Abstract
Purpose of Review
Diamond Blackfan anemia (DBA) is an inherited bone marrow failure syndrome characterized by erythroid failure, congenital anomalies and predisposition to cancer. Recently, the notion of DBA as a disorder of ribosome biogenesis has been clarified. Correlations between molecular underpinnings and disease pathophysiology while elusive are beginning to emerge. Advances in these areas will be explored in this review
Recent Findings
All known genes mutated in DBA encode ribosomal proteins associated with either the small (RPS) or large (RPL) subunit and in these cases ribosomal protein haploinsufficiency gives rise to the disease. The number of genes affected, their potential interactions with the environment and modifier genes, and the myriad of potential signaling pathways linking abortive ribosome synthesis to cell cycle regulators may all contribute to disease heterogeneity. Genotype/phenotype relationships emerging over the past year promise to shed light on these complex interrelationships and their role in DBA pathophysiology.
Summary
The nosology of DBA has recently expanded to include two distinct disease categories; a classical inherited bone marrow failure syndrome and a “ribosomopathy”. The description of DBA as a ribosomopathy has provided a context for scientific inquiry analogous to the description of Fanconi anemia as a disorder of DNA repair.
Keywords: Diamond Blackfan anemia (DBA), pure red cell aplasia, ribosome biogenesis, inherited bone marrow failure syndrome (IBMFS), cancer predisposition
Introduction
The ribosome once thought to be too large and too complicated for a complete understanding of its molecular underpinnings and too ubiquitous and too vital to cellular processes to be a primary factor in disease etiology, has undergone a remarkable transformation over the past decade. In the past year alone we have witnessed the Nobel Prize in Chemistry awarded to three scientists involved in deducing the three dimensional structure of the ribosome (nobelprize.org/nobel_prizes/chemistry/laureates). Furthermore there is now widespread recognition that defects in the synthesis of this extraordinary organelle are the underlying cause of the disease Diamond Blackfan anemia and also contribute to the pathophysiology of a number of other human diseases.
Diamond Blackfan anemia (DBA; MIM #105650) is one of a rare group of genetic disorders, known as the inherited bone marrow failure syndromes (IBMFS)1. These disorders have in common pro-apoptotic hematopoiesis, bone marrow failure, birth defects 2 and in the majority a predisposition to cancer 3. The study of the IBMFS has revealed significant new and in many cases unanticipated molecular events in development and cellular function. In particular DBA has revealed itself as a “ribosomopathy” (reviewed in 4).
The purpose of this review is not to provide a nearly 75 year chronology of DBA discovery from the initial description by Josephs in 1936 5 and eponymously by Diamond and Blackfan in 1938 6. We will not dwell on, but mention, the clinical aspects of DBA only as they relate to improvements in diagnosis, corticosteroid therapy, hematopoietic stem cell transplantion and the management of transfusions and transfusion acquired iron overload, all of which having been recently reviewed 7. Rather, we will concentrate on scientific advances that have moved the field over the past year. These advances are slowly connecting the dots between ribosomal protein haploinsufficiency, faulty ribosome biogenesis, diminished translational capacity, nucleolar stress, cell cycle regulatory events, and accelerated apoptosis that ultimately result in erythroid failure, birth defects and cancer predisposition.
The diagnostic criteria for classical DBA published in 1976 consist of: presentation of anemia prior to the first birthday with near normal or slightly decreased neutrophil counts, variable platelet counts, reticulocytopenia, macrocytosis and normal marrow cellularity with a paucity of red cell marrow precursors 8. These criteria have, until recently, remained the accepted standard 7. With the discovery of the first gene mutated in DBA 9 it became evident that the penetrance of autosomal dominant DBA is quite variable with regard to both hematologic and non-hematologic manifestations. Indeed, as a consequence of more recent RPS19 mutational analysis in family members of probands the estimated incidence of familial, autosomal dominant DBA has increased from approximately 10–15% to 45% 10. Thus, although the diagnostic criteria for classical DBA remain unchanged, there are numerous patients not meeting these criteria for whom a “non-classical” DBA diagnosis is appropriate 11 (Table 1). Indeed, in 2008 a consensus document developed by a cadre of international experts carefully re-defined DBA on the basis of new laboratory and clinical research 7. Consequently, a diagnosis of DBA (or if a re-appellation were ever to be considered, Diamond Blackfan syndrome) may now be suitable for example, in individuals with tri-lineage hypoplasia, little or no anemia, macrocytosis only, a presentation in adulthood, a phenotypically normal parent of an affected offspring, as well as individuals with congenital anomalies or short stature consistent with DBA and minimal or no evidence of abnormal erythropoiesis 7,11,12. In the absence of all four classical criteria (Table 1) a patient may be diagnosed as having non-classical DBA if a known DBA gene mutation is confirmed. In the absence of an identifying mutation a diagnosis of probable DBA, with variable degrees of certitude, is appropriate11. Thus, recent advances in the understanding of DBA, in part as a result of data from international Diamond Blackfan anemia registries, are resulting in more sophisticated diagnostic criteria and improvements in clinical care.
