Abstract
The vacuole of Saccharomyces cerevisiae has been a seminal model for studies of lysosomal trafficking, biogenesis, and function. Several yeast mutants defective in such vacuolar events have been unable to grow at low levels of hygromycin B, an aminoglycoside antibiotic. We hypothesized that such severe hypersensitivity to hygromycin B (hhy) is linked to vacuolar defects and performed a genomic screen for the phenotype using a haploid deletion strain library of non-essential genes. Fourteen HHY genes were initially identified and were subjected to bioinformatics analyses. The uncovered hhy mutants were experimentally characterized with respect to vesicular trafficking, vacuole morphology, and growth under various stress and drug conditions. The combination of bioinformatics analyses and phenotypic characterizations implicate defects in vesicular trafficking, vacuole fusion/fission, or vacuole function in all hhy mutants. The collection was enriched for sensitivity to monensin, indicative of vacuolar trafficking defects. Additionally, all hhy mutants showed severe sensitivities to rapamycin and caffeine, suggestive of TOR kinase pathway defects. Our experimental results also establish a new role in vacuolar and vesicular functions for two genes: PAF1, encoding a RNAP II-associated protein required for expression of cell cycle-regulated genes, and TPD3, encoding the regulatory subunit of protein phosphatase 2A. Thus, our results support linkage between severe hypersensitivity to hygromycin B and vacuolar defects.
Keywords: Yeast vacuole, Lysosome, Hygromycin B, Vesicular trafficking
Introduction
The yeast vacuole is a membrane-bounded organelle analogous to the mammalian lysosome. As such, it has multiple functions associated with degradation, receptor down regulation, ion and pH homeostasis, and stress survival (for comprehensive reviews see Jones et al. 1997; Katzmann et al. 2002; Pelham 2002; Bowers and Stevens 2005; Luzio et al. 2007; Mijaljica et al. 2007; Li and Kane 2009). Most recently, vacuoles/lysosomes have been viewed more as regulatory organelles responsible for fine tuning of cellular processes and less as simple storage and degradation compartments. Therefore, vacuole trafficking, biogenesis, and function remain intensively explored fields. Like the mammalian lysosome, the yeast vacuole maintains an acidic environment containing vacuolar hydrolases that degrade macromolecules, structural debris, and waste products (Thumm 2000; Martínez-Muñoz and Kane 2008). As with lysosomes, several pathways are responsible for delivery of cargo to the vacuole. The biosynthetic pathway involves sorting of newly synthesized vacuolar proteins away from the secretory pathway in the trans-Golgi network (TGN) and their vesicular delivery to the vacuole; the pathway includes vacuolar delivery both through the late endosome (CPY-pathway) and independent of it (ALP-pathway) (Nothwehr and Stevens 1994; Horazdovsky et al. 1995; Bryant and Stevens 1998; Conibear and Stevens 1998; Bowers and Stevens 2005). The endocytic pathway involves the vesicular delivery of external and cell surface components through the early endosome to late endosome, and through the maturation product of late endosome—the multivesicular body (MVB)—to the vacuole (Katzmann et al. 2002; Pelham 2002; Piper and Katzmann 2007). The autophagic and cytoplasm-to-vacuole targeting (Cvt) pathways involve the transport of cytoplasmic components directly to the vacuole through vesicular delivery. Under starvation conditions, the autophagy pathway is stimulated in order to provide supplementary reserves for the cell; this pathway is also a housekeeping and stress-responsive process and involves the autophagy (ATG) genes (for latest reviews see Shintani and Klionsky 2004; Levin 2005; Huang and Klionsky 2007; Mijaljica et al. 2007; Li and Kane 2009).
Yeast genetics and molecular genetics have been instrumental in deciphering the vesicular trafficking machinery conserved in eukaryotic cells. Genetic screens have resulted in collections of mutants defective in various stages of vesicular trafficking. Over the past two decades, several genetic screens have uncovered mutant collections defective in the CPY-pathway of vacuolar localization including pep (Jones 1977), vps (Bankaitis et al. 1986; Rothman and Stevens 1986; Bonangelino et al. 2002), vam (Wada et al. 1992), and env (Takahashi et al. 2008) mutants. Some mutants in these collections have shown sensitivities to low levels of the antibiotic hygromycin B, an aminoglycoside translation inhibitor isolated from Streptomyces hygroscopicus (Mcgaha and Champney 2007). Hygromycin B is an atypical aminoglycoside antibiotic in that it has dual inhibitory effects on translation; it interferes with both ribosomal translocation and with aminoacyl-t-RNA recognition (Cabañas et al. 1978). As such, at 200 µg/mL or more, the drug inhibits the growth of most yeast strains. At 50 µg/mL, hygromycin B has been shown to inhibit growth of yeast mutants defective in glycosylation functions (Ali et al. 2004). At hygromycin B concentrations below 50 µg/mL, several vacuolar trafficking and function mutants have shown degrees of growth sensitivities, including vps1, vps11/pep5, vps15, vps27, and vps44/nhx1, while vps45 and vps54/luv1 have shown complete growth inhibition (Conboy and Cyert 2000; Ali et al. 2004; Mukherjee et al. 2006). Finally, env1 mutants are unable to grow in the presence of 25 µg/mL hygromycin B (Takahashi et al. 2008).
We hypothesized that severe hypersensitivity to hygromycin B may be associated with a specific set of vacuolar defects and may be used to identify new genes involved in vacuolar events. Several comprehensive yeast genomic screens for various drug sensitivities including hygromycin B have been reported in homozygous, heterozygous, or haploid deletion strain collections (Ross-Macdonald et al. 1999; Parsons et al. 2006; Hillenmeyer et al. 2008; for review see Hoon et al. 2008). The screens have not revealed a strong relationship between hygromycin B sensitivity and vacuolar events, yet none of the screens narrowed the scope of the screened phenotype to complete lack of growth. In this study, we performed a genomic screen for growth hypersensitivity to hygromycin B (hhy mutants) as scored by complete growth inhibition at 25 µg/mL hygromycin B. Here, we report identification of 14 hhy mutants. The corresponding genes include four VPS genes—VPS34, VPS45, SAC2 (VPS52), and LUV1 (VPS54); five genes with known functions in vesicular trafficking—DRS2, SBH2, ARF1, CHC1, and SAC1; two genes that have been classified as VPS genes in a genomic screen—DHH1 and BUD32; and two genes with published functions in cell cycle-related events—TPD3 and PAF1. The screen also uncovered hhy1, a mutant deleted in YEL059W which is classified as a dubious hypothetical open reading frame (ORF). A combination of nomarski and fluorescent microscopy, growth sensitivity studies, biochemical assays, and bioinformatics implicate defects in vacuolar function, morphology, and/or trafficking in all the uncovered mutants.
Materials and methods
Media, yeast strains, and antibodies
Yeast cells were grown in yeast extract-peptone-dextrose (YPD) or synthetic complete (SC) medium; YPD or SC media were supplemented with selective components as specified; all yeast and supplemental chemicals were purchased from Sigma Chemicals (St. Louis, MO, USA). The MAT-α haploid S. cerevisiae deletion strain library, parental strains BY4742 (MAT-α, his3Δl, leu2Δ0, lys2Δ0, and ura3Δ0) and BY4741 (MAT-a, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0), and MAT-ahaploids of the 14 hhy strains were gifts fromDr. Greg Payne (UCLA). The collection of 4,828 strains was developed by the Saccharomyces Genome Deletion project and contains 80% of the Saccharomyces cerevisiae genome; it was generated by PCR-based disruption of all open reading frames by chromosomal integration of a KanMX4 module through homologous recombination (Wach et al. 1994). Mutants were stored in glycerol in fifty-two 96-well plates at −80°C. The mouse monoclonal anti-proCPY antibody was custom ordered from U. Oregon and was initially developed in T. Stevens Laboratory (U.O.). The secondary goat anti-mouse-HPR antibody was purchased from Pierce (Rockford, IL, USA). The parental supersensitive strain used for halo assays was GPY1796 which was generated from GPY404.2 (MAT-a, ura3-52, leu2-3,112, his3-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, and GAL-MEL sst1::LYS).
Yeast genomic screen for hypersensitivity to hygromycin B (hhy)
MAT-α yeast deletion strains in the BY4742 background were transferred from thawed 96-well plates onto YPD and YPD + hygromycin B (25 µg/mL) plates with a pinning tool. Plates were incubated at 30°C for 72 h. hhy phenotype was scored as absence of growth on YPD + hygromycin B plates in comparison to growth on parallel YPD plates. A secondary screen was conducted in order to confirm absence of growth by streaking putative hhy strains onto YPD and YPD + hygromycin B (25 µg/mL) plates. Positive results were verified using the corresponding MAT-a knockout strain in the BY4741 background. The deletion of YEL059W ORF in hhy1 strain was confirmed by PCR. YF 5′-TTTAACTATTGGTATATGTGTCCGTGA-3′ binds upstream of the YEL059W ORF, YR 5′-ACTTCTAACAAAAGTGACCATGACG-3′ binds within the YEL059W ORF, and KR 5′-CTGCAGCGAGGAGCCGTAAT-3′ binds within the kanamycin cassette. As expected, PCR product of primers YF and YR was a 522-bp band from WT samples while no band was observed in hhy1 samples; PCR product of YF and KR was a 554-bp band in hhy1 samples, while no band was observed in WT samples (data not presented).
Reintroduction of deleted ORF’s into tpd3Δ, paf1Δ, and hhy1Δ
To determine whether the hygromycin B sensitivity of tpd3Δ, paf1Δ, and hhy1Δ resulted from the absence of the deleted ORF, corresponding ORF’s were reintroduced by means of CEN and/or 2-µ vectors. YCp-TPD3 (van Zyl et al. 1992) was a gift from Dr. James Broach (Princeton University). pJJ1371 (unpublished) is a uracil selectable 2-µ vector containing PAF1 and was a gift from Dr. Joan Betz (Regis University). PWO 0577 contains PRB1 in pRS426 and was a gift from Dr. Dieter Wolf (University of Stuttgart, Germany). pSEY8-PRB1 (Moehle et al. 1986) was a gift from Dr. Scott Emr (Cornell University). PCR-based subcloning was carried out to construct the vectors containing YELO59W sequences. The YELO59W ORF from BY4742 genomic DNA was amplified using the following primers containing restriction sites on the 5′ ends. F1 (BamHI) 5′-GGGGGGATCCTTGGCGATGAAGCTAATTG-3′; R1 (EcoRI) 5′-GGGGGAATTCTGACCTTTGCCTTGGTCCT-3′; R2 (EcoRI) 5′-GGGGGAATTCGCCAACTCAACACGAAATTC-3′; R3 (KpnI) 5′-GGGGGGTACCTTTAGCTGCAACACTGCATG-3′. pDOG360 was constructed using F1 and R1 which resulted in a 1.2-kb insert containing 661-bp upstream and 273-bp downstream of YELO59W and includes SOM1. pDOG361 was constructed using F1 and R2 which resulted in a 1.6-kb insert containing 661-bp upstream and 635-bp downstream of YELO59W and includes SOM1. pDOG362 was constructed using F1 and R3 which resulted in a 3.4-kb insert containing 661-bp upstream and 2.48-kb downstream of YELO59W and includes SOM1 and PCM1. PCR reactions were carried out in a final volume of 50 µL containing genomic DNA (100 ng) by using Phusion DNA polymerase (New England Biolabs) according to the manufacturer’s instructions. The PCR products were cloned into the uracilselectable CEN vector pRS316 (Sikorski and Hieter 1989) and expanded in Top 10 chemically competent E. coli (Invitrogen). Preparation of plasmid DNA was carried out using the Zippy Plasmid Maxiprep and Midiprep Kits (Zymo Research). The yeast deletion strains were transformed using the Frozen EZ Yeast Transformation II kit (Zymo Research). Parent vectors were cloned into parallel cultures to serve as controls. Transformed cells were selected on synthetic minimal media (SM) without uracil. For complementation studies, colonies were streaked on ammonium sulfate free SM-URA + hygromycin B (50 µg/mL) plates.
Invertase secretion assays
Quantitative liquid invertase assays have been described previously (Johnson et al. 1987). The assay was repeated a minimum of four times for each of the strains, and standard deviations were denoted as error bars.
Halo pheromone assays
A single colony of GPY1796 was added to 3 ml of YPD + 0.5% agar; the mixture was poured onto YPD plates and allowed to solidify for 10 min. Single colonies of hhy and wild type control strains were patched onto the seeded plates with sterile wooden applicators. Plates were incubated at 30°C overnight. Halo assays were repeated six times.
Immunodetection assays
Single colonies of hhy and wild-type strains were grown and processed for lysis/no lysis and immunodetection with monoclonal anti-pro-CPY antibody as previously described (Takahashi et al. 2008). Immunodetection assays were repeated a minimum of three times for each of the hhy strains.
Nomarski and fluorescence microscopy
FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl)pyridium dibromide] fluorescence microscopy and Nomarski microscopy were performed as previously described (Vida and Emr 1995).
Quinacrine [N′-(6-chloro-2-methoxy-acridin-9-yl)- N,N-diethyl-pentane-1,4-diamine] fluorescence microscopy was performed as previously described (Weisman et al. 1987) in YPD plus 50 mM NA2HPO4, pH 7.6.
Stained cells were viewed with a Nikon Eclipse E600 fluorescence microscope using corresponding filters. For each strain, prominent phenotypes were determined by scoring 200–300 cells in random fields.
Growth assays in selective media and at high/low temperatures
Selective media were made by addition of NaCl, KCl, CaCl2, MnCl2, ZnCl2, rapamycin, caffeine, sorbitol, or by adjusting pH during preparation of YPD agar plates to achieve final concentrations or pH values specified in figures and tables. Monensin plates were prepared by addition of monensin to synthetic complete media (SC) to a final concentration of 50 µM. Serial tenfold dilutions up to 1:10,000 were made in 96-well microtiter plates starting with OD600 = 1.00 of mid-log cells. A pinning tool was used to transfer cells from wells onto selective and replica YPD or SC plates. Plates were incubated at 30°C for 24, 48, or 72 h based on growth rate of strains on YPD or SC plates. For temperature sensitivity studies, two sets of YPD plates were streaked with hhy strains and wild type parental BY4742 and were incubated at 15, 30, and 37°C. Growth was observed every 24 h for 4 days for 37°C and for 14 days for 15°C studies. Growth assays were repeated a minimum of three times.
Bioinformatics/statistical significance calculation
To determine significance of Gene Ontology annotation, genes of interest were queried in GO::Term Finder Saccharomyces Genome Database (SGD), which is comprised of object-oriented Perl models (Boyle et al. 2004). The genes were queried under the three different classes available. Default settings for background, feature type, ORF qualifier, annotation methods, annotation source, and evidence codes were used. Statistical significance (P value) is calculated using hypergeometric distribution with a cutoff value set at ≤0.01.
Biostatistical analyses
Liquid invertase assay results and microscopic scoring of mutants were subjected to two sample t test to assess statistically significant differences from wild type. The statistical tests were conducted using MINITAB software and the alpha P value was set at ≤0.05.
Results
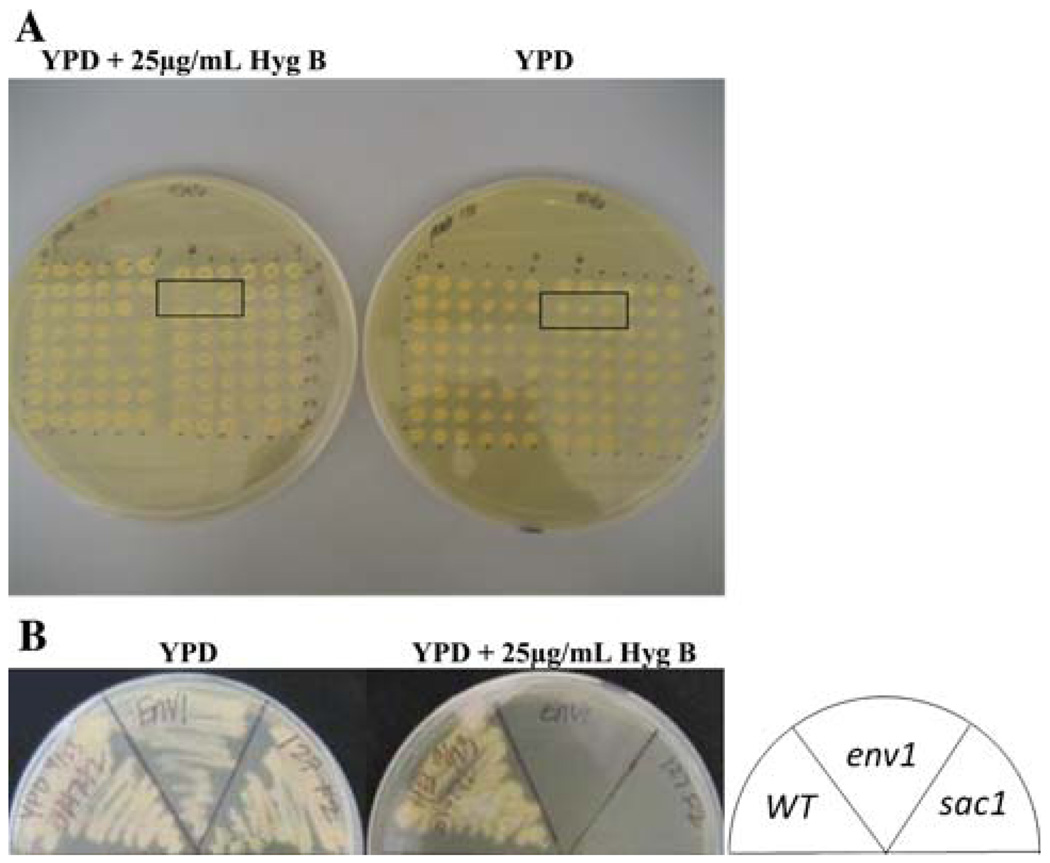
In order to test the hypothesis that severe hypersensitivity to hygromycin B is indicative of defects in vacuolar trafficking and/or function, the yeast haploid deletion strain collection of non-essential genes was subjected to a hygromycin B survival screen. The hygromycin B concentration for the screen was established at 25 µg/mL based on growth sensitivity assays within a range of hygromycin B concentrations performed for env1 mutant, a novel allele of VPS35 isolated in our laboratory (Takahashi et al. 2008). At this concentration, env1 consistently showed no growth upon prolonged incubations, while glycosylation defective mutants with growth defects at 50 µg/mL hygromycin B, grew comparable to wild type (data not shown). The primary screen assayed for lack of growth of stamped strains on YPD plates supplemented with 25 µg/mL hygromycin B (Fig. 1a). Hygromycin B hypersensitive mutants (hhy mutants) were confirmed by a secondary screen of streaked strains on the same selective media (Fig. 1b). Screening of the complete collection resulted in the identification of 14 mutant strains (<0.3% of the total) with complete growth inhibition in the presence of 25 µg/mL hygromycin B. The 14 strains showed no growth under the assay conditions upon prolonged incubations of 1–2 weeks; additionally, they continued to exhibit severe—but not complete—growth sensitivity in hygromycin B concentrations as low as 10 µg/mL (data not shown). Throughout the primary and secondary screens, the putative strains were identified only by their well/plate numbers to minimize any bias for or against specific genes. HHY genes were further verified in the MAT-a background. Table 1 lists the HHY genes, their biological process, molecular function, and cellular component as gleaned from Saccharomyces Genome Database (http://www.yeastgenome.org). 24 additional mutant strains (0.5% of the total) showed slowed growth under the screen condition as well as at hygromycin B concentrations up to 50 µg/mL. They are listed at the bottom of Table 1, but were not pursued further since this study was focused on no growth at 25 µg/mL hygromycin B.
Fig. 1.
Primary and secondary hygromycin B hypersensitivity screens. a Strains of haploid Mat-α deletion collection were replica plated from 96-well plates onto YPD and YPD + 25 µg/ml hygromycin B plates, and were incubated for 3 days at 30°C. Rectangles depict strains hypersensitive to hygromycin B. b Putative hhy strains from the primary screen were streaked on YPD and YPD + 25 µg/ml hygromycin B plates, and were incubated for 3 days at 30°C. In this representative plate, sac1 was confirmed as an hhy mutant. env1 serves as a hypersensitive control
Table 1.
HHY genes and their known biological processes, molecular functions, and cellular components (SGD)
| Gene name | Biological process | Molecular function | Cellular component |
|---|---|---|---|
| YEL059W | Unknown | Unknown | Integral to membrane |
| DRS2 | Endocytosis, post-Golgi vesicle- mediated transport, ribosomal small unit assembly | ATPase activity, Phospholipid- translocating ATPase activity | Trans-Golgi network |
| VPS54/LUV1 | Ascospore wall assembly, Golgi to vacuole transport, retrograde transport | Unknown | GARP complex, Golgi apparatus, mitochondrion |
| SBH2 | Cotranslational protein targeting to membrane | ARF guanyl-nucleotide exchange factor activity | Endoplasmic reticulum, translocon complex |
| SAC1 | Phosphoinositide dephosphorylation | Phosphatase activity | Integral to ER membrane and Golgi, mitochondrial outer membrane |
| ARF1 | ER to Golgi and intra-Golgi vesicle-mediated transport, Golgi to plasma membrane transport | GTPase activity | Cytosol, Golgi-associated vesicle |
| PAF1 | DNA recombination, histone methylation, negative regulation of transposition (RNA-mediated) | RNA polymerase II transcription elongation factor activity | Cdc73/Paf1 complex, nucleus, transcription elongation factor complex |
| DHH1 | Cellular morphogenesis (cellular fusion), cytoplasmic mRNA- processing body assembly, deadenylation-dependent deccaping (mRNA), stress granule assembly | Protein binding, RNA helicase activity | Cytoplasm, cytoplasmic mRNA- processing body |
| VPS45 | Golgi to vacuole transport, protein complex assembly, vacuole organization | Unfolded protein binding | Cytosol, extrinsic to membrane, Golgi membrane |
| VPS52/SAC2 | Actin-filament base process, Golgi to vacuole transport, retrograde transport | Protein binding | GARP complex, Golgi apparatus |
| VPS34 | Phosphoinositide phosphorylation, protein amino acid phosphorylation | Protein kinase activity | Endosome, vacuole membrane, phosphatidylinositol 3-kinase complex (I and II), pre-autophagosomal structure |
| TPD3 | Actin filament organization, cell bud growth, mitotic cell cycle spindle assembly checkpoint, translation, | Contributes to serine/threonine phosphatase activity | |
| CHC1 | Endocytosis, vesicle-mediated transport | Structure molecule activity | Clathrin vesicle coat |
| BUD32 | Positive regulation of transcription from RNA polymerase II promoter, telomere maintenance, cellular bud site selection | Serine/threonine kinase activity | EKC/KEOPS protein complex, cytoplasm, nucleus |
Strains deleted in the following genes exhibited slow growth under the screen conditions: ERG6, YM1010W-A, CPA1, CIN5, MRP51, MSY1, TAF14, PET100, YDR065W, RVS167, RMD7, GLN3, BUR2, YGR043C, YOR200W, MCT1, MRPL15, ISM1, DBP7, YDL129W, NAM2, TUP1, VAN1, and YPR022C
Published information on HHY gene products allowed classification into one major and one minor category prior to our characterization studies. Eleven genes have known vesicular and/or vacuole trafficking functions and include four original VPS genes VPS34, VPS45, SAC2 (VPS52), and LUV1 (VPS54) (Banta et al. 1988; Robinson et al. 1988; Bowers and Stevens 2005) and three genes that were uncovered in a genomic vps screen: ARF1, DHH1, and BUD32 (Bonangelino et al. 2002). The remaining genes in this category are DRS2, SBH2, CHC1, and SAC1 (Sewell and Kahn 1988; Toikkanen et al. 1996; Chen et al. 1999; Hughes et al. 2000; Gruenberg and Stenmark 2004). The second category is comprised of two genes with established roles in cell cycle regulation and progression. Paf1p, a nuclear RNA polymerase II-associated factor, is required for the expression of cell cycle-regulated genes (Shi et al. 1996, 1997; Porter et al. 2002). TPD3 encodes the regulatory subunit A of protein phosphatase 2A (PP2A); PP2A is required in multiple cell cycle regulation and budding events (Van Zyl et al. 1992; Wang and Burke 1997; Koren et al. 2004). Finally, one previously uncharacterized dubious ORF, YEL059W, was uncovered by our screen; the mutant is named hhy1.
