form helix bundles. β-Peptide 13-mer 2 (0.1 mM) displays [θ]205/[θ]214 ≈ 0.7 in pure methanol, a solvent expected to disfavor self-association. However, 0.1 mM 2 shows [θ]205/[θ]214 ≈ 1.8 in aqueous Tris buffer, pH 7.3, conditions demonstrated by AU to promote extensive self-assembly. Under similar conditions, [θ]205/[θ]214 ≈ 2.0 for β-peptide 16-mer 3. We conclude that these large [θ]205/[θ]214 values represent a high degree of self-association. Previously reported AU data for 1.6 mM 10-mer 1 in Tris buffer, pH 8, indicated partial self-assembly,2a and this sample displays [θ]205/[θ]214 ≈ 0.95, a value between the extremes.
Figure 2.
CD spectra of 1, 2, 3, and iso-1 under various solution conditions. (A) 0.1 mM, Tris pH 7.3, (3, black10 and 2, blue), and 10–100% MeOH (2, purple to red, dashed lines); (B) iso-1, 1.6 mM, Tris pH 8.0 (black) 1, 1.6 mM MeOH (blue), 1, and 1.6 mM, Tris pH 8.0 (red).
Table 1.
[θ]205/[θ]214 Ratios of β-Peptides 1–3 and iso-1 under Various Solution Conditions
| β-peptide | solvent | [θ]205/[θ]214 |
|---|---|---|
| 1 | 1.6 mM, Tris pH 8.0 | 0.95 |
| 1 | 0.1 mM, Tris pH 7.3 | 0.75 ± 0.03 |
| iso-1 | 1.6 mM, Tris pH 8.0 | 0.62 ± 0.01 |
| 2 | 0.1 mM, Tris pH 7.3 | 1.78 ± 0.08 |
| 2 | 0.1 mM, MeOH | 0.70 ± 0.02 |
| 3 | 0.1 mM, Tris pH 7.3 | 2.04 ± 0.05 |
CD data indicate that the associated state of 13-mer 2 can be disassembled, without disruption of 14-helicity, by addition of methanol (Figure 2A). This cosolvent is known to promote helical secondary structure, relative to pure water, in both α- and β-peptides;6b,11 however, methanol disfavors tertiary and quarternary structure among proteins, because this cosolvent diminishes the drive for burial of hydrophobic surfaces.12 Figure 3 shows how [θ]205/[θ]214 changes when an increasing proportion of methanol is added to an aqueous solution of 0.1 mM 2. Small methanol proportions exert little effect, but self-assembly appears to be completely suppressed for >50 vol % methanol. The midpoint for the transition from self-assembled to nonassembled states occurs near 30 vol % methanol, which is similar to the methanol content necessary to denature small globular proteins such as α-chymotrypsinogen.12c NMR data support the conclusion that methanol prevents the self-association, because the amide resonances are sharp for 0.1 mM 2 in pure CD3OH, while, as noted above, these resonances are extensively broadened for 0.1 mM 2 in aqueous solution. Similar behavior was observed for 1 at higher concentrations.9
Figure 3.
Plot of [θ]205/[θ]214 of 2 as a function of MeOH concentration.