Table I.
Diagnosing Diamond Blackfan Anemia
| Classical Diagnostic criteria |
| Age less than 1 year |
| Macrocytic anemia with no other significant cytopenias |
| Reticulocytopenia |
| Normal marrow cellularity with a paucity of erythroid precursors |
| Supporting criteria |
| Major |
| Gene mutation described in “classical” DBA (currently RPS19, 24 and17 and RPL35a, 5 and 11) |
| Positive family history |
| Minor |
| Elevated erythrocyte adenosine deaminase (eADA) activity |
| Congenital anomalies described in “classical” DBA |
| Elevated fetal hemoglobin |
| No evidence of another inherited bone marrow failure syndrome |
Robust gene discovery as a consequence of modern technology and well-characterized patient databases is not without clinical pitfalls. Not all polymorphisms, or sequence changes from a reference norm, represent pathogenic mutations responsible for disease pathology. The identification of causative mutations is particularly challenging for a rare disease like DBA which typically lack extensive familial transmission that permit investigators to show the segregation of a particular polymorphism with disease state. This analysis is further complicated by the low penetrance of disease causing mutations as outlined above. Thus extreme care is still warranted in trying to establish the underlying genetic lesion in DBA except for a small number of well characterized genes. Mutational analysis for these genes is routinely provided by CLIA approved laboratories. For those patients lacking mutations in these well characterized genes the best approach to understand the underlying genetic basis for the their disease is to enter into a gene discovery research study where potential pathogenic mutations can be analyzed at a number of levels to establish their potential role in disease etiology. When included in such a study, the patient and family should be notified in a timely fashion that a strong potential candidate gene has been identified. Further studies may however be necessary before such a mutation may be used for clinical and reproductive decision making. Providing clinically useful information to the patient and family is the responsibility of the primary hematologist drawing upon available information from the research community. An understanding of the responsibility of the research scientist, both laboratory and clinical, to the patient has become quite complex and must be under constant re-evaluation.
Genetics and Pathophysiology
With the discovery of three new DBA genes over the past year 13,14, the total number of ribosomal protein genes affected in DBA was raised to six 9,15,16, accounting for approximately 50% of DBA cases. The finding that the refractory anemia associated with 5q− syndrome is linked to haploinsufficiency for a ribosomal protein gene leaves little doubt that ribosome synthesis is the target of pathogenic lesions in DBA 17. A common feature of all these studies is that reduced expression of structural proteins of the ribosome interferes with the assembly of ribosomal subunits. Recent work by Robledo and colleagues 18 demonstrates that ribosome synthesis can be affected when expression of many of the 80 human ribosomal proteins is reduced. Thus, there are many candidate ribosomal protein genes that have not as yet been analyzed which could account for DBA in patients where the affected gene is currently unknown.
These findings do not however provide insight into how disrupting the synthesis of ribosomes, a ubiquitous feature of all cell types, gives rise to the defining clinical characteristics of DBA. Animal models of DBA described over the past year promise to shed light on the tissue specific manifestations of ribosomal protein haploinsufficiency. Zebrafish models relevant to DBA have been published from three laboratories. The model closest genotypically to DBA, where one copy of 28 different ribosomal genes is inactivated by insertional mutagenesis, give rise to fish with growth impairment and in many instances a specific class of neural sheath tumors 19. The growth impairment observed in these fish is reminiscent of Drosophila minute mutations which also fall in ribosomal protein genes 20 and could potentially phenocopy the poor growth characteristics of DBA patients. The lack of a hematological phenotype in this zebrafish model limits its utility in understanding the erythroid failure in DBA. In contrast, zebrafish models where ribosomal protein expression is reduced by antisense oligonucleotides have elicited hematological phenotypes in addition to other developmental abnormalities 21,22. Although these models are limited by the transient nature of the way gene expression is reduced and the extent to which the expression of ribosomal proteins expression is reduced, they have led to the important observation that the aberrant phenotypes observed can be rescued by mutations in p53. A similar result was reported for a recent mouse model for DBA which arose unexpectedly from a screen for genes affecting skin pigmentation in mice 23. RPS19 and RPS20 were both identified as genes that when mutated give rise to a dark skin phenotype in mice. More importantly from the DBA perspective is that these mice also exhibit a mild erythroid hypoplasia. Both phenotypes reported for these mice were rescued by mutations in p53 23.
Supplied with better investigative tools there are two testable mechanisms we can reasonably hypothesize to describe the DBA phenotype. It seems quite logical that ribosomal protein haploinsufficiency will affect protein synthesis through a global translational defect, but it remains to be determined if there are selective effects on the synthesis of one or more critical proteins required for erythroid development 24 (Fig. 1). Supporting this viewpoint, a remarkable treatise published by Lodish provided evidence that reductions in general components of the translational machinery could have selective effects on the translation of specific mRNAs 25. One such candidate mRNA relevant to DBA encodes the feline leukemia, subgroup C, receptor (FLVCR) whose loss of function in mice gives rise to many clinical features observed in DBA patients 26. FLVCR is responsible for the export of toxic free heme from developing red cells and FLVCR null mice have reduced erythropoiesis as well as limb and craniofacial anomalies. The search for mutations in the FLVCR genes in DBA patients has so far proved negative 27, suggesting that affects on FLVCR expression in DBA may be more indirect. In this regard, Rey et al. 28 have recently shown that splicing of FLVCR transcripts is affected in cells derived from the bone marrow of DBA patients with mutations in RPS19. One explanation for this aberrant splicing is that alterations in translation brought about by RPS19 haploinsufficiency affect the expression of one or more factors that regulate FLVCR splicing.
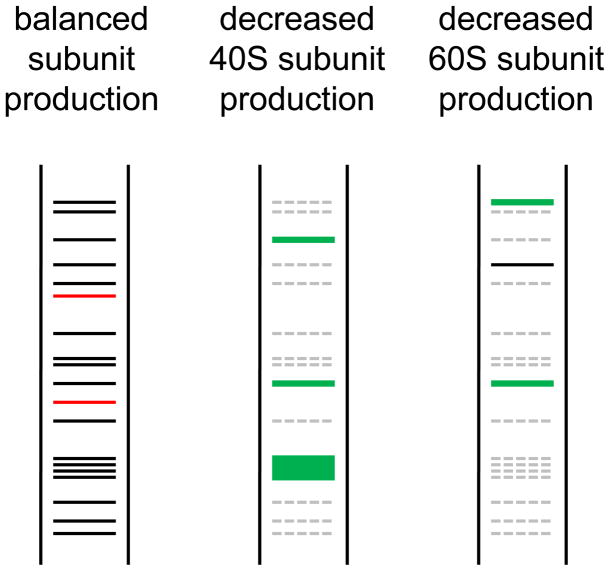
Figure 1. Hypothetical features of the translational readout hypothesis for DBA pathophysiology.
Left panel, proteins translated when subunit synthesis is balanced. Proteins labeled in red show a severe reduction in synthesis under conditions of increased mRNA competition for translation when either subunit level is decreased (middle and right panel). Middle panel, decreased 40S production. The general reduction in protein synthesis brought about by reduced amounts of 40S subunits is shown by dashed grey lines. Translation of some mRNAs (TOP mRNAs) is specifically enhanced when 40S subunit production is reduced. Proteins in red in left panel are severely reduced in amount when 40S subunits are depleted. Right panel, decreased 60S subunit production. It is expected that the translation of certain mRNA may be preferentially affected by the subunit affected. Here certain proteins with enhanced expression when 40S subunits are depleted show reduced expression when 60S subunits are depleted. Other proteins may exhibit enhanced expression when 60S subunits are depleted relative to 40S subunit depletion. Finally, mRNAs for proteins shown in red in the left panel may be preferentially affected by reduced amounts 60S as well as 40S subunits. These proteins could be of interest as critical factors responsible for generic features of DBA common to haploinsufficiency for proteins of either ribosomal subunit.