HHY genes were queried with the use of controlled vocabulary and hierarchical organization provided by Gene Ontology Term Finder from the Saccharomyces Genome Database. The results showed a cluster frequency of five out of 14 genes with GO terms of “post-Golgi vesicle-mediated transport” and “Golgi vesicle transport”, with final P values of 1.21e–05 and 1.25e–03, respectively. The five genes for both biological processes were DRS2, ARF1, VPS45, VPS52, and VPS54. The same five genes and CHC1 were included in a cluster of six out of 14 genes in the GO term “vesicle-mediated transport” with a final P value of 2.35e–03. VPS45, VPS52, and VPS54—three genes shared by all three clusters—are involved in direct “Golgi to vacuole transport” with a final P value of 1.21e–03. However, this does not discard the remaining genes that were not in the GO Term Finder results; it suggests that they have not been annotated yet to the particular gene ontology. The total genes in the background distribution were 7,163; the total number of genes annotated within the distribution—directly and indirectly—varied depending on the GO term (24–345 genes). We may say that the relationship of at least six out of 14 genes is statistically significant based on the P values; the significant relationship is in vesicular and vacuolar trafficking. The same bioin-formatic analyses were carried out for the combination of 14 HHY genes and the 24 genes that showed slow growth under the screen conditions. No additional functional enrichments were uncovered; however, RVS167, encoding an actin-associated protein involved in regulation of endocytosis (Lombardi and Riezman 2001), also fell within the “vesicle-mediated transport” GO term.
Thus, based on published literature as well as bioinformatic analyses, genes involved in vacuole function and/or trafficking were overrepresented in hhy mutants. We set out to further characterize the hhy mutants with respect to vacuole trafficking, morphology, and function, with a focus on genes that have not been linked extensively to vacuolar events in published literature.
Secretion in hhy mutants
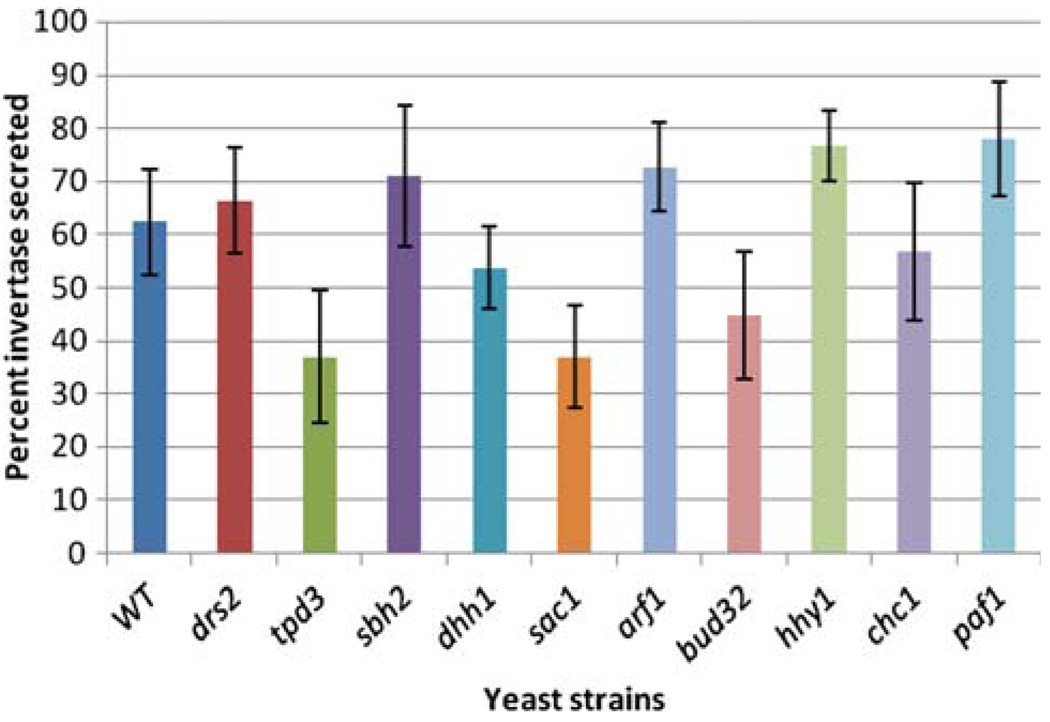
The state of secretion was assessed in hhy mutants, excluding the four vps mutants. vps mutants have been well established as lacking secretion defects (Robinson et al. 1988; Bonangelino et al. 2002). A quantitative liquid assay for invertase secretion was performed to assess the vesicular trafficking functions required for secretion (Fig. 2). Statistical analysis of repeated results indicate secretion levels that are significantly lower than wild type in tpd3Δ and sac1Δ mutants (P = 0.008 and 0.003, respectively). This suggests that vesicular trafficking step(s) including ER to Golgi, intra-Golgi, or Golgi to plasma membrane may be compromised in the two strains but are intact in the remaining hhy mutants.
Fig. 2.
State of secretion of invertase in hhy mutants. Invertase secretion was assayed in yeast mutants by quantitative liquid invertase assays at 30°C. Strains tpd3 and sac1 showed statistically significant defects in secretion relative to WT (BY4742). Two-sample t test biostatistical analysis was used
α-factor processing in hhy mutants
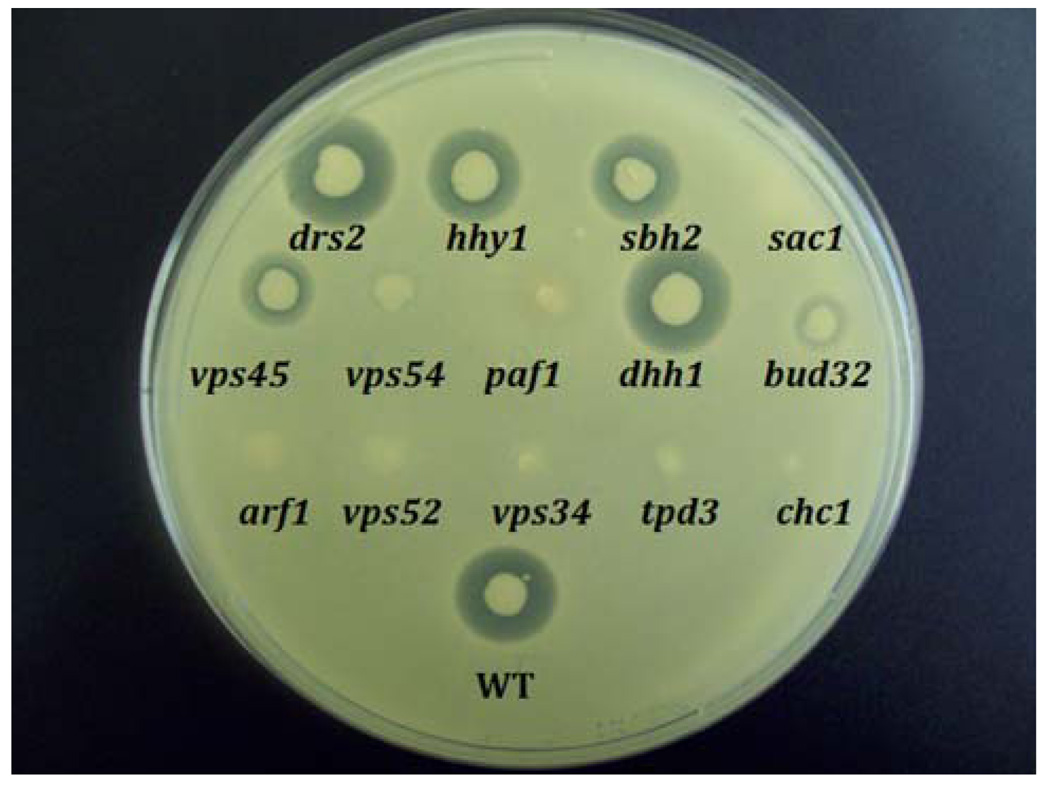
In order to assess the secretion of active α-factor, pheromone halo assays were conducted on the 14 hhy strains. The halo assay measures the responsiveness of lawn cells of the opposite mating type to mating pheromones. Correct anterograde/retrograde trafficking of Kex2p at the late endosome and Golgi interface is essential for processing and activation of α-factor pheromone prior to its secretion. Thus, in the presence of wild type α-factor, lack of halos is indicative of either defects in secretion of the pheromone, or defects in Kex2p trafficking events at the Golgi and late endosome interface. As expected, all four vps mutants resulted in minimal or no halos due to established defects at Golgi and late endosome interface (Fig. 3). Also as expected, sac1Δ, tpd3Δ, and bud32Δ strains which showed a range of reduced invertase secretion resulted in minimal or no halos, as did chc1Δ and arf1Δ strains whose gene products are involved in Golgi and late endosome trafficking events (Sewell and Kahn 1988; Redding et al. 1996). paf1Δ, which had not demonstrated any defects in invertase secretion, was completely defective in generating halos. Results of the invertase secretion and halo assays combined are suggestive of defects affecting Golgi and late endosome interface in paf1Δ. Finally, drs2Δ, hhy1Δ, sbh2Δ, and dhh1Δ resulted in halos equivalent to wild type; these results combined with the intact invertase secretion in the four mutants indicate intact vesicular trafficking at ER to Golgi, intra-Golgi, Golgi to plasma membrane, and Golgi and late endosome interface stages.
Fig. 3.
Analysis of active α-factor secretion in hhy mutants. Active α-factor secretion was assessed with standard halo assays using a lawn of GPY-1796 cells. This experiment was repeated six times with reproducible results
Carboxypeptidase Y processing in hhy mutants
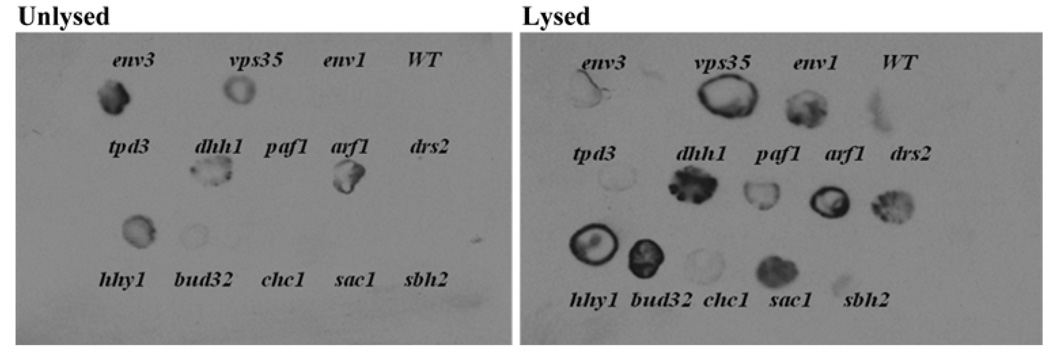
The vacuolar enzyme carboxypeptidase Y (CPY) transits through the ER, Golgi, and late endosome before arriving at the vacuole. During transit, it is processed from pro-forms (p1CPY and p2CPY glycosylated forms) to the mature CPY through proteolytic cleavage by proteinase A. Vesicular trafficking defects and/or vacuolar function defects lead to persistent forms of pro-CPY. An immunodetection experiment was performed on intact and lysed hhy cells using a monoclonal antibody specific for pro-CPY (Fig. 4). Mislocalized and secreted pro-CPY can be detected without lysis (e.g., vps35Δ), while internal pro-CPY can only be detected in lysed cells (e.g., env1). vps mutants were excluded as they have already been established as defective in CPY processing and localization (Robinson et al. 1988; Bonangelino et al. 2002). In repeated experiments, tpd3Δ, chc1Δ, and sbh2Δ showed no significant pro-CPY signal suggesting CPY processing equivalent to wild type; all three showed wild type levels of mature CPY on westerns (data not shown). The results suggest that the three genes are not essential for correct CPY processing. All other hhy mutants assayed showed persistent pro-CPY. Three hhy mutants showed mislocalization and secretion of pro-CPY. They include deletion strains of DHH1 and ARF1 which have been previously classified as vacuolar protein sorting (VPS) genes in a genomic screen (Bonangelino et al. 2002) and hhy1Δ. The remaining hhy strains, paf1Δ, drs2Δ, bud32Δ, and sac1Δ, showed internal accumulation of pro-CPY. Thus, 11 of the l4 HHY genes affect the CPY pathway to or CPY processing in the vacuole including PAF1 whose product has not previously been associated with vacuolar events.
Fig. 4.
State of p2CPY processing in hhy mutants. Patched cells were transferred to nitrocellulose membranes for overnight growth, and were probed with anti-proCPY-specific monoclonal antibody either without lysing cells or after lysis. Under the assay conditions, wild type has minimal signal due to efficient processing of proCPY forms into mCPY, while env1 and vps35 controls show persistent p2CPY either inside cells or secreted, respectively
Vacuole morphology and endocytosis in hhy strains
In order to examine vacuole morphology and integrity of endocytosis pathway in hhy strains, Nomarski light microscopy and fluorescent microscopy of cells labeled with FM4-64 were performed (Fig. 5). The lipophylic vital dye FM4-64 is incorporated into the plasma membrane and serves as an endocytic marker that localizes to the vacuole membrane within 60 min in wild type cells at 30°C (Vida and Emr 1995). 200–300 cells from each strain were scored 60 min after staining as summarized in Fig. 5b. In the wild type BY4742 strain, FM4-64 staining and Nomarski optics showed 1–3 prominent vacuoles as the predominant phenotype.
Fig. 5.
Microscopic characterization of hhy mutants. a Cells were stained with FM4-64 for 60 min and viewed using fluorescence and Nomarksi optics microscopy. The predominant vacuole morphology observed is represented. b Percentage of fragmented versus prominent versus no vacuoles in FM4-64 stained cells. 200–300 cells from each strain were scored in random fields. Two-sample t test biostatistical analysis was used
Of the ten hhy strains subjected to microscopic studies, six showed vacuole morphology variations from wild type that were statistically significant (P = 0.0001–0.027). sac1Δ, arf1Δ, luv1Δ, hhy1Δ, and paf1Δ strains exhibited a significant increase in fragmented vacuoles compared to wild type. paf1Δ and hhy1Δ strains had the highest percentage of cells with fragmented vacuoles at 58% and 66%, respectively. Mutants with fragmented vacuoles have been previously categorized as Class B, and those lacking a visible vacuole have been categorized as Class C (Banta et al. 1988; Raymond et al. 1992). Phenotypic scoring did not reveal any Class C hhy strains. The mutants were also examined for vacuole acidification using quinacrine staining; hhy1Δ was the only mutant with vacuole acidification defect (data summarized in Table 3). The four original vps mutants were not included in microscopic studies as they have been thoroughly classified with respect to their vacuole morphologies in previous studies. vps54 and vps52 are Class B with fragmented vacuoles; vps45 and vps34 are Class D with enlarged vacuoles (Banta et al. 1988; Raymond et al. 1992).
Table 3.
Summary of various characterizations of hhy mutants (this study)
| Gene name |
Secretion | Halo assays |
CPY processing |
Ion/pH sensitivity |
Caff./Rap. sensitivity |
Vacuole morphology |
Acidification | Temperature sensitivity at 37°C |
Temperature sensitivity at 15°C |
|---|---|---|---|---|---|---|---|---|---|
| tpd3 | Defective | Defective | WT | Yes | Yes | WT | WT | Yes | Yes |
| drs2 | WT | WT | Defective | Slight | Yes | WT | WT | No | Yes |
| sbh2 | WT | WT | WT | Yes | Yes | WT | WT | No | Yes |
| sac1 | Defective | Defective | Defective | Yes | Yes | Fragmented | WT | No | Yes |
| arf1 | WT | Defective | Defective | Yes | Yes | Fragmented | WT | No | Yes |
| chc1 | WT | Defective | WT | Yes | Yes | WT | WT | Slight | Yes |
| paf1 | WT | Defective | Defective | Yes | Yes | Fragmented | WT | Yes | Yes |
| dhh1 | WT | WT | Defective | Yes | Yes | Fragmented | WT | No | Yes |
| bud32 | WT | Defective | Defective | Yes | Yes | WT | WT | Yes | Yes |
| hhy1 | WT | WT | Defective | Yes | Yes | Fragmented | Defective | Yes | No |
Finally, FM4-64 localization to either prominent or fragmented vacuoles in all examined hhy mutants indicates normal endocytic trafficking from plasma membrane, through early and late endosomes, to the vacuole. Thus, none of the l4 hhy mutants exhibit defects in bulk endocytosis nor lack of visible vacuoles, while ten of the 14 exhibit vacuole morphology defects including a clustering of eight as Class B mutants with fragmented vacuoles.
Growth of hhy mutants under environmental and chemical stress conditions
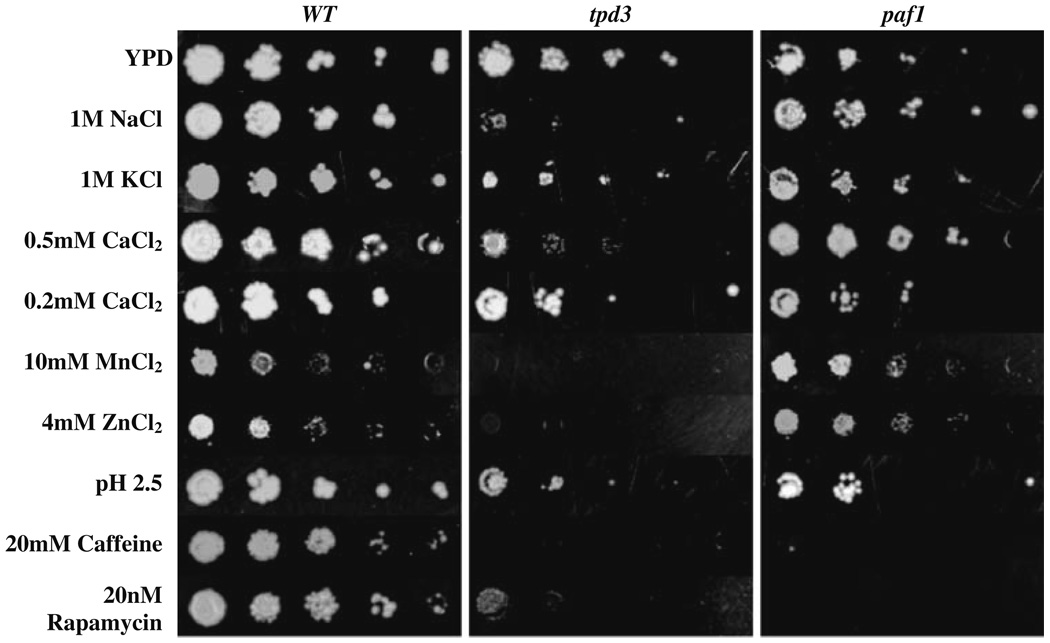
Growth sensitivities in the presence of certain ions and drugs have been suggestive of defective vacuole function (reviewed in Luzio et al. 2007; Li and Kane 2009). Additionally, several vps mutants are sensitive to 37°C, pH extremes, osmotic pressure, and caffeine (Robinson et al. 1988; Conboy and Cyert 2000). hhy mutants were assayed for growth under various ionic, pH, hyperosmotic, temperature, and drug conditions associated with vacuolar defects as represented in Fig. 6 and summarized in Tables 2 and 3. All hhy mutants exhibited strong growth sensitivities under at least three of the tested stress conditions. Several strains showed increased sensitivities to the divalent ions zinc and/or manganese indicating defects in the vacuolar sequestration of those ions; these included vps54/luv1Δ, vps45Δ, vps52/sac2Δ, vps34Δ (zinc only), chc1Δ, bud32Δ, and tpd3Δ. Most showed increased sensitivities to both low and high pH, and cold sensitivity. Most significantly, all hhy mutants showed strong sensitivities to rapamycin and caffeine, drugs indicative of a compromised TOR kinase pathway.
Fig. 6.
Ion and drug sensitivities of hhy mutants. Serial tenfold dilutions of log-phase cells were stamped with pinning tool on YPD plates or selective YPD plates with ion or drug concentrations or pH as indicated. Incubations were at 30°C for 48 h for YPD plates and for 72 h for selective YPD plates. Wild type, tpd3, paf1 strains are represented in the figure. Data for all hhy mutants are presented in Table 2
Table 2.
Drug and ion sensitivities of uncovered hhy mutants
| Strain | YPD | 1M NaCl |
1M KCl |
0.5 M KCl |
0.5 mM CaCl2 |
0.2 mM CaCl2 |
10 mM MnCl2 |
4 mM ZnCl2 |
pH 2.5 |
pH 8.0 |
20 nM rapamycin |
20 mM caffeine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| hhy1 | +++ | +++ | +++ | +++ | ∓ | ∓ | ∓ | ∓ | ∓ | ∓ | ∓ | ∓ |
| drs2 | +++ | +++ | +++ | +++ | ++ | ++ | ++ | +++ | +++ | ++ | ∓ | ∓ |
| luv1 | +++ | +++ | +++ | +++ | +++ | +++ | ∓ | ∓ | ∓ | ∓ | − | − |
| sbh2 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ∓ | ∓ | ∓ |
| sac1 | +++ | ++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | ∓ | ∓ | − |
| arf1 | ++ | ++ | ++ | ++ | ++ | ++ | ∓ | ++ | ++ | ++ | ++ | ∓ |
| paf1 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | + | ++ | − | − |
| dhh1 | +++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | ++ | ∓ | − |
| vps45 | +++ | ++ | +++ | +++ | +++ | +++ | ∓ | ∓ | ++ | ∓ | ∓ | − |
| sac2 | +++ | ++ | +++ | +++ | +++ | +++ | ∓ | ∓ | − | ∓ | ∓ | − |
| vps34 | ++ | ∓ | ++ | ++ | ++ | ++ | ++ | − | ∓ | − | − | − |
| tpd3 | ++ | ∓ | + | + | + | ++ | − | − | + | ∓ | − | − |
| chc1 | +++ | +++ | +++ | +++ | +++ | +++ | − | ∓ | ∓ | − | − | − |
| bud32 | +++ | +++ | +++ | +++ | +++ | +++ | ∓ | ∓ | ∓ | +++ | ∓ | − |
BY4742 (WT) and 14 mutants were grown on YPD and selective media with the indicated concentrations. Following 48–96-h growth, colony number and size on plates were scored as follows: +++ strong growth; ++ moderate growth; ∓ weak growth; − no growth
In order to further assess vacuole trafficking, we investigated the growth of hhy mutants in the presence of monensin, an ionophore that interferes with intracellular trafficking. A recent genomic screen found strong correlation between monensin sensitivity and known genes involved in vacuole trafficking and morphology (Gustavsson et al. 2008). We also found a high correlation between monensin sensitivity and HHY genes (Fig. 7). Ten of the 14 hhy mutants showed sensitivity to monensin, suggestive of defects in vacuole events. Notably, hhy1Δ and paf1Δ, both of which had no established connection to vacuolar events prior to this study, were sensitive to monensin.
Fig. 7.
Monensin sensitivity assay of hhy mutants. Serial tenfold dilutions of log-phase cells were stamped with pinning tool onto synthetic complete (SC) plates in absence or presence of monensin (50 µM). Plates were incubated at 30°C for 48 and 72 h
Phenotypic complementation of tpd3Δ, paf1Δ, and hhy1Δ
TPD3, PAF1, and the hypothetical YEL059W ORF have not been reported to be involved in vesicular trafficking and/or vacuolar events in previous studies. In order to confirm that the observed vacuolar/vesicular phenotype of the corresponding mutants is due to the deleted ORF, each of the genes were reintroduced into the corresponding mutant (Fig. 8). PAF1 and TPD3 were each able to complement the hhy phenotype of paf1Δ and tpd3Δ, respectively. YEL059W sequences in single copy or multi copy did not complement hhy1Δ in neither mating type, despite confirmation of YEL059W deletion and KAN cassette insertion by PCR approaches as detailed in “Materials and methods” (Fig. 8; data not presented). Furthermore, a construct containing YEL059W as well as adjacent upstream (SOM1) and downstream (PCM1) ORF’s along with a minimum of 500 bases of flanking sequences also failed to complement hhy1Δ (Fig. 8). PCM1 is an essential gene involved in GlcNAC synthesis, and SOM1 encodes a mitochondrial inner membrane peptidase essential for respiratory growth. As such, deletion strains of the two genes are not included in the collection. Finally, PRB1 which encodes the vacuolar protease proteinase B and lies two ORF’s upstream of YEL059W, also failed to complement hhy1Δ; prb1Δ of the deletion strain collection did not exhibit hhy nor vacuole fragmentation phenotype upon reexamination (data not presented).
Fig. 8.
Complementation tests for hhy1, tpd3, and paf1 mutant strains. a Strains were transformed with empty vector or vector containing the corresponding deleted ORF. Transformed cells were streaked onto SM-URA plates with or without 50 µg/ml hygromycin B and grown at 29°C. WT exhibited comparable growth with or without hygromycin B as did tpd3 and paf1 strains transformed with their corresponding deleted ORFs. hhy1 continued to exhibit severe hypersensitivity to hygromycin B when transformed with pDOG362 containing YEL059W ORF flanked by its two adjacent ORF’s as detailed in b. b Diagrammatic representation of constructs used for hhy1 complementation tests and summary of results
Discussion
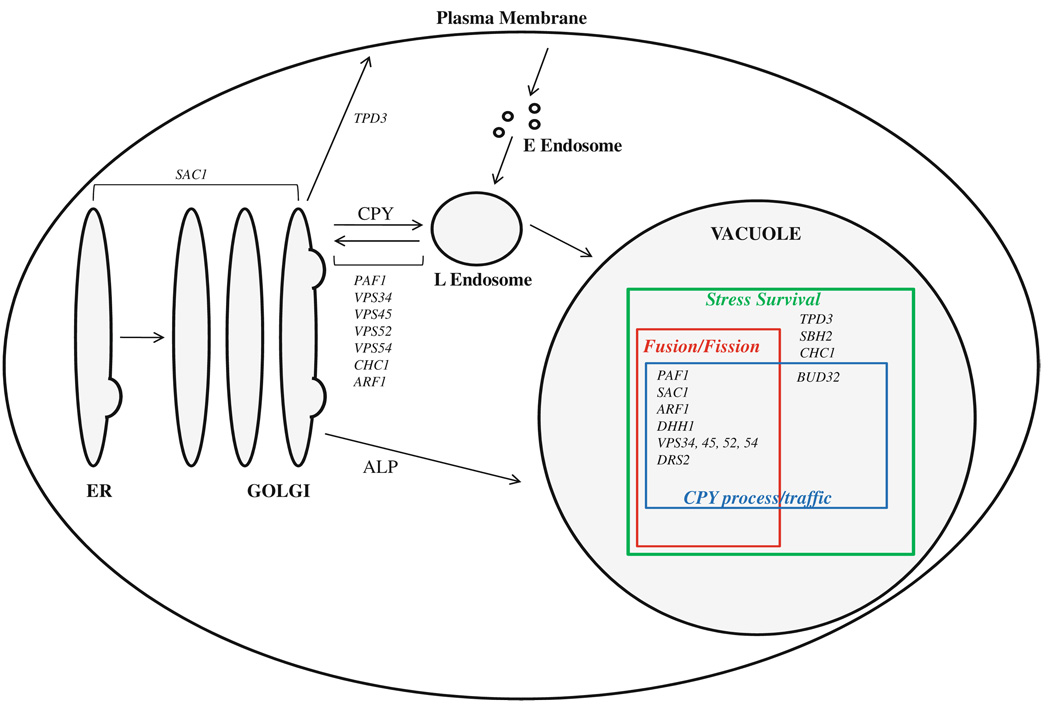
Several yeast mutants with vacuole trafficking or function defects have recently been shown to exhibit sensitivity to <50 µg/mL hygromycin B (Conboy and Cyert 2000; Ali et al. 2004; Mukherjee et al. 2006; Takahashi et al. 2008). Specifically, vps54 and env1 mutants show no growth at such low hygromycin concentrations (Mukherjee et al. 2006; Takahashi et al. 2008). We hypothesized that such severe hypersensitivity to hygromycin B may be indicative of specific vacuole trafficking or function defects. To test this hypothesis and to potentially identify additional genes involved in vacuolar events, a genomic screen of 4,828 yeast haploid deletion strains was conducted for growth sensitivities in 25 µg/mL hygromycin B. 38 mutant strains showed a range of growth sensitivities. Fourteen of the uncovered mutant strains showed no growth upon prolonged incubation times and under a range of hygromycin B concentrations; they were denoted as hypersensitive to hygromycin B (hhy mutants) and were pursued in this study (Table 1). Hypersensitivity of these mutants to hygromycin B is not likely to be due to the effect of the drug on protein translation since the mutants did not show increased sensitivity to the translation inhibitor cycloheximide used in our immunodetection screen. Conboy and Cyert (2000) also reached the same conclusion with their mutants which were sensitive to hygromycin B, but did not show additional sensitivity to translation inhibition. Bioinformatics analysis established that the genes showed a statistically significant clustering with GO terms associated with post-Golgi vesicle mediated transport to the vacuole. Microscopic, biochemical, and environmental stress assays established that vacuole morphology, trafficking, and stress survival defects were also highly enriched in the hhy collection. Figure 9 summarizes the proposed functions of HHY genes in vesicular trafficking and vacuole events based on our results.
Fig. 9.
Overview of vesicular trafficking in yeast with the proposed site/mode of action of HHY genes as suggested by this study. ER endoplasmic reticulum; CPY carboxypeptidase Y pathways; ALP alkaline phosphatase pathway
Four of the hhy mutants were null alleles of the original vps collection: luv1 (vps54), vps45, sac2 (vps52), and vps34. These vps mutants have previously been shown to display a defect in protein sorting to the vacuole (Banta et al. 1988; Robinson et al. 1988). Vps52p and Vps54p are members of the Golgi-associated retrograde protein (GARP) and their mutants have class B fragmented vacuoles (Conibear and Stevens 2000). Vps45p is required for Golgi vesicle fusion with the late endosome and its mutant form results in enlarged vacuoles, categorized as class D (Raymond et al. 1992; Bryant and James 2001). Vps34p is a phosphoinositide kinase required for multiple vacuolar trafficking events including vacuole segregation, vacuolar protein localization, multivesicular body formation, and autophagy (Herman and Emr 1990; Schu et al. 1993; Munn and Riezman 1994; Kihara et al. 2001; Wurmser and Emr 2002; Katzmann et al. 2003). Its mutation results in class D enlarged vacuoles (Raymond et al. 1992). Thus, all four uncovered established VPS genes affect membrane fission/fusion events at the late endosome and the vacuole.
Three hhy mutants have been designated as vacuolar protein sorting mutants through a genomic screen for additional VPS genes: arf1Δ, dhh1Δ, and bud32Δ (Bonangelino et al. 2002). Arf1p has been established as a GTPase of the Ras superfamily involved in regulation of coated vesicle formation in vesicular trafficking (Sewell and Kahn 1988). Dhh1p, a dead box protein, regulates Ste12p translation and has a role in recovery from G1/S checkpoint (Bergkessel and Reese 2004; Ka et al. 2008). The dhh1 mutant displays abnormal cellular morphology and defects in cytokenesis and mitosis, including aberrant “shmoo” formation at mating and abnormal budding morphology (Moriya and Isono 1999). Bud32p is involved in bud site selection and interacts with the KEOPS complex for transcription regulation and telomere maintenance/homeostasis; BUD32 mutation results in decreased telomere size and defects in cell cycle (Ni and Snyder 2001; Facchin et al. 2007). Our results showing CPY processing and vesicular trafficking defects and temperature sensitivity for the three mutants are consistent with those reported by Bonangelino et al. (2002). Our microscopy results indicate excessive vacuole fragmentation in all three; Bonangelino et al. (2002) also classified bud32Δ and arf1Δ as having vacuole morphology defects. Additionally, all three mutants showed growth sensitivities to multiple stress conditions tested. Thus, the three mutants show defects in vacuolar trafficking, fusion, and stress survival. Interestingly, DHH1 and BUD32 are also involved in cell cycle and cell growth events.
Four of the remaining hhy mutants are known to be involved at various stages of vesicular trafficking; those are drs2Δ, chc1Δ, sac1Δ, and sbh2Δ. Drs2p is an amino-phospholipid translocase (flippase) that maintains post-Golgi secretory vesicles and contributes to clathrin-coated vesicle formation (Ripmaster et al. 1993; Chen et al. 1999). Deletion mutants of the gene have been reported to accumulate membrane structures analogous to Berkeley bodies and to exhibit a block in endosome-to-vacuole stage of trafficking at non-permissive temperature (Chen et al. 1999; Hua et al. 2002). Interestingly, drs2 mutant alleles exhibit synthetic lethality with arf1Δ (Chen et al. 1999). Chc1p is a clathrin heavy chain coat protein involved in post-Golgi-coated vesicle transport and endocytosis (Gruenberg and Stenmark 2004). Sac1p is a lipid phos-phatase localized to the endoplasmic reticulum and Golgi membranes; it is involved in several vesicular trafficking processes, including cell wall maintenance, Golgi trafficking, and secretion (Hughes et al. 2000; Strahl and Thorner 2007). Additionally, it functions in the control of vacuolar morphology and actin cytoskeleton (Foti et al. 2001). Sbh2p is a component of the Ssh1p-Sss1p-Sbh2p complex involved in protein translocation into the endoplasmic reticulum (Toikkanen et al. 1996). Of the four vesicular trafficking genes, only SBH2 has an established role limited to the endoplasmic reticulum; consistent with that, its deletion did not result in defects in pro-CPY processing in our studies. It is conceivable that hypersensitivity of sbh2Δ to hygromycin B was due to direct effects of the drug on the translation machinery and the post-translational ER translocation of proteins in yeast. However, neither liquid invertase assays nor halo assays revealed compromised secretion—an expected phenotype if the drug was significantly interfering with protein translation/translocation. Instead, sbh2Δ cells showed severe growth sensitivities to pH/ionic/osmotic stresses, and multidrug sensitivities suggestive of an essential role in vacuolar function. Taken together, our results suggest a likely role for all four genes in vacuole morphology and/or function; the role of Chc1p and Sbh2p in this process does not appear to affect CPY processing within the limit of our assays.
Two of the uncovered HHY genes had no previously established roles in vesicular trafficking—TPD3 and PAF1. Both genes have known rolesincell cycle regulation. Tpd3p is one of two regulatory subunits of the heterotrimeric protein phosphatase 2A (PP2A), a ser/thr phosphatase required for cell morphogenesis, cell cycle progression and cytokinesis, and for transcription by RNA polymerase III (Van Zyl et al. 1992; Wang and Burke 1997). Koren et al. (2004) have suggested that Tpd3p is not essential for the cytokinesis functions of PP2A, but enhances the enzyme activity. We detected multiple defects associated with vesicular trafficking and vacuole function. tpd3Δ mutants were defective in both invertase secretion and halo assays, but not in CPY processing, suggesting a defect at pre-endosomal stages of vesicular trafficking and/or secretion (Fig. 8). However, the mutants exhibited multi drug sensitivities and severe growth defects under various stress conditions suggestive of compromised vacuole function. These results reinforce the notion that Tpd3p regulation of protein phosphatase 2A affects a variety of biological processes in the cell including secretion and stress survival functions.
The second HHY gene with no previously established vacuolar function is PAF1. Paf1p is a nuclear RNA polymerase II-associated protein that is required for full expression of cell cycle-regulated genes; it acts in the Pkc1p-Slt2p MAP kinase cascade and its human homolog, hPaf1, is associated with tumorigenesis (Shi et al. 1996, 1997; Chang et al. 1999; Porter et al. 2002; Moniaux et al. 2006). Consistent with Shi et al.’s (1996) observations, paf1Δ strains in our studies showed larger cell size and severe temperature sensitivity. Additionally, we established severely fragmented vacuoles, CPY processing and halo induction defects, and multi drug sensitivities. Taken together, these results are suggestive of a defect at Golgi to vacuole stage of trafficking affecting vacuole morphology and stress function. Paf1p may regulate the expression of protein(s) affecting vacuolar events including vacuole fusion. We are currently exploring the feasibility of microarray and/or proteomic approaches to investigate global transcriptional and/or translational consequences of PAF1 and TPD3 deletion.
Our screen also uncovered one previously uncharacterized dubious ORF, YEL059W, whose deletion in hhy1 mutant was experimentally confirmed. The mutant strain exhibited no defects in secretion or retrograde trafficking at late endosome and Golgi interface based on invertase secretion and halo assays. It exhibited defective CPY processing, extreme sensitivities to pH extremes, divalent cations, and multiple drugs indicative of severely compromised vacuole function. Both mating types of the mutant exhibited enlarged cells and highly fragmented vacuoles in addition to hypersensitivity to hygromycin B. The deleted ORF is located on chromosome V and is not conserved in most yeast or higher eukaryotes. Single copy or multi copy reintroduction of YEL059W ORF and its flanking sequences in trans, as well as reintroduction of sequences that include YEL059W and its two flanking ORF’s, did not rescue hhy1. Reintroduction of PRB1, an ORF approximately 800 bases upstream of YEL059W that encodes vacuolar protease B, also did not rescue the mutant. At this point, we have not identified the locus responsible for the hhy phenotype of hhy1 mutant. Alternatively, YEL059W sequences may have a role in distant cis-acting regulation of other gene(s) involved in vacuolar morphology and function.
FM4-64 localization to vacuoles in hhy mutants indicates normal bulk endocytic trafficking. The late endosome to vacuole stage of trafficking is shared between endocytic and biosynthetic CPY pathways, and normal bulk endocytic trafficking to the vacuole would suggest normal late endosome to vacuole trafficking. Therefore, the CPY-processing defects of drs2Δ, sac1Δ, bud32Δ, and hhy1, which have no pre-endosomal trafficking defects, are more likely due to defective vacuole function than defective postendosomal vacuolar trafficking.
The combination of bioinformatic and experimental characterizations of hhy mutants implicate defects in vacuole trafficking, morphology, and/or stress survival function for all uncovered mutants, including the two mutants deleted in cell cycle regulatory genes PAF1 and TPD3 with no previously established connection to vacuolar events. Thus, in agreement with our hypothesis, severe hypersensitivity to hygromycin B was indicative of vacuolar defects and the screen uncovered two genes with no previous known connection to vesicular and/or vacuolar events. The identification of PAF1 and TPD3 suggests that there are yet unidentified gene functions in vacuolar events. The fact that all hhy mutants were sensitive to a minimum of three stress conditions suggests that hygromycin B hypersensitivity may be due to compromised stress tolerance in these mutants. In one of the most recent large scale genomic screens for drug sensitivities, multi drug sensitivity to at least 20% of the unique treatments was significantly enriched in deletions of genes associated with vesicular trafficking, and specifically genes associated with vacuole trafficking and function (Hillenmeyer et al. 2008).
Despite >100 genes identified in vacuolar trafficking, function, and morphology, only 14 hhy mutants were uncovered. Moreover, the limited number and the functional clustering of mutants that show no growth under low concentrations of hygromycin B versus those that merely show slowed growth suggest that the two phenotypes are associated with functionally distinct classes of mutants. Stress conditions have been documented to result in vacuole fragmentation (Li and Kane 2009; Luzio et al. 2007). Our results support this connection since vacuole fusion/fission defects as well as defects in stress survival are enriched in hhy mