Abstract
The MLL (Mixed Lineage Leukemia) proto-oncogene encodes a histone methyltransferase that creates the methylated histone H3K4 epigenetic marks, commonly associated with actively transcribed genes. In addition to its canonical histone methyltransferase SET domain, the MLL protein contains three plant homeodomain (PHD) fingers that are well conserved between species but whose potential roles and requirements for MLL function are unknown. Here we demonstrate that the third PHD domain of MLL (PHD3) binds histone H3 trimethylated at lysine 4 (H3K4me3) with high affinity and specificity, and H3K4me2 with eight-fold lower affinity. Biochemical and structural analysis using NMR and fluorescence spectroscopy identified key amino acids essential for the interaction with H3K4me3. Site-directed mutations of the residues involved in recognition of H3K4me3 compromised in vitro H3K4me3 binding but not in vivo localization of full-length MLL to chromatin sites in target promoters of MEIS1 and HOXA genes. Whereas intact PHD3 finger was necessary for MLL occupancy at these promoters, H3K4me3 binding was critical for MLL transcriptional activity. These results demonstrate that MLL occupancy and target gene activation can be functionally separated. Furthermore, these findings reveal that MLL not only “writes” the H3K4me3 mark, but also binds the mark as well, and this binding is required for the transcriptional maintenance functions of MLL.
Keywords: MLL, PHD finger, histone, chromatin, methylation
INTRODUCTION
Distinct histone modifications (e.g. methylation, acetylation) are crucial for epigenetic mechanisms associated with regulating gene expression 1. Overproduction or mutations of key enzymes that create histone marks are frequent in neoplasia, yet relatively little is known of the molecular mechanisms linking aberrant chromatin dynamics to neoplastic diseases.
The MLL proto-oncogene encodes a histone methyltransferase (reviewed in 2) that is involved in chromosomal translocations and tandem duplications associated with leukemogenesis. The MLL gene encodes a large protein of 430 kDa that is the human homolog of Drosophila trithorax (TRX) (reviewed in 3), a founding member of a group of transcriptional regulators that positively maintain expression of homeobox (HOX) transcription factor genes and contribute to cellular memory. The highest similarity between MLL and TRX occurs in the SET domain and three PHD finger domains 4. The SET domain contains a catalytic site responsible for methylating histone H3K4 within chromatin of target genes, including MEIS1 and the HOX gene cluster whereas functions mediated by the MLL PHD domains are unclear.
PHD modules exist throughout the eukaryotic proteome, and are often found in chromatin-associated proteins 5. The PHD domain coordinates two zinc ions bound in a cross-braced topology, and is structurally related to FYVE and RING finger modules (reviewed in 6). Accumulating evidence shows that PHD fingers mediate physiological functions via associations with chromatin. Mutations of PHD domains in numerous proteins are associated with carcinogenesis as well as pathogenesis of immune and genetic disorders 6; 7. Due to the strong conservation between TRX and MLL PHD domains, it is likely that these modules serve crucial MLL-dependent functions 4. Of the three MLL PHD domains, only one (abbreviated MLL PHD3) displays significant sequence similarity with PHD domains that recognize transcriptionally active histone marks (e.g H3K4me2/3, H3K36me3) 8; 9; 10; 11; 12. Here we show that MLL PHD3 selectively associates with trimethylated histone H3K4, which is crucial for expression at a set of MLL target genes. Interestingly, MLL occupancy on select gene loci does not depend on H3K4me3 recognition but requires the presence of structurally folded PHD3. Taken together, these results suggest that MLL-specific recognition of the H3K4me3 mark is not sufficient for localization of MLL at target chromatin sites, however it is necessary for MLL-mediated maintenance of gene expression.