A proviso to this “translational readout hypothesis” is that the critical protein or proteins whose translation is affected would have to be affected by reduced levels of either ribosomal subunit as genes for proteins of both subunits have now been identified in DBA. This stipulation, if true, may guide proteomic approaches to identify these proteins. A more global effect of ribosomal protein haploinsufficiency on translation may also explain other aspects of DBA pathophysiology. The provocative anecdote that DBA patients accumulate tissue iron disproportionately to their transfusion burden when compared to equally transfused patients with thalassemia has been buttressed with more data29. This observation is counter-intuitive as thalassemia is characterized by markedly ineffective erythropoiesis, not a feature of DBA, suggesting a defect in a specific and as yet undermined protein critical to iron transport in DBA.
However the affect of ribosomal protein haploinsufficiency on translation cannot readily account for the observation that phenotypes reported for zebrafish and mouse models of DBA can be partially or fully rescued by mutations in p53, a finding which strongly suggests that p53 stabilization and activation plays an important role in the pro-apoptotic phenotype of cells with ribosomal protein haploinsufficiency. Thus considerable attention has been directed toward a second theory, the “ribosomal stress hypothesis” reviewed recently by Dianzani and Loreni 4 in which disruption of ribosome assembly by ribosomal protein haploinsufficiency activates p53 or other cell cycle regulatory proteins, inducing downstream events resulting in cell cycle arrest and/or apoptosis 30. This in turn could result in the DBA phenotype of anemia, poor growth and congenital malformations. Furthermore, the relationship of the nucleolus and defective ribosome synthesis to p53-mediated apoptosis suggests a role for interdicting mutations in p53 and distal pathways in oncogenesis in DBA 31.
As mentioned, in addition to RPS19 five other ribosomal protein genes have been shown to be mutated in patients with DBA 11–14. Mutations in these genes account for approximately 50% of DBA cases. Work being done by investigators in collaboration with the NHLBI Resequencing and Genotyping Service (http://rsng.nhlbi.nih.gov/scripts/about.cfm) to sequence each of the 80 ribosomal protein genes in patients from the North American DBA registry (DBAR) will no doubt identify additional DBA genes. The identified genes published to date encode proteins of both the 40S ribosomal subunit (RPS17, RPS19, and RPS24) and the 60S subunit (RPL5, RPL11, and RPL35A). Boria et al. 32 have recently created a database for mutations in the 40S ribosomal genes listed above. This database includes both the nature of mutations linked to DBA and clinical outcomes in the patients affected. Although no genotype/phenotype correlations were made from this data set, more recent data suggest that certain proteins affected in DBA may indeed be linked to distinct phenotypes. In this regard, the majority of patients with mutations in RPL5 have cleft palates or lip whereas patients with mutations in RPS19 lack these malformations 13. These emerging relationships may be a reflection of alternative mechanisms by which different ribosomal protein deficiencies signal to cellular stress responses suggesting perhaps that both theories, operative to a greater or lesser degree, determine important phenotypic differences between genotypes.
