Abstract
The Notch-Delta signaling pathway enables communication between neighboring cells during development1. It plays a critical role in the formation of ‘fine-grained’ patterns, generating distinct cell fates among groups of initially equivalent neighboring cells, and in sharply delineating neighboring regions in developing tissues 2,3,4,5. The Delta ligand has been shown to have two activities: it trans-activates Notch in neighboring cells, and cis-inhibits Notch in its own cell. However, it remains unclear how Notch integrates these two activities and how the resulting system facilitates pattern formation. To address these questions, we developed a quantitative time-lapse microscopy platform for analyzing Notch-Delta signaling dynamics in individual mammalian cells. By controlling both cis- and trans-Delta levels, and monitoring the dynamics of a Notch reporter, we measured the combined cis-trans input-output relationship for the Notch-Delta system. The data revealed a striking difference between the response of Notch to trans- and cis-Delta: While the response to trans-Delta is graded, the response to cis-Delta exhibits a sharp, switch-like response at a fixed threshold, independent of trans-Delta. We developed a simple mathematical model that shows how these behaviors emerge from the mutual inactivation of Notch and Delta proteins. This interaction generates an ultrasensitive switch between mutually exclusive sending (high Delta / low Notch) and receiving (high Notch / low Delta) signaling states. At the multicellular level, this switch can amplify small differences between neighboring cells even without transcription-mediated feedback. This Notch-Delta signaling switch facilitates the formation of sharp boundaries and lateral inhibition patterns in models of development, and provides insight into previously unexplained mutant behaviors.
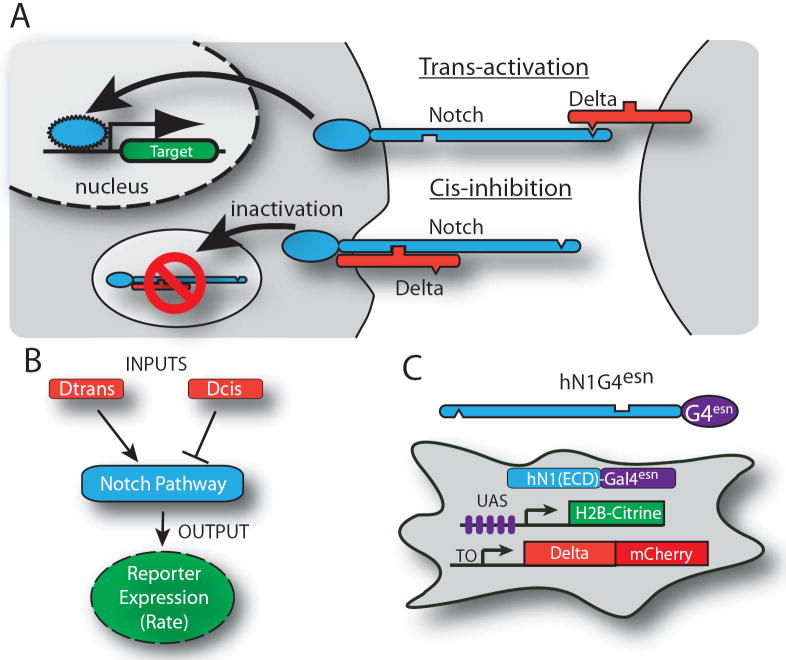
Notch and Delta are single-pass transmembrane protein families found in metazoan species. Delta in one cell can bind to, and transactivate, Notch in a neighboring cell. This interaction results in proteolytic release of the Notch intracellular domain (ICD), which translocates to the nucleus and activates target genes6 (Fig. 1A). Delta also plays a second role, inhibiting Notch activity in its own cell (cis-inhibition) 7,8,9,10. Cis-inhibition has been shown to involve direct interaction of the two proteins11, but current understanding is incomplete12.
Figure 1. System for analyzing signal integration in the Notch-Delta pathway.
(A) Notch (blue) and Delta (red) interactions are indicated schematically. (B) Notch activity integrates cis- and trans-Delta. (C) CHO-K1 cell line for analyzing Notch activity. (C) The hN1G4esn cell line stably incorporates a variant of hNotch1 in which the activator Gal4esn replaces Notch ICD. This cell line also contains genes for Histone 2B (H2B)-Citrine (YFP) reporter controlled by a UAS promoter, a Tet-inducible Delta-mCherry fusion protein, and a constitutively expressed H2B-Cerulean (CFP) for image segmentation (not shown). A similar cell line expressing full length hNotch1 (hN1 cell line) was also analyzed (Figs. S1, S2). These cells exhibit no detectable endogenous Notch or Delta activities. Notch-Delta interactions are indicated schematically and do not represent molecular interaction mechanisms11.