RESULTS AND DISCUSSION
The ability of MLL PHD3 to bind modified histone peptides was examined by pull-down experiments and peptide microarray analyses (Fig. 1). The GST-PHD3 fusion was incubated with biotinylated histone H3 peptides (either mono-, di- or tri-methylated at Lys 4, Lys 9, Lys 27 or Lys 36) in the presence of streptavidin sepharose beads, and the peptide-bound protein was detected using anti-GST antibodies. As shown in Fig. 1A, GST-PHD3 recognized H3K4me3 peptide and, to a lesser degree, H3K4me2. Minimal or no interaction was observed with peptides methylated at other lysine residues (Fig. 1A). Strong preference of the PHD3 finger for trimethylated Lys 4 was also seen in a microarray assay, in which GST-PHD3 and biotinylated peptides immobilized on a streptavidin coated chip were used (Fig. 1B and Suppl. Fig. 1). The MLL PHD3 finger bound to H3K4me3/me2 but did not associate with H3 and H4 peptides containing other post-translational histone modifications, including acetylation and phosphorylation marks, or methylated p53 peptides. Mono-methylation and asymmetrical di-methylation at Arg2 disrupted the MLL PHD3-H3K4me3 interaction whereas symmetrical di-methylation of Arg2 did not significantly alter the interaction. To determine whether the specificity of the PHD3 finger was preserved within intact histones, it was tested in a pull down assay using calf thymus histones. GST-PHD3 associated with intact histone H3 containing methylated marks on Lys 4 (Fig. 1C). Thus, the PHD3 finger of MLL selectively binds to the histone H3 tail poly-methylated at Lys 4.
Figure 1. The MLL PHD3 finger binds histone H3K4me3.
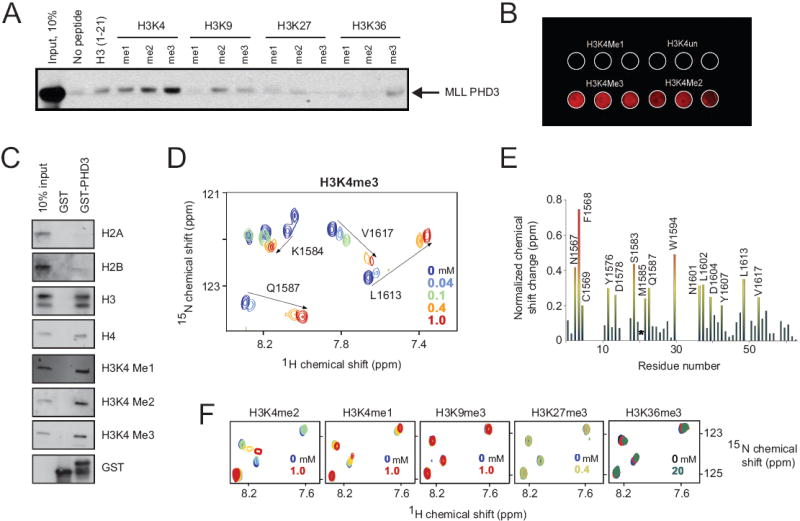
(A, B) Immuno-detection (anti-GST antibody) shows binding of GST-PHD3 to the indicated biotinylated histone peptides in a peptide pull-down assay (A) and peptide microarray (B). (C) Western blot using antibodies to the indicated epitopes shows binding of GST-PHD3 to a calf thymus histone mixture. (D, F) Shown are superimposed 1H, 15N HSQC spectra of 0.2 mM PHD3, collected while the indicated histone peptides were titrated in. The spectra are color-coded according to the peptide concentrations (inset). (E) The histogram displays normalized 1H, 15N chemical shift changes observed in backbone amides (or the side chain of W1594) in the corresponding (D) spectra of the PHD3 finger. Colored bars indicate significant change being greater than an average plus one-half standard deviation. Asterisk indicates loss of M1585 resonance.
The molecular basis of H3K4me3 recognition by MLL PHD3 was investigated by NMR spectroscopy. The sequence-specific 1H, 13C and 15N resonance assignments of PHD3 were obtained using a set of triple-resonance NMR experiments. Subsequently, histone peptide binding was characterized by collecting 1H,15N HSQC (heteronuclear single quantum coherence) spectra of the 15N-labeled protein. Substantial chemical shift changes were observed in the MLL PHD3 finger when H3K4me3 peptide was titrated in (Fig. 1D, E). Addition of a small amount of peptide caused line broadening of many NMR resonances revealing an intermediate-to-fast exchange regime on the NMR time scale and a high-affinity interaction. A large number of the amide resonances were significantly perturbed, including that of N1567, F1568, C1569, Y1576, D1578, S1583, M1585, M1586, Q1587, W1594, N1601, L1602, D1604, Y1607, L1613, and V1617, suggesting an extensive binding site (Fig. 1D and 1E) as was also seen in the case of the ING2 PHD finger 9. We note that some PHD3 residues were in conformational exchange that precluded observation of their amide resonances in NMR spectra.