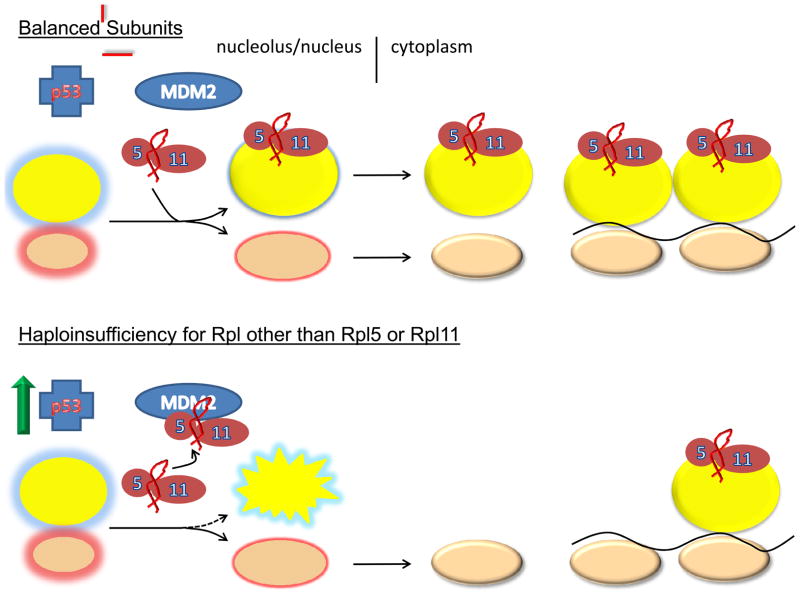
Much of the initial interest in the ribosome stress hypothesis centered on the observed interaction of certain ribosomal proteins with MDM2 (Murine Double Minute) a potent regulator of p53 levels and activity. MDM2 is a RING finger ubiquitin ligase that interacts with and promotes the degradation of p53. Three proteins of the 60S subunit, RPL5, RPL11, and RPL23 have been shown to bind to MDM2 reducing its ubiquitin ligase activity, which in turn results in p53 stabilization 33–37. Thus, a defect that interferes with the incorporation of these proteins into 60S subunits could promote their interaction with MDM2 resulting in p53 activation and downstream effects on cell cycle progression and apoptosis. Intriguingly, p53 activation as a result of deficiencies in 40S ribosomal proteins also requires Rpl11, even though the assembly of 40S and 60S subunits occurs relatively independently of one another 38. In this case, signaling through Rpl11 is somewhat more indirect through an effect mediated at the translational level. In their studies, Fumagali et al. 38 show that when 40S subunit biogenesis is disrupted TOP (Terminal OligoPyrimidine tract) mRNAs are translationally up-regulated. TOP mRNAs are known to encode components of the translational machinery including many ribosomal proteins 39. Translational up-regulation of Rpl11 mRNA presumably results in the synthesis of excess Rpl11 making it (and possibly Rpl5) available for interactions with MDM2. This pathway (Figure 2) makes the point that the “translational readout” and “ribosome stress” mechanisms are not mutually exclusive and that components of each likely contribute to the diverse clinical phenotypes observed in DBA.
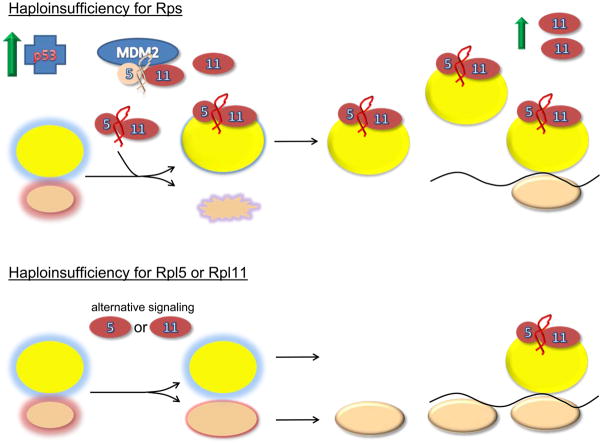
Figure 2. Models of ribosomal stress.
2a Balanced subunits, schematic represents the normal state with the balanced production of 40S and 60S subunits. Immature subunits are shown in the nucleolus/nucleus with colored glows. Mature subunits in the cytoplasm are either free or engaged in translation as polysomes on the black line representing mRNA. Under these conditions there is insufficient free ternary complexes of Rpl5, Rpl11 and 5S rRNA to bind MDM2 and block its interaction with p53. Thus, p53 levels are relatively low. 2b shows the situation when essential large subunit ribosomal proteins other than Rpl5 and Rpl11 are depleted. Under these conditions large subunit assembly is disrupted and the Rpl5/Rpl11/5S rRNA ternary complex is free to interact with MDM2. Interaction of the ternary complex with MDM2 interferes with the ability of MDM2 to interact with p53 resulting in p53 stabilization. The third panel shows the situation when essential 40S ribosomal subunit proteins are depleted. Under these conditions it is shown that TOP mRNAs including Rpl11 are upregulated. Excess Rpl11, presumably in complex with Rpl5 and 5S rRNA is then available for interaction with MDM2 resulting in p53 activation. In this manner, signaling through large subunit ribosomal components is possible even though it is the small subunit, whose assembly is disrupted. Finally, the last panel shows the situation when either Rpl5 or Rpl11 is depleted. Here accumulation of the ternary complex signaling module is lost creating a situation where alternative means of signaling ribosome stress must be employed. These alternative signaling mechanisms may be less effective and give rise to distinct and more severe clinical phenotypes.
The pathways outlined above employ Rpl11 and Rpl5 as central regulators of MDM2 function linking ribosome stress to p53 activation. If these pathways are critical elements of DBA pathophysiology one might therefore predict that mutations in these genes would not give rise to DBA. And yet, both have been shown to be DBA genes 13. This paradox is apparently resolved by the association of specific types of physical anomalies with mutations in these genes implying one or more additional pathways may participate in ribosome stress signaling to other cellular processes. In this regard, recent studies have identified ribosomal protein S7 as another interacting partner with MDM2 that mediates cellular responses to a wide range of cellular stresses 40. The view that ribosomal proteins liberated as a consequence of defects in ribosome assembly are critical signaling components of the ribosome stress hypothesis raises the specter that genes encoding many of the non-structural factors involved in ribosome assembly may also harbor pathogenic mutations in DBA making the already daunting task of gene discovery for this disease even more challenging.
Relationship of DBA to other diseases linked to defects in ribosome synthesis
The difficulties encountered in establishing genotype/phenotype relationships in DBA is further complicated when considering other human diseases linked to defects in ribosome synthesis. DBA exhibits certain phenotypic overlap with Treacher Collins Syndrome (TCS), a disease characterized by craniofacial anomalies 41. The gene affected in TCS is TCOF1, which encodes a protein involved in ribosomal RNA transcription and base modification 42. Mouse models of TCS, like their counterparts for DBA, are rescued by mutations in p53 43. Despite these similarities only a subset of DBA patients exhibit the characteristic craniofacial abnormalities observed in TCS, and individuals with TCS do not exhibit any hematological abnormalities. Understanding the molecular underpinnings linking the craniofacial anomalies observed in DBA patients with mutations in RPL5 with TCS patients with mutations in TCOF1 promise to shed light on these issues. Similarly, a recent publication compared 60S subunit biogenesis defects in yeast models of DBA and Shwachman Diamond Syndrome with an eye towards understanding how differences in the mechanisms by which ribosome synthesis is affected can give rise to distinct clinical phenotypes 44. This theme of comparing and contrasting different disease states linked to ribosome biogenesis defects was also evident in studies examining telomere length changes in different bone marrow failure syndromes 45.
Conclusions
The growing identification of non-classical cases of DBA driven by low penetrance of disease-causing mutations and the potential influence of environment factors and modifier genes strongly suggests that classical features of DBA are really only the tip of the iceberg in terms of phenotypic outcomes when ribosome synthesis is disrupted. The finding that 5q− syndrome may be considered as an acquired form of DBA indicates that abortive ribosome synthesis may play a much broader role in disease pathophysiology and not be restricted to rare congenital disorders. Since many of the pathways involved in signaling abortive ribosome synthesis to cell fate decisions have also been implicated in tumorigenesis, a more thorough description of their involvement in DBA and their contribution to cancer predisposition in this disorder should open up new avenues in cancer research.
Many features of DBA pathophysiology remain unanswered. In particular, the unusual sensitivity of the erythron to defects in ribosome synthesis and why primitive erythropoiesis is spared and erythroid failure is not, in most cases, a feature of fetal development. Likewise unexplained, is that as many as 20% of patients with DBA will remit from either transfusion support or steroid treatment? Finally, it remains to be seen whether mutations in genes encoding ribosomal proteins will account for all clinically defined cases of DBA or if the identification of non-ribosomal genes will help clarify some of these outstanding issues.
In addition to DBA, other hematologic disorders involving defects in ribosome assembly and or function; cartilage hair hypoplasia, Shwachman Diamond syndrome and dyskeratosis congenita have also been described (reviewed in 46) and a new nosology of human disease is emerging. Motivated by newly recognized clinical relevance the study of ribosome biosynthesis and function is attracting more interested scientists.
Our understanding of the molecular basis of DBA has also pointed the way to new potential therapies. And while p53 inhibitors would appear to pose an unacceptable risk for cancer in patients with DBA perhaps small molecules inhibitors of the signaling pathways linking abortive ribosome assembly to p53 activation could provide clinical benefit without increasing cancer risk. The recent observation that the loss of a small subunit ribosomal protein gene, RPS14, is responsible for the refractory anemia in 5q− syndrome has also raised the possibility the lenalidomide may have some potential clinical benefit in DBA 17. A recent study has shown that leucine stimulates protein synthesis in lymphoblasts derived from DBA patients 47. This study led to the only published work on the effects of leucine supplementation in a DBA patient 48. Administration of leucine correlated with the resolution of the anemia for this DBA patient. This finding suggests that an improvement in global translation may ameliorate the red cell failure in DBA. A review of the effects of leucine administration on the protein synthetic machinery has been published 49. A clinical trial has been designed to test the hypothesis that leucine administration will provide clinical benefits to DBA patients. This trial will open in 2010. Finally, preliminary work setting the stage for gene therapy trials is ongoing 50.
The study of DBA, since its first description nearly 75 years ago, has yielded extraordinary opportunities to understand the biology of hematopoiesis, oncogenesis and morphogenesis. As important, significant improvements in patient care have emerged and should continue with a greater knowledge of disease mechanism.
Acknowledgments
This work was supported by grants from the Daniella Maria Arturi Foundation, Diamond Blackfan Anemia Foundation, Pediatric Cancer Foundation, National Institutes of Health R01 HL 079571 (JML) and R01HL79583 (SRE) and the Feinstein Institute for Medical Research at the NSLIJ General Clinical Research Center M01 RR018535.
References
- 1.Young NSAB. Inherited Bone Marrow Failure Syndromes: Introduction. Philadelphia: WB Saunders Company; 1994. [Google Scholar]
- 2.Gripp KW, McDonald-McGinn DM, La Rossa D, et al. Bilateral microtia and cleft palate in cousins with Diamond-Blackfan anemia. Am J Med Genet. 2001;101:268–274. doi: 10.1002/ajmg.1329. [DOI] [PubMed] [Google Scholar]
- 3.Lipton JM, Federman N, Khabbaze Y, et al. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23:39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Dianzani I, Loreni F. Diamond-Blackfan anemia: a ribosomal puzzle. Haematologica. 2008;93:1601–1604. doi: 10.3324/haematol.2008.000513. [DOI] [PubMed] [Google Scholar]
- 5.Josephs H. Anemia of infancy and early childhood. Medicine. 1936;15:307–451. [Google Scholar]
- 6.Diamond LK, Blackfan KD. Hypoplastic Anemia. American Journal of Diseases of Children. 1938;56:464–467. [Google Scholar]
- 7**.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. This article is a consensus document from Internationally recognized experts relating to recent developments in DBA diagnosis and patient care. The concept of a non-classical presentation of DBA is diacussed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond LK, Wang WC, Alter BP. Congenital hypoplastic anemia. Adv Pediatr. 1976;22:349–378. [PubMed] [Google Scholar]
- 9.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 10.Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125:243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 11*.Anur P, Nemecek ER, Kurre P. The evolving spectrum of ‘non-classical’ Diamond-Blackfan anaemia--a case of eADA positive pancytopenia in a young adult. Br J Haematol. 2009;145:428–430. doi: 10.1111/j.1365-2141.2009.07590.x. This report emphasizes the need to recognize non-classical presentations of DBA. [DOI] [PubMed] [Google Scholar]
- 12.Kuze M, Matsubara H, Uji Y. Ocular hypertelorism and exotropia as presenting signs in Diamond-Blackfan anemia. Jpn J Ophthalmol. 2009;53:67–68. doi: 10.1007/s10384-008-0610-2. [DOI] [PubMed] [Google Scholar]
- 13**.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue M-F, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal Protein L5 and L11 Mutations Are Associated with Cleft Palate and Abnormal Thumbs in Diamond-Blackfan Anemia Patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. This study is the first to show a correlation between the gene affected in DBA and a distinct clinical presentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Farrar JE, Nater M, Caywood E, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. This study is the first to demonstrate that genes encoding large subunit, not only small subunit, ribosomal proteins are mutated in DBA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazda HT, Grabowska A, Merida-Long LB, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 17**.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. This report shows that the refractory anemia associated with 5q− syndrome is linked to haploinsufficiency for ribosomal protein S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn. 2009;238:76–85. doi: 10.1002/dvdy.21815. This study confirms and expands upon previous work by this group demonstrating an association between tumor predisposition in zebrafish and ribosomal protein haploinsufficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- 21**.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008 doi: 10.1182/blood-2008-01-132290. One of the seminal demonstrations that hematological abnormalities in animal models of DBA can be reversed by inactivating p53. [DOI] [PubMed] [Google Scholar]
- 22.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17:3204–3211. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- 23**.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. The first mammalian model of DBA with a hematological phenotype that can be rescued by mutations in p53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109:3152–3154. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- 25**.Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. A mathematical model of translational regulation. [DOI] [PubMed] [Google Scholar]
- 26**.Keel SB, Doty RT, Yang Z, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. This work provides strong evidence that reduced expression of FLVCR may be involved in DBA pathophysiology. [DOI] [PubMed] [Google Scholar]
- 27.Quigley JG, Gazda H, Yang Z, Ball S, Sieff CA, Abkowitz JL. Investigation of a putative role for FLVCR, a cytoplasmic heme exporter, in Diamond-Blackfan anemia. Blood Cells Mol Dis. 2005;35:189–192. doi: 10.1016/j.bcmd.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Rey MA, Duffy SP, Brown JK, et al. Enhanced alternative splicing of the FLVCR1 gene in Diamond Blackfan anemia disrupts FLVCR1 expression and function that are critical for erythropoiesis. Haematologica. 2008;93:1617–1626. doi: 10.3324/haematol.13359. [DOI] [PubMed] [Google Scholar]
- 29.Roggero S, Quarello P, Vinciguerra T, Longo F, Piga A, Ramenghi U. Severe iron overload in Blackfan-Diamond anemia: A case-control study. Am J Hematol. 2009 doi: 10.1002/ajh.21541. [DOI] [PubMed] [Google Scholar]
- 30.Badhai J, Frojmark AS, EJD, Schuster J, Dahl N. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbadis.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–241. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 32*.Boria I, Quarello P, Avondo F, et al. A new database for ribosomal protein genes which are mutated in Diamond-Blackfan Anemia. Hum Mutat. 2008;29:E263–270. doi: 10.1002/humu.20864. A useful database of RPS19 mutations linked to clinical phenotypes. [DOI] [PubMed] [Google Scholar]
- 33.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Fumagalli S, Di Cara A, Neb-Gulati A, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. This study shows how a defect in the assembly of 40S ribosomal subunits could signal to p53 activation through up-regulation of a large subunit ribosomal protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Y, Poyurovsky MV, Li Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon MJ. Treacher Collins syndrome. Hum Mol Genet. 1996;5(Spec No):1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- 42.Sakai D, Trainor PA. Treacher Collins syndrome: unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41:1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Jones NC, Lynn ML, Gaudenz K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. This study demonstrates certain parallels between the molecular underpinnings of DBA and TCS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore JBt, Farrar JE, Arceci RJ, Liu JM, Ellis SR. Distinct ribosome maturation defects in yeast models of Diamond Blackfan anemia and Shwachman Diamond syndrome. Haematologica. 2009 doi: 10.3324/haematol.2009.012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavesi E, Avondo F, Aspesi A, et al. Analysis of telomeres in peripheral blood cells from patients with bone marrow failure. Pediatr Blood Cancer. 2009;53:411–416. doi: 10.1002/pbc.22107. [DOI] [PubMed] [Google Scholar]
- 46.Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583–4588. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- 47.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 48*.Pospisilova D, Cmejlova J, Hak J, Adam T, Cmejla R. Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica. 2007;92:e66–67. doi: 10.3324/haematol.11498. The study reports a positive hematological outcome for a DBA patient treated with leucine supplements. [DOI] [PubMed] [Google Scholar]
- 49.Kimball SR, Jefferson LS. Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J Nutr. 2006;136:227S–231S. doi: 10.1093/jn/136.1.227S. [DOI] [PubMed] [Google Scholar]
- 50*.Flygare J, Olsson K, Richter J, Karlsson S. Gene therapy of Diamond Blackfan anemia CD34(+) cells leads to improved erythroid development and engraftment following transplantation. Exp Hematol. 2008;36:1428–1435. doi: 10.1016/j.exphem.2008.06.012. Recent developments in gene therapy for DBA. [DOI] [PubMed] [Google Scholar]