In order to understand how levels of cis- and trans-Delta are integrated by the Notch pathway (Fig. 1B), we constructed cell lines that allowed us to independently modulate the levels of cis- and trans-Delta, and quantitatively monitor the transcriptional response of a Notch reporter (Figs. 1C, S1, supplementary). These cell lines stably expressed Notch receptors and corresponding Citrine fluorescent protein (YFP) reporters of Notch activity (Figs. S1, S2). These cell lines also contained a doxycycline-inducible chimeric Delta(rDll1)-mCherry fusion gene (Fig. S3). In our main cell line, hN1G4esn, the intracellular domain of hNotch1 was replaced with a minimal variant of the transcriptional activator Gal4, denoted Gal4esn (ref. 13), to avoid activation of endogenous Notch targets14,15,16. A second cell line, hN1, containing the full length hNotch1 was analyzed as a control (Figure S1). Notch mRNA expression levels in these cells were comparable to those observed in early T-cell progenitors where Notch is active17 (supplementary).
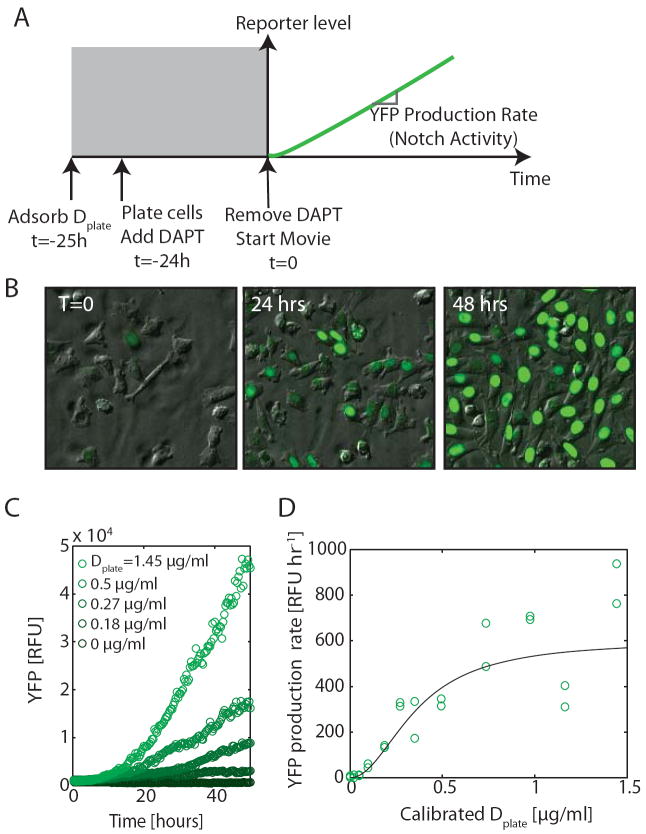
We first asked how Notch activity depends on the level of trans-Delta. We adsorbed IgG-Deltaext fusion proteins to the surface of plates at different concentrations, denoted Dplate (Fig. 2A, S4)18,19, and recorded time-lapse movies of Notch activation. Prior to the start of each movie (t<0), we inhibited Notch activation using the γ-secretase inhibitor N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT). At t=0, DAPT was washed out, allowing the fluorescent reporter to accumulate at a rate determined by Notch activity (Fig. 2B,C, Movie S1). The YFP production rate showed a graded response to Dplate well fit by Hill functions with modest Hill coefficients (Fig. 2D). A similar response was observed in the hN1 cell line (Figure S1). This graded response was not due to the use of plate-bound ligands: When cells expressing only Delta were co-cultured with cells expressing only Notch, we observed a similarly graded dependence of Notch activity on Delta expression level, but with greater variability (Fig. S5).
Figure 2. Trans-activation of Notch occurs in a graded fashion.
(A) Schematic of experimental design. The rate of increase of fluorescence (slope of green line) measures Notch activity. (B) Typical hN1G4esn filmstrip, with Dplate=1.16 μg/ml, and frame times as indicated (Movie S1, cf. Fig. S6). (C) hN1G4esn cells respond in a graded manner to varying Dplate concentrations. Curves show the median fluorescence of individual cells within a single field of view for selected Dplate levels (see Fig. S15 for distributions). (D) The relationship between Dplate and Notch activity (in Relative Fluorescence Units per hour, from the linear regime in (C)). Hill function fit is indicated by black lines, with Hill coefficient n=1.7 (95% confidence interval (CI): 0.8-2.7). Similar results were obtained using the hN1 cell line (Fig. S1). Note that doxycycline does not directly affect Notch activation or cell growth, nor does Dplate affect cell growth (Fig. S12).
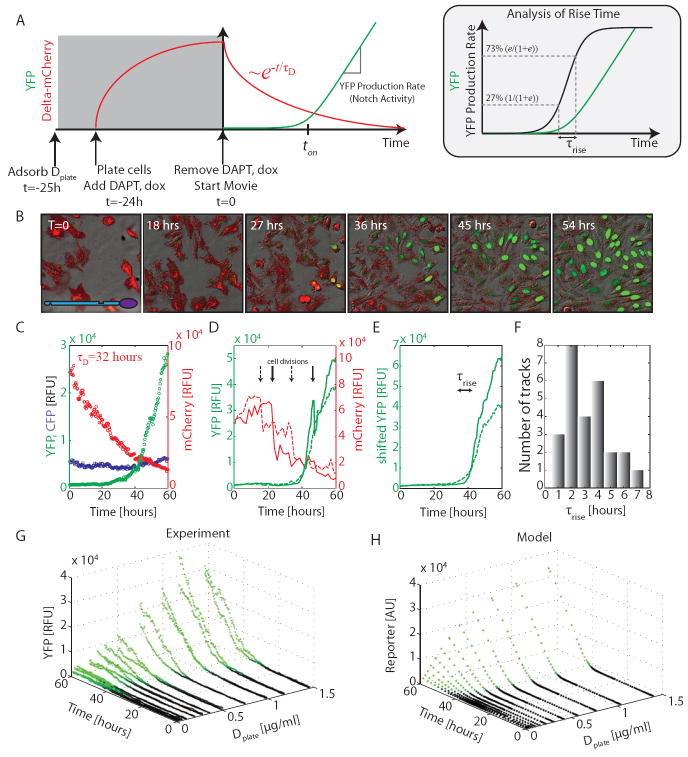
We next set out to quantify the response of Notch to varying levels of cis-Delta in the hN1G4esn cell line. We used a scheme in which Delta-mCherry was expressed in a pulse prior to the start of the movie and subsequently allowed to dilute, effectively titrating its concentration (Fig. 3A)20. These experiments were performed at low cell density, where relatively weak intercellular activation of Notch is observed (Fig. S6), and trans-activation was induced predominantly by Dplate. At the beginning of the movie Notch reporter expression was fully inhibited by high Delta-mCherry levels (Fig. 3B, Movie S2). Subsequently, Delta-mCherry levels gradually declined with a timescale of τD=32±2.5 hours, consistent with dilution by cell growth and division (Fig. 3C). At ton≈40 hours, we observed a sharp onset of reporter expression in the median response of the population (Fig 3C). Even sharper responses were evident in individual cell lineages (Fig. 3D,E,F, S13). Similar behavior was observed in the hN1 cell line (Fig. S7).
Figure 3. Cis/trans signal integration by Notch.
(A) Schematic of the experimental protocol. Inset: Rise time, τrise, is the time required for Notch activity (black line, or slope of green line) to change by a factor of e . (B) Filmstrip of hN1G4esn cells, with Dplate=1.45 μg/ml (Movie S2), showing Delta-mCherry fluorescence (red) and concomitant activation of Notch reporter (green); times as indicated (cf. Fig. S6). (C) Population average (median) response for the same movie shows a slow decay of Delta-mCherry fluorescence (red points), but a sharp response of reporter expression (green points). Constitutively expressed pCMV-H2B-Cerulean (blue) remains constant (control). Compare single-cell tracks in Fig. S13, and response to modulation of dox in Fig. S14. (D) Single-cell response for two individual cells (solid and dashed lines, colors as in C). Black arrows mark cell divisions. (E) Single cell traces in (D) replotted, but shifted up after each cell division event to ‘add back’ sister cell fluorescence, in order to show the continuity of Notch activity (see also Fig. S13). (F) Histogram of τrise from 26 non-overlapping cell lineages (Fig. S13). (G) Notch response to both cis and trans Delta. Data shown are from 2 duplicate movies acquired at each of 12 Dplate values for hN1G4esn cells. Green coloring indicates points that exceed a detection threshold. Note that turn-on (black to green transition) occurs at approximately the same time in all movies. (H) Simulations based on the model in Box 1 are qualitatively similar to data in (G) (see supplementary and Fig. S16 for model details).
To quantify the sharpness of cis-inhibition, we computed the rise time, denoted τrise, required for Notch activity to increase by a factor of e in individual cells (Figs. 3E, and 3A, inset). The distribution of τrise showed a median of 2.6 hours, considerably shorter than τD (Fig. 3F). For comparison, an equivalently sharp Hill function of cis-Delta would require a Hill coefficient .