Alignment of the MLL PHD3 amino acid sequence with the sequences of other H3K4me3-binding PHD fingers revealed a number of conserved residues (Fig. 2A) To determine the role of the most conserved and perturbed F1568, Y1576, M1585 and W1594 residues in binding to H3K4me3, each of these residues was substituted by alanine, and the corresponding mutant proteins were assessed by pull-down experiments, peptide microarrays and NMR spectroscopy. An additional mutant, in which the acidic residue D1592 was replaced with Ala, was generated. Among the five mutants, F1568A and D1592A retained the ability to associate with the histone H3 tail in a methylation dependent manner in peptide binding assays (Fig. 2B) and peptide microarray (Fig. 2C) although slightly weaker than the wild type protein, indicating that these residues are not essential for binding (Fig. 2D). The remaining mutants (Y1576A, M1585A, W1594A) were more severely compromised in their abilities to recognize H3K4me3. Although the Y1576A and M1585A mutants remained structured (Suppl. Fig. 2), small to negligible changes in their 1H,15N HSQC spectra, induced by the H3K4me3 peptide, indicated that binding was essentially abolished (data not shown). In contrast, the Ala substitution of W1594 caused unfolding of PHD3, impeding the ability to bind H3K4me3 and suggesting the critical role of this residue for structural stability (Suppl. Fig. 2). Taken together, these data indicate that the aromatic residues Y1576 and W1594, and the hydrophobic residue M1585 are required for high affinity interaction of the MLL PHD3 finger with H3K4me3.
Figure 2. Binding affinities of the MLL PHD3 finger.
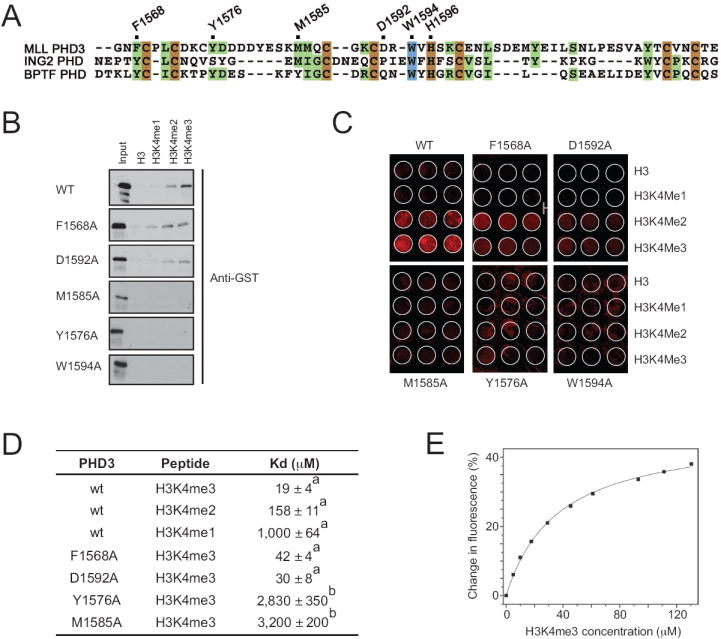
(A) Alignment of PHD domain sequences: zinc-coordinating and absolutely and moderately conserved residues are colored brown, blue and green, respectively. Residues of the MLL PHD3 finger that were mutated are marked with a black dot above the sequences and labeled. (B) Immuno-detection (anti-GST) shows binding of wt or mutant GST-PHD3 proteins indicated on the left to biotinylated histone peptides in a peptide pull-down assay. (C) Peptide microarray results are shown for binding of wt or mutant GST-PHD3 to the indicated biotinylated histone peptides. (D) The Kd values for interaction of wild type or mutant MLL PHD3 fingers with the indicated histone peptides were measured by tryptophan fluorescence (a) or NMR (b). (E) A representative binding curve used to determine Kds for wild type PHD3 by tryptophan fluorescence.