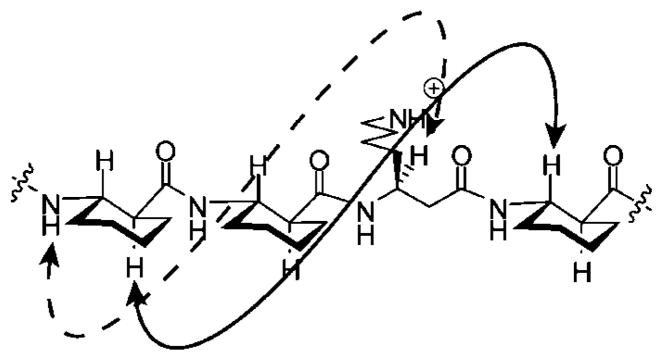
Our results show that a shift in CD minimum from 214 to 205 nm is correlated with a shift from monomeric to self-assembled states of β-peptides, but is there evidence to indicate that 14-helical folding persists upon self-assembly? Addressing this question is critical, because an alternative explanation for the CD shift is that self-assembly causes a change in β-peptide secondary structure. Indirect evidence for persistence of 14-helical folding in the self-assembled state may be found in recent work of Schepartz et al.,2d who observed a CD minimum at 205 nm for the self-associated state of a β-peptide containing exclusively β3-residues; this β-peptide crystallizes as a bundle of eight 14-helical molecules. To gain direct evidence against induction of alternative folding patterns upon self-association in solution, we conducted two-dimensional NMR studies of 10-mer 1 at 2.3 mM in aqueous buffer, conditions that allow partial self-association but that provide sufficiently resolved NMR resonances to enable conformational analysis. All backbone proton resonances of 1 could be assigned, and we could observe medium-range NOEs characteristic of the 14-helix (Figure 4, Table S2, Supporting Information). The strongest such NOEs occur between CβH(i) and CβH(i+3). Four of the seven possible NOEs of this type could be assigned for 1; the other three may have been present but were ambiguous because of resonance overlap. NH(i) → CβH(i+2) NOEs are characteristic of the 14-helix as well, although they are weaker than the CαH(i) →CβH(i+3) NOEs (Figure 4).13 Three of the seven possible NH(i) → CβH(i+2) NOEs were assigned, and two others were ambiguous. Additionally, one weak 14-helical NH(i) → CβH(i+3) was assigned. Most importantly, no NOEs inconsistent with 14-helical folding were detected, which suggests that self-association does not cause the population of a different secondary structure.
Figure 4.
Representative 14-helical NOEs: NH(i) → CβH(i+2); dashed line, CαH(i) →CβH(i+3) solid line.
Our findings indicate that self-assembly of 14-helical β-peptides causes a diagnostic change in CD signature relative to monomeric 14-helices. This change seems to be characteristic of a specific mode of 14-helix assembly, because the formation of liquid crystalline phases by 14-helical β-peptides, which involves assembly into nanofi-bers,14 does not cause the characteristic shift from 214 to 205 nm. Schepartz et al. have identified β-peptides that do not contain ACHC but that form helical bundles in aqueous solution.2d,f,g These β-peptides display a CD minimum at 205 nm in the self-assembled state, which was attributed to 14-helix formation. However, our results show that the 205 nm minimum is not reporting on secondary structure, but instead is a distinctive indicator of helix bundle self-assembly. The structural changes that give rise to this CD shift appear to be relatively subtle, since 14-helicity is maintained. Lau and Hodges made related CD observations with α-helical coiled-coils.8 The ratio of the n–π * (~220 nm) and π–π * (~209 nm) transition minimum intensities is 0.8 for a monomeric α-helix, but after α-helical coiled-coil formation [θ]220/[θ]209 = 1.0. This α-peptide precedent supports our hypothesis that the electronic transitions underlying β-peptide far-UV CD are affected in a characteristic manner by 14-helix-bundle formation.
Our results will facilitate the screening of new β-peptide sequences for the propensity to form either a quaternary or tertiary structure because CD is a more rapid technique for initial analysis than is NMR or AU. In addition, these results raise the possibility that far-UV CD will be useful for detecting higher-order structure in other oligoamide backbones that are preorganized to display a high degree of secondary structure.
Supplementary Material
Acknowledgments
This research was supported by the UW-Madison NSEC (NSF DMR-042588) and NIH grant GM56414. T.L.R.G. was supported in part by a graduate fellowship from the National Science Foundation, and J.R.L. was supported in part by a PGS B scholarship from the Natural Sciences and Engineering Research Council (NSERC) of Canada. We thank PepTech for providing Fmoc β3-amino acids at discounted prices.
Footnotes
Supporting Information Available: NMR, CD, and AU data in addition to experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]; (b) Cheng RP, Gellman SH, DeGrado WF. Chem Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; (c) Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev. 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]; (d) Goodman CM, Choi S, Shandler S, Degrado WF. Nature Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Raguse TL, Lai JR, LePlae PR, Gellman SH. Org Lett. 2001;3:3963–3936. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]; (b) Cheng RP, Degrado WF. J Am Chem Soc. 2004;124:11564–11565. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]; (c) Qiu JX, Petersson J, Matthews EE, Schepartz A. J Am Chem Soc. 2006;128:11338–11339. doi: 10.1021/ja063164+. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J Am Chem Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Goodman JL, Petersson EJ, Daniels DS, Qiu JX, Schepartz A. J Am Chem Soc. 2007;129:14746–14751. doi: 10.1021/ja0754002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Petersson EJ, Craig CJ, Daniels DS, Qiu JX, Schepartz A. J Am Chem Soc. 2007;129:5344–45. doi: 10.1021/ja070567g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Petersson EJ, Schepartz A. J Am Chem Soc. 2008;130:821–823. doi: 10.1021/ja077245x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Burkoth TS, Beausoleil E, Kaur S, Tang D, Cohen FE, Zuckermann RN. Chem Biol. 2002;9:647–654. doi: 10.1016/s1074-5521(02)00140-0. [DOI] [PubMed] [Google Scholar]; (i) Lee YC, Zuckermann RN, Dill KA. J Am Chem Soc. 2005;127:10999–11009. doi: 10.1021/ja0514904. [DOI] [PubMed] [Google Scholar]; (j) Delsuc N, Leger JM, Massip S, Huc I. Angew Chem, Int Ed. 2007;46:214–217. doi: 10.1002/anie.200603390. [DOI] [PubMed] [Google Scholar]; (k) Horne WS, Price JL, Keck JL, Gellman SH. J Am Chem Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]; (l) Price JL, Horne WS, Gellman SH. J Am Chem Soc. 2007;129:6376–6377. doi: 10.1021/ja071203r. [DOI] [PubMed] [Google Scholar]; (m) Bradford VJ, Iverson BL. J Am Chem Soc. 2008;130:1517–1524. doi: 10.1021/ja0780840. [DOI] [PubMed] [Google Scholar]
- 3.Crick FHC. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 4.(a) Woolfson DN. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]; (b) Lupas AN, Gruber M. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 5.(a) Degrado WF, Wasserman ZR, Lear JD. J Am Chem Soc. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]; (b) Degrado WF, Lear JD. J Am Chem Soc. 1985;107:7684–9. [Google Scholar]
- 6.(a) Appella DH, Barchi JJ, Jr, Durell SR, Gellman SH. J Am Chem Soc. 1999;121:2309–2310. [Google Scholar]; (b) Lee MR, Raguse TL, Schinnerl M, Pomerantz WC, Wang X, Wipf P, Gellman SH. Org Lett. 2007;9:1801–1804. doi: 10.1021/ol070511r. [DOI] [PubMed] [Google Scholar]
- 7.Kritzer JA, Tirado-Rives J, Hart SA, Lear JD, Jorgensen WL, Schepartz A. J Am Chem Soc. 2005;127:167–178. doi: 10.1021/ja0459375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SYM, Taneja AK, Hodges RS. J Biol Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 9.Please see Supporting Information.
- 10.For comparison, the molar ellipticity of 3 was normalized to the intensity of 2 at 214 nm to account for difficulties in accurately determining the concentration of 3 by UV.
- 11.(a) Conio G, Patrone E, Brighetti S. J Biol Chem. 1970;245:3335–3340. [PubMed] [Google Scholar]; (b) Gung BW, Zou D, Stalcup AM, Cottrell CE. J Org Chem. 1999;64:2176–2177. [Google Scholar]
- 12.(a) Bianchi E, Rampone R, Tealdi A, Ciferri A. J Biol Chem. 1970;245:3341–3345. [PubMed] [Google Scholar]; (b) Herskovitz TT, Gadegbeku B, Jaillet H. J Biol Chem. 1970;245:2588–98. [PubMed] [Google Scholar]; (c) Kamatari YO, Konno T, Kataoka M, Akasaka K. J Mol Biol. 1996;259:512–523. doi: 10.1006/jmbi.1996.0336. [DOI] [PubMed] [Google Scholar]
- 13.Appella DH, Barchi JJ, Jr, Durell SR, Gellman SH. J Am Chem Soc. 1999;121:2309–2310. [Google Scholar]
- 14.Pomerantz WC, Yuwono VM, Pizzey CL, Hartgerink JD, Abbott NL, Gellman SH. Angew Chem, Int Ed. 2008;47:1241–1244. doi: 10.1002/anie.200704372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.