We repeated the experiment at varying levels of Dplate, allowing us to directly measure the integrated response of Notch across the two-dimensional input space of cis- and trans-Delta levels (Fig. 3G, S14). Activation occurred at a similar ton, and therefore cis-Delta level, regardless of Dplate, as indicated by the fixed position of the transition from black to green points in Fig. 3G. In addition, the activation remained sharp at all Dplate values where it could be clearly measured.
Thus, an explanation for the observed cis and trans signal integration must simultaneously account for the three key features of the experimental data: (1) A graded response to trans-Delta (Fig. 2D), (2) A sharp response to cis-Delta (Fig. 3C-F), and (3) a fixed threshold for cis-inhibition across varying levels of trans-Delta (Fig. 3G). We show here that a simple model can explain these observations in a unified way (Box 1, Fig. 3H). The model’s key assumption is that Notch and Delta in the same cell mutually inactivate each other. As shown in Box 1, strong enough mutual inactivation can produce an ultrasensitive switch between two mutually exclusive signaling states: Cells can be in a predominantly “sending” state, with high Delta and low Notch, or a “receiving” state, with high Notch / low Delta (“receiving”), but cannot be in both states at the same time. Alternative models that do not include mutual inactivation fail to account for the observed data (Fig. S8).
Box 1: Model of mutual inactivation of Notch and Delta.
Here we describe a simple model of Notch-Delta interactions that explains the experimental data and provides insight into developmental patterning processes. The model involves several reactions: First, during intercellular signaling Notch in one cell binds to extracellular Delta, Dtrans, leading to release of the Notch intracellular domain and degradation of its extracellular domain6. Similarly, Notch in a neighboring cell, Ntrans, can bind to Delta. Second, Notch binds irreversibly to Delta in the same cell to form a stable, inactive, complex, which is effectively removed from the system12. Finally, Notch and Delta are produced at constant rates, and degraded and/or diluted at a constant rate, in addition to their removal through the interactions described above.
These reactions can be expressed as a set of ordinary differential equations for free Notch, N, and free Delta, D, in an individual cell. An additional equation represents the intracellular domain of Notch, denoted S, which activates expression of the fluorescent reporter gene:
Here, Dtrans represents Dplate in Figs 2-3, but could also represent Delta levels in one or more neighboring cells (supplementary). Similarly, D in these equations corresponds to Dcis in the experiments. βN and βD denote the production rates of Notch and Delta, respectively. The combined degradation and dilution rate, γ, is assumed for simplicity to be the same for Notch and Delta. γS is the rate of decay for S. kc and kt determine the strengths of cis-inhibition and trans-activation, respectively. See supplementary for a more detailed description.
At steady-state, mutual inactivation leads to a switch between two qualitatively distinct behaviors, depending on the relative production rates of Delta and Notch. When βD > βN, excess Delta effectively inactivates most Notch, allowing cells to send, but not to efficiently receive, signals. Conversely, when βD < βN, excess Notch effectively inactivates Delta, allowing cells to receive, but not efficiently send. Thus, the system approaches two mutually exclusive signaling states: high Delta / low Notch (“sending,” pink shading in Box Fig.), and high Notch / low Delta (“receiving,” blue shading). Note that this switch is not bistable.
At steady-state the transition between the two regimes is ultrasensitive: Near the threshold, a small relative change in βD (or βN) can lead to a much larger change in signaling (Fig. S11). Related biochemical kinetics occur in bacterial sRNA, and protein sequestration27,28,29. In Fig. 3, ultrasensitivity occurs dynamically in response to the decay of the total Delta concentration (supplementary).
The three features described above emerge naturally in this model. First, in the absence of cis-Delta, Notch activation is proportional to trans-Delta concentration, generating a graded response. Second, a sharp response to cis-Delta results from mutual inactivation, which causes an excess of either protein to strongly diminish the activity of the other. Finally, the switching point occurs when Notch and cis-Delta levels are comparable, and is therefore only weakly dependent on trans-Delta.
The mutual inactivation model predicts cis-inhibition, not just of Notch by Delta, but also of Delta by Notch. This interaction is supported by results in other systems12,21,22. We tested this prediction in our system using a transactivation assay based on co-culture of Delta-expressing “sending” cells with Notch reporter cells. Expression of Notch in the Delta-expressing cells reduced their ability to transactivate, as predicted (Fig. S9). While the exact biochemical mechanism of mutual inactivation remains unclear, we observed no sharp drop in total cellular Delta-mCherry fluorescence during switching, suggesting that the inactive complex may be stable in these conditions (Fig. 3C,D).
What implications does the signaling switch have for multicellular patterning? Consider two neighboring cells which produce Notch and Delta at constant rates (Fig. 4A). A slight excess of Notch production in one cell and a slight excess of Delta production in its neighbor can generate a strong signaling bias in one direction: the first cell becomes a receiver and the second becomes a sender. In this way, a small difference in production rates between cells is amplified into a much larger difference in Notch activity (Fig. 4B). This amplification does not require transcriptional regulation or feedback.
Figure 4. The mutual inactivation model in multicellular patterning.

(A) Signal amplification (schematic). Two interacting cells with the same amount of Notch (here, 2 molecules), but different amounts of Delta (1 or 3 molecules). Due to the cis-interaction between Notch and Delta, signaling is strongly biased to cell1. (B) Notch amplifies differences between cells. Signal amplification, defined as shown, for two interacting cells, with different Delta production rates, βD(2) = 1.35 βD(1) (see model in supplementary). The x-axis specifies the average Delta production rate, . Maximum amplification occurs when Delta production rates flank βN (vertical line). Stronger mutual inactivation (smaller kc/kt) increases signal amplification. (C-D) Sharp boundary formation in response to a gradient of Delta production. (C) Simulation of a field of interacting cells in which Delta production rates decay exponentially from the center, according to βD (x) = βD0exp (−x/x0) with x0 =7 cells (dashed red line). Notch production rate, βN, is constant (dashed blue line). Resulting free Notch and Delta protein levels are indicated (solid lines). Notch activation occurs in two sharply defined columns of cells (green line in plot and green cells in cellular diagram). (D) This model explains suppression of mutant phenotypes. Gray lines indicate positions where βN = βD (x), leading to Notch activity peaks. Simultaneous reduction of both Notch and Delta production rates by half maintains boundary positions (dotted lines) (Fig. S10). (F-G) Mutual inactivation facilitates lateral inhibition patterning, shown schematically in (F). (G) In the absence of cooperativity in regulatory feedback, a standard lateral inhibition model 24 cannot pattern (first panel) while a model of lateral inhibition with mutual inactivation can (second panel).
The send-receive signaling switch can facilitate formation of sharp boundaries. For example, in Drosophila, Notch and Delta sharply delineate wing vein boundaries 4,5. In this system, Delta production is initially expressed in a graded profile transverse to the vein. Eventually, Notch signaling is restricted to two sharp side bands on either side of the vein axis.
As a simplified model, we simulated the development of a field of cells with a graded rate of Delta production and a uniform rate of Notch production (Fig. 4C). The mutual inactivation model generated sharply defined side-bands of Notch signaling at positions where the two production rates intersect, i.e. where ‘sender’ and ‘receiver’ cells are next to each other (Fig. 4C). Moreover, this model explains a striking mutant behavior occurring in the Drosophila wing vein system. While Notch and Delta are individually haploinsufficient (causing thicker veins), the Notch+/- Delta+/- double mutant restores the wild-type phenotype23. This suppression of the single mutant phenotypes in the double mutant emerges automatically in the model because proportional rescaling of the Notch and Delta production rates does not move their intersection points (Fig. 4D). This suppression is maintained across a broad range of parameter values and persists even with additional feedbacks (Fig. S10f), but is difficult to explain in other models (Fig. S10, supplementary).
The send-receive signaling switch can also facilitate lateral inhibition patterning. When Notch transcriptionally downregulates Delta expression, the resulting intercellular positive feedback loop can generate ‘checkerboard’ patterns of Notch activity (Fig. 4E)24,25. Without mutual inactivation, pattern formation requires a minimum Hill coefficient of n > 2, or higher, in the regulatory feedback loop (Fig. 4F, left, supplementary). Although we cannot rule out such cooperativity, or additional feedback loops, no evidence for strongly cooperative trans-activation was observed here or previously (Figs. 2D, S1). In contrast, mutual inactivation enables patterning even without cooperativity, by introducing a sharp response to changes in Delta expression, (Fig. 4F, right panel). In addition, for strong enough cis-inhibition, mutual inactivation allows cells with high Delta levels to co-exist next to one another at steady-state, leading to a broader range of possible patterns (Fig. S17). Finally, we note that low levels of free Notch (Delta) exist in sender (receiver) cells at finite mutual inactivation strength (Fig. S11). The resulting signaling between like cells (senders or receivers) can play a role in lateral inhibition patterning dynamics.
Different signal transduction pathways are optimized to encode and transmit information in different ways, depending on the tasks they perform in the organism. Our results show that mutual inactivation between Notch and Delta in the same cell forces cells into predominantly sending or receiving states (Box 1 Figure). In a multicellular context, this mechanism amplifies small initial differences between neighboring cells, and facilitates pattern formation (Fig. 4). This signaling switch thus appears to optimize the Notch-Delta pathway for directional signaling, and may explain why it is employed in specific developmental processes. Moreover, this mechanism could also provide other advantages such as faster dynamics 26,27. We note that interactions between Notch and Delta are typically embedded in more complex dynamic regulatory networks that involve additional feedbacks. It will be important to explore how this signaling switch functions in the context of larger regulatory circuits.
Methods Summary
Genetic constructs and cell lines were assembled by standard methods (Table S1). All cell lines used in the main text (Table S2) were derived from T-REx-CHO-K1 (Invitrogen). Cell lines were constructed by sequential rounds of Lipofectamine 2000 (Invitrogen) transfection and selection. Stably transfected clones were isolated by limiting dilution or fluorescence-activated cell sorting (FACS). Time-lapse microscopy was performed with cells plated on 24-well glass-bottom plates (Mattek). For plate-bound Delta experiments, IgG-Deltaext was adsorbed to the plate together with 5 μg/ml hamster fibronectin (Innovative Research) prior to cell plating. Before imaging, cells were switched to a low-fluorescence medium, consisting of 5% FBS in αMEM lacking riboflavin, folic acid, phenol red, and vitamin B12. Movies were acquired using an Olympus IX-81 ZDC microscope, equipped with a 37°C environmental chamber supplying 5% CO2, a 20X 0.7 NA objective, and automated acquisition software (MetaMorph). Western blots for Gal4 were obtained using standard protocols. Blots were probed with rabbit anti-Gal4 DBD primary antibody (sc-577, Santa Cruz Biotechnology, 1:200) followed by incubation with horseradish peroxidase-labeled anti-rabbit IgG secondary antibody (Amersham, 1:2000). Bands were quantified using a VersaDoc gel imaging system. qRT-PCR was performed using standard protocols based on the RNeasy kit (Qiagen) and iScript cDNA synthesis kit (Bio-Rad). Co-culture experiments were analyzed for YFP fluorescence using a FACScalibur flow cytometer (Becton Dickinson) and standard protocols. Movies were analyzed in several stages. First, individual cell nuclei were identified on CFP images using a custom Matlab-based algorithm based on edge detection and thresholding of constitutively expressed H2B-Cerulean fluorescence. Then, for analysis of single-cell expression trajectories, individual nuclei were tracked across frames using custom software (Matlab, C) based on the SoftAssign algorithm (supplementary). All single-cell trajectories were validated manually. For further details see supplementary.
Methods
Genetic constructs
We used standard molecular biology techniques to assemble all constructs used in this paper (Table S1). The construct used to generate the hN1 cell line, pcDNA3-hN1-mCherry was constructed by fusing the coding sequence of mCherry to hN1, provided by Jon Aster7. The construct used to generate the hN1G4esn cell line, pcDNA3-hNECD-Gal4esn was constructed by replacing amino acids 1742 to 2556 of hNotch1 with the amino acids 1-147 and 768-881 of Gal4. The Delta-mCherry fusion consists of the entire coding sequence of rDll1 concatenated directly to the coding sequence of mCherry. The reporter for hNotch1 activation was constructed from the 12XCSL construct provided by Urban Lendahl6. The reporter for hNECD-Gal4esn activation was constructed from the UAS construct provided by Scott Fraser5. Both reporters used a protein fusion of H2B-Citrine in order to localize fluorescence to cell nuclei where it could be more accurately quantified. Doxycycline-inducible constructs were based on the T-REx system (Invitrogen).
Generation of stable cell lines
All cell lines used in the main text (Table S2) were based on the cell line T-REx-CHO-K1 (Invitrogen). Cells were grown in Alpha MEM Earle’s Salts (Irvine Scientific) supplemented with 10% Tet System Approved FBS (Clontech), 100 U/ml penicillin -100 ug/ml streptomycin − 0.292 mg/ml L-glutamine (Gibco), and 10 μg/ml Blasticidin (InvivoGen) at 37°C in the presence of 5% CO2 under a humidified atmosphere. Cell lines incorporating multiple transgenes were constructed by sequential rounds of Lipofectamine 2000 (Invitrogen) transfection and selection. Stably transfected clones were generated by limiting dilution or fluorescence-activated cell sorting (FACS) of single cells. hN1 and hN1G4esn cell lines were first created by stably integrating the 12xCSL-H2B-Citrine or UAS-H2B-Citrine reporters, respectively, into T-REx-CHO-K1 cells. After selection with media containing 400 μg/ml Zeocin (Invitrogen) and 10 μg/ml Blasticidin (InvivoGen), individual clones were obtained. Clones with the best dynamic range of reporter induction were identified and used in subsequent stages. pcDNA3-hN1-mCherry or pcDNA3-hNECD-Gal4esn was transfected into the 12xCSL-H2B-Citrine or UAS-H2B-Citrine reporter cell lines, respectively. Cells were selected with media containing 400 μg/ml Zeocin, 10 μg/ml Blasticidin, and 600 μg/ml Geneticin (Invitrogen). Individual clones were obtained and tested for Notch activity by plating on 2.5 ug/ml IgG-Deltaext. Clones with minimal background levels and high reporter activation when exposed to Delta were selected and transfected with a plasmid expressing Delta-mCherry under a doxycycline inducible promoter (pcDNA5-TO-Dl-mCherry). These cells were selected in media containing 400 μg/ml Zeocin, 10 μg/ml Blasticidin, 600 μg/ml Geneticin, and 500 μg/ml Hygromycin (InvivoGen). Clonal cell populations were obtained, and the clone with the lowest mCherry background expression in the absence of doxycycline, as well as good inducibility of mCherry expression when exposed to 1 μg/ml doxycycline, was selected for experiments. hN1 and hN1G4esn cell lines also contain an integrated H2B-Cerulean under constitutive CMV promoter. A separate cell line containing only inducible Delta-mCherry was created by transfecting T-REx-CHO-K1 cells with pcDNA5-TO-Dl-mCherry. Clones were generated as above, but selection media contained only Blasticidin and Hygromycin. This cell line was then used to generate the TO-DMC+hN1G4esn cell line by stably transfecting with pcDNA3-hNECD-Gal4esn construct (600 μg/ml Geneticin).
Experimental techniques and imaging protocols
Surface preparation: All time-lapse microscopy experiments were performed with cells plated on 24-well glass bottom plates (Mattek). IgG-Deltaext constructs were generously provided by Irwin Bernstein. For plate-bound Delta experiments, IgG-Deltaext was adsorbed together with 5 μg/ml hamster fibronectin (Innovative Research) to the glass plate surface by incubation for 1 hour at 4°C prior to cell plating. Cells were diluted to 2×104 cells/ml (1×105 cells/ml for coculture experiments). A calibration was performed for determining the dependence of active IgG-Deltaext concentration on the concentration added during incubation (see supplementary). Preparation of cells for imaging: Before imaging, cells were switched to a low-fluorescence medium, consisting of 5% FBS in αMEM lacking riboflavin, folic acid, phenol red, and vitamin B12. Time-lapse microscopy: Movies were acquired using an Olympus IX-81 ZDC microscope, equipped with a 37°C environmental chamber supplying 5% CO2, a 20X 0.7 NA objective, and automated acquisition software (MetaMorph). For each movie, fluorescence images were acquired in CFP, YFP, and RFP channels, as well as Differential interference contrast (DIC).
Western blot
Western blots were performed using standard protocols. For detection of Gal4, TO-Gal4esn cells were either uninduced or induced for 24 hr with 100 ng/ml of doxycycline. 4 ×106 cells were lysed with 200ul 1.5x complete SDS loading buffer at 0 hr, 1 hr, 2 hr, 4 hr and 6 hr post-doxycycline removal. 10ul of cell lysate was run in triplicate on a NuPAGE Novex 4-12% Bis-Tris Midi Gel (Invitrogen) and transferred to a 0.2 μm nitrocellulose membrane using the iBlot from Invitrogen. The blot was probed with rabbit anti-Gal4 DBD primary antibody (sc-577, Santa Cruz Biotechnology, 1:200) followed by incubation with horseradish peroxidase-labeled anti-rabbit IgG secondary antibody (Amersham, 1:2000). Bands were quantified using a VersaDoc gel imaging system.
Real-time qRT-PCR
qRT-PCR was performed using standard protocols based on the RNeasy kit (Qiagen) and iScript cDNA synthesis kit (Bio-Rad). RNA was isolated from hN1 and hN1G4esn cells. cDNA was subsequently synthesized from 1ug of RNA. From a 20ul reaction, 2ul of cDNA was used to assess Notch and β-Actin mRNA levels.
Flow cytometry analysis of co-cultures
TO-DMC or TO-DMC+ hN1G4esn cells were co-cultured with hN1G4esn-No-Delta cells. 1×105 cells were plated at a ratio of 20% Delta cells and 80% Notch reporter cells. For each set of co-cultures, a 12 hr pulse of 1.6 ng/ml and 100 ng/ml doxycycline was performed (a well with no doxycycline served as a control). 24hr after doxycycline removal, co-cultured cells were trypsinized and then analyzed for YFP fluorescence using a FACScalibur flow cytometer (Becton Dickinson) and standard protocols.
Image and data analysis
Movies were analyzed in several stages: (1) Segmentation. Individual cell nuclei were identified on CFP images using a custom Matlab-based algorithm based on edge detection and thresholding of constitutively expressed H2B-Cerulean fluorescence. (2) Tracking. For analysis of single-cell expression trajectories, individual nuclei were tracked across frames using custom software (Matlab, C) based on the SoftAssign algorithm– see supplementary materials for details. (3) Verification. All single-cell trajectories were validated using a semi-automated custom software system (Matlab). (4) More detailed analysis of the resulting data is described in supplementary material.
Supplementary Material
Acknowledgments
We would like to thank Irwin Bernstein for the Delta ligands, Urban Lendahl for the 12XCSL reporter construct, J. Aster for hNotch1 and other constructs, and Gerry Weinmaster for the rat Dll1 construct and advice. We also thank Roger Tsien and Kurt Thorn for mCherry, Sean Megason and Scott Fraser for H2B-Citrine and other constructs, Urban Lendahl for the 12xCSL constructs, Rochelle Diamond and Diana Perez for assistance with FACS, and Fred Tan and John Yong for help with cloning some of the constructs. We thank Avigdor Eldar, James Locke, Georg Seelig, Roy Kishony, Boris Shraiman, Andrew C. Oates, and members of the Elowitz lab for fruitful discussions and advice. This work was supported by NIH Fellowship F32GM77014 to D.S., the Caltech Center for Biological Circuit Design, and the Packard Foundation.
Footnotes
Author Contributions DS and MBE designed the research. DS, LAS, MEF, and GAA built cell lines and performed experiments. DS, AL, LL, JGO, and MBE performed data analysis and mathematical modeling. DS and MBE wrote the manuscript with substantial input from the other authors.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Goodyear R, Richardson G. Pattern formation in the basilar papilla: evidence for cell rearrangement. J Neurosci. 1997;17:6289–6301. doi: 10.1523/JNEUROSCI.17-16-06289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. doi:0092-8674(91)90263-X [pii] [DOI] [PubMed] [Google Scholar]
- 4.Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 5.de Celis JF, Bray S, Garcia-Bellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 6.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. doi:nrm2009 [pii] 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 7.de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 8.Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 9.Klein T, Brennan K, Arias AM. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. doi:S0012-1606(97)98564-X [pii] 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- 10.Miller AC, Lyons EL, Herman TG. cis-Inhibition of notch by endogenous delta biases the outcome of lateral inhibition. Curr Biol. 2009;19:1378–1383. doi: 10.1016/j.cub.2009.06.042. doi:S0960-9822(09)01319-0 [pii] 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordle J, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. doi:nsmb.1457 [pii] 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda M, Chitnis AB. Interaction with Notch determines endocytosis of specific Delta ligands in zebrafish neural tissue. Development. 2009;136:197–206. doi: 10.1242/dev.027938. doi:dev.027938 [pii] 10.1242/dev.027938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakidani H, Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. doi:0092-8674(88)90504-1 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. doi:S0092-8674(00)81193-9 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Aster JC, et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang LT, et al. Fringe glycosyltransferases differentially modulate Notch proteolysis induced by Delta and Jagged1. Mol Biol Cell. 2005;16:927–942. doi: 10.1091/mbc.E04-07-0614. doi:E04-07-0614 [pii] 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. doi:nri2232 [pii] 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varnum-Finney B, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(Pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. doi:S0896-6273(00)80515-2 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. doi:307/5717/1962 [pii] 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen TL, Brennan K, Arias AM, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125:4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- 22.Shaye DD, Greenwald I. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development. 2005;132:5081–5092. doi: 10.1242/dev.02076. doi:dev.02076 [pii] 10.1242/dev.02076. [DOI] [PubMed] [Google Scholar]
- 23.de Celis JF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development. 2000;127:1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]
- 24.Collier JR, Monk NA, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. doi:S0022-5193(96)90233-7 [pii] 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- 25.Plahte E. Pattern formation in discrete cell lattices. J Math Biol. 2001;43:411–445. doi: 10.1007/s002850100105. [DOI] [PubMed] [Google Scholar]
- 26.Melen GJ, Levy S, Barkai N, Shilo BZ. Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol Syst Biol. 2005;1:0028. doi: 10.1038/msb4100036. doi:msb4100036 [pii] 10.1038/msb4100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. doi:06-PLBI-RA-1518 [pii] 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchler NE, Louis M. Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. doi:S0022-2836(08)01248-5 [pii] 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 29.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. doi:10.1016/j.cell.2004.06.009 S0092867404005732 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Hansson EM, et al. Recording Notch signaling in real time. Dev Neurosci. 2006;28:118–127. doi: 10.1159/000090758. doi:DNE20060281_2118 [pii] 10.1159/000090758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.