H3K4me2 and H3K4me1 peptides induced a similar pattern of chemical shift perturbations in 1H,15N HSQC spectra of the MLL PHD3 finger, suggesting that these peptides were bound in the same binding site as H3K4me3, albeit much more weakly (Fig. 1F). The affinities of the PHD3 finger for di- and mono- methylated H3K4 were eight- and fifty three-fold lower (Fig. 2D), corroborating the data obtained by pull-down experiments. Addition of histone H3 peptides methylated at Lys 9, Lys 27 or Lys 36 to the PHD3 NMR sample caused no significant chemical shift changes, demonstrating again that MLL PHD3 is unable to recognize these methylation marks and displays a clear preference for H3K4me3 (Fig. 1F).
The dissociation constant (Kd) for the interaction of MLL PHD3 with H3K4me3 was determined to be 19 μM as measured by tryptophan fluorescence (Fig. 2D, E). Comparable affinities, in the low μM range, are exhibited by other histone-binding modules including bromodomains, chromodomains, Tudor and MBT, which have been shown to be functionally relevant, suggesting that similarly the MLL PHD3-H3K4me3 interaction might be functionally significant.
To determine the potential role of MLL PHD3 recognition of H3K4me3 for MLL function, point mutations (F1568A, M1585A, D1592A, W1594A and H1596A) were engineered into full-length MLL. Following transfection into 293T cells, MLL targeting was assessed by chromatin immunoprecipitation (ChIP). Occupancy of exogenous wild type MLL was substantially enriched at the MEIS1 promoter, a physiologic MLL target, compared to the MEIS1 3’ genomic (Fig. 3) and coding (data not shown) regions (occupancy of 12 versus 4). MLL mutants F1568A and D1592A that maintained their ability to bind H3K4me3 in vitro also occupied the MEIS1 promoter. In contrast, MLL proteins harboring either the W1594A (shown to be unstructured in vitro) or H1596A (zinc coordinating His residue, mutation of which should completely disrupt the structure of PHD3) substitutions displayed virtually no binding to the MEIS1 genomic sites (occupancy of ~2-4). Furthermore, MLL containing the “loss-of-function” M1585A mutation, which essentially eliminated H3K4me3 binding in vitro, also occupied the MEIS1 promoter. A similar ChIP analysis of four distinct regions within the HOXA9 promoter showed that MLL proteins harboring the “loss-of-structure” W1594A or H1596A mutations consistently displayed no or minimal localization at the HOXA9 promoter sites in contrast to M1585A, F1568A and D1592A mutants, which occupied most HOXA9 sites. Reduced occupancy observed for the W1594A mutant likely reflected at least in part its reduced stability in vivo (Fig. 3B). These observations indicate that high affinity binding of PHD3 to H3K4me3 is not necessary for MLL association with promoter chromatin, which is likely mediated by alternative motifs (e.g. CXXC, AT hooks) implicated in MLL DNA binding, however the structural integrity of PHD3 is essential.
Figure 3. MLL PHD3-to-histone H3K4me3 association is not required for MLL occupancy on select gene loci.
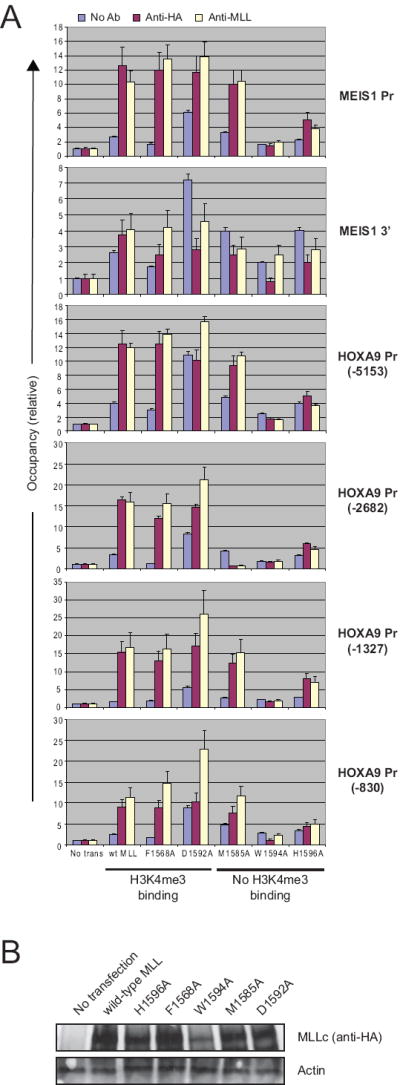
(A) Quantitative real-time PCR was used to analyze chromatin immunoprecipitation results in 293T cells expressing wild type or mutant MLL proteins. (B) MLL protein levels were analyzed by western blot analysis.
The consequences of PHD3 mutation were assessed for expression of MLL target genes using real-time PCR (Fig. 4). Exogenous wild type MLL increased expression of HOXA9, HOXA10, and MEIS1, whereas MLL proteins containing the “loss-of-structure” mutations H1596A and W1594A were unable to promote target gene expression, consistent with the effects of these mutations on chromatin localization and in vitro binding to H3K4me3. MLL containing the F1568A and D1592A mutations that bind H3K4me3 in vitro induced target gene expression similar to wild type MLL, which is consistent with their abilities to associate with chromatin in target genes. Conversely, the ability of the “loss-of-function” M1585A mutant to induce HOXA9, HOXA10 or MEIS1 expression was substantially impaired despite an ability to associate with promoter chromatin, but nevertheless consistent with an inability to bind H3K4me3 in vitro. Thus, binding of H3K4me3 by PHD3 is dispensable for localization at target chromatin sites, however it is necessary for the transcription-promoting effects of MLL.
Figure 4. MLL PHD3-to-histone H3K4me3 association is critical for MLL dependent target gene expression.
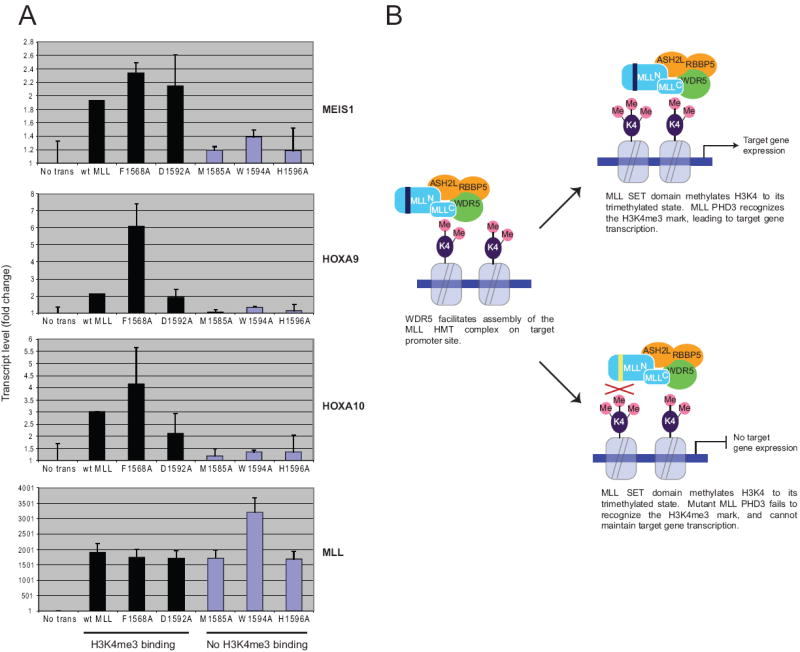
(A) Quantitative real-time RT-PCR was used to analyze gene expression levels in 293T cells expressing wild-type or mutant MLL proteins. (B) Model for MLL PHD3-mediated functions.
Our studies demonstrate that MLL not only “writes” the H3K4me3 histone code, but also “reads” it as well, which contributes to its transcriptional function. Although recognition of H3K4me3 by PHD3 is required, its specific role in the cascade of molecular events underlying transcriptional maintenance by MLL remains unclear. One possibility is that processive reading of the newly deposited H3K4me3 mark may contribute to sliding of the MLL complex along the gene to establish a broad, methylated chromatin domain. Alternatively, PHD3 may facilitate presentation or protection of the mark for competitive binding by heterologous proteins that facilitate translation of the histone code into a transcriptional output. Future studies will focus on distinguishing among these possibilities.
MATERIALS AND METHODS
Protein purification
Wild type and mutant MLL PHD3 finger (residues 1565-1627) constructs (in pGEX-2T or pGEX-6P3 vectors) were expressed in E. coli BL21(DE3) pLysS cells grown in LB, or minimal medium supplemented with 15NH4Cl or 15NH4Cl/13C-glucose. Bacteria were harvested by centrifugation after induction with IPTG (1 mM) and lysed by sonication or by French press at 4°C. The unlabeled, 15N- and 15N/13C-uniformly labeled glutathione S-transferase (GST)-fusion proteins were purified on a glutathione sepharose 4B column. GST was cleaved (using Thrombin or PreScission protease) or left for the purposes of western blot assays, in which case the GST-fusion protein was eluted off the column with 0.05 M reduced L-glutathione (Sigma Aldrich). The proteins were further purified by size exclusion chromatography and concentrated into 25 mM d11-Tris pH 6.8 buffer containing 150 mM KCl, 10 mM perdeuterated dithiothreitol (DTT) and 7% 2H2O.
Peptide binding assays
MLL PHD3 GST-fusion proteins were incubated with carboxy-terminal biotinylated peptides (synthesized at Stanford PAN facility or purchased from Upstate Biotechnology) corresponding to unmodified H3 (residues 1-21) and singly modified H3K4me1/2/3 (residues 1-21), H3K9me1/2/3 (residues 1-21), H3K27me1/2/3 (residues 21-44) and H3K36me1/2/3 (residues 21-44) histone tails in the presence of streptavidin Sepharose beads (GE Healthcare) in binding buffer [50 mM Tris (pH 7.5), 150 mM NaCl, and 0.05% Nonidet P-40]. The beads were collected via centrifugation and washed five times with peptide binding buffer. Bound protein was detected by western blot using an anti-GST HRP conjugated monoclonal antibody (GE Healthcare). GST-fusion PHD3 in the absence of peptides was run in parallel as negative control.
Calf thymus histone binding assays
Protein (2 μg) was incubated with 1-10 μg of calf thymus total histones (Worthington) in binding buffer [(50 mM Tris-HCl 7.5, 0.5 or 1M NaCl, 1% NP-40, 0.5 mM EDTA, 1 mM phenylmethyl sulphonyl fluoride (PMSF) plus protease inhibitors (Roche)] at 4 °C for 4 h, followed by an additional 1 h with glutathione beads (Amersham). Bound proteins were analyzed by SDS-PAGE and detected by western analysis.
NMR spectroscopy and sequence specific assignments
NMR experiments were performed at 298K on Varian INOVA 800, 600 and 500 MHz spectrometers using pulse field gradients to suppress artifacts and eliminate water signal. The NMR samples contained 1-2 mM unlabeled or uniformly 15N- and 15N/13C-labeled MLL PHD3. The 1H,15N heteronuclear single quantum coherence (HSQC), 15N-edited NOESY-HSQC 13; 14, HSQC-NOESY-HSQC 15, TOCSY-HSQC 16, 13C-edited NOESY-HMQC 17, HNCACB 18, CBCA(CO)NH 19, HNCO 19, C(CO)NH 20, and H(CCO)NH 20 were recorded and analyzed for sequential and spin system assignments. The NMR data were processed with nmrPipe 21 and analyzed with CCPN 22, nmrDraw and in-house software programs on Sun and Silicon Graphics workstations.
NMR titrations of histone peptides
The 1H,15N HSQC spectra of 0.2 mM uniformly 15N-labeled wild type or mutant PHD3 fingers were collected using 1024 t1 increments of 2048 data points, 112 number of increments and spectral widths of 7510 and 1367 Hz in the 1H and 15N dimensions, respectively. The binding was characterized by monitoring chemical shift changes in 1H,15N HSQC spectra of PHD3 as 12-mer histone tail peptides (synthesized by the University of Colorado Denver Peptide Core Facility) were added stepwise. The dissociation constants (KDs) were determined by a nonlinear least-squares analysis in Kaleidagraph using the equation:
where [L] is concentration of the peptide, [P] is concentration of the protein, Δδ is the observed chemical shift change, and Δδmax is the normalized chemical shift change at saturation. Normalized 23 chemical shift changes were calculated using the equation , where Δδ is the change in chemical shift in parts per million (ppm).
Fluorescence spectroscopy
Spectra were recorded at 25°C on a Fluoromax3 spectrofluorometer. The samples containing 10 μM MLL PHD3 and progressively increasing concentrations of the histone peptide were excited at 295 nm. Emission spectra were recorded between 305 and 405 nm with a 0.5 nm step size and a 1 s integration time and averaged over 3 scans. The Kd values were determined using a nonlinear least-squares analysis and the equation: ΔI=ΔImax((([L]+[P]+KD)-sqrt(sqr([L]+[P]+KD)-(4*[P]*[L])))/(2*[P])), where [L] is the concentration of the histone peptide, [P] is the concentration of MLL PHD3, ΔI is the observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states of the PHD3 finger. The Kd value was averaged over three (two for H3K4me2/me1) separate experiments, with error calculated as the standard deviation between the runs.
RT-PCR assays
293T cells were transfected with wildtype or mutant HA-tagged MLL constructs, and after two days RNA was prepared using Trizol (Invitrogen) and reverse-transcribed using First Strand Synthesis kit (Invitrogen). Quantitative RT-PCR was performed in duplicate on the ABI PRISM 7900 Sequence Detection System. HOXA9, HOXA10, MEIS1 and MLL expression was calculated following normalization to GAPDH levels by the comparative Ct (Cycle threshold) method. Taqman probes and primers used in the study are available upon request.
ChIP assays
ChIP assays were performed according to published protocols (http://www.stanford.edu/group/gozani/protocols.html). Briefly, 293T cells were transfected with wildtype or mutant HA-tagged MLL construct for two days, followed by ChIP assays with the indicated antibodies. Quantitative PCR reactions were performed with ChIP-bound and input DNA.
Supplementary Material
Table 1.
Summary of functional properties of MLL PHD3 domains.
| Construct | In vitro H3K4me3 binding | Chromatin occupancy | Transcription |
|---|---|---|---|
| WT | + | + | + |
| F1568A | +/- | + | + |
| D1592A | +/- | + | + |
| M1585A | - | + | - |
| W1594A | - | - | - |
| H1596A | - | - | - |
Acknowledgments
P-Y. C. was supported by a research fellowship from the Hope Street Kids Foundation. These studies were supported by the Children’s Health Initiative of the Packard Foundation (to M. L. C.) and grants from the National Institutes of Health (CA55029, CA116606, CA113472 and GM079641) to M. L. C. and T. G. K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.David Allis C, Danny Reinberg TJ. Marie-Laure Caparros. Epigenetics [Google Scholar]
- 2.Slany RK. When epigenetics kills: MLL fusion proteins in leukemia. Hematol Oncol. 2005;23:1–9. doi: 10.1002/hon.739. [DOI] [PubMed] [Google Scholar]
- 3.Slany RK. Chromatin control of gene expression: mixed-lineage leukemia methyltransferase SETs the stage for transcription. Proc Natl Acad Sci U S A. 2005;102:14481–2. doi: 10.1073/pnas.0507401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol. 2001;21:3589–97. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutherland HG, Mumford GK, Newton K, Ford LV, Farrall R, Dellaire G, Caceres JF, Bickmore WA. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum Mol Genet. 2001;10:1995–2011. doi: 10.1093/hmg/10.18.1995. [DOI] [PubMed] [Google Scholar]
- 6.Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–23. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–5. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuiderweg ER, Fesik SW. Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry. 1989;28:2387–91. doi: 10.1021/bi00432a008. [DOI] [PubMed] [Google Scholar]
- 14.Marion D, Kay LE, Sparks SW, Torchia DA, Bax A. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J Am Chem Soc. 1989;111:1515–7. [Google Scholar]
- 15.Zhang O, Forman-Kay JD. NMR studies of unfolded states of an SH3 domain in aqueous solution and denaturing conditions. Biochemistry. 1997;36:3959–70. doi: 10.1021/bi9627626. [DOI] [PubMed] [Google Scholar]
- 16.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989;28:6150–6. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 17.Ikura M, Kay LE, Tschudin R, Bax A. 3-Dimensional NOESY-HMQC spectroscopy of a C-13-labeled protein. J Magn Reson. 1990;86:204–9. [Google Scholar]
- 18.Wittekind M, Mueller L. HNCACB, A high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J Magn Reson. 1993;101:201–5. [Google Scholar]
- 19.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J Magn Reson. 1992;96:432–40. [Google Scholar]
- 20.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson. 1993;101:114–9. [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 23.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–61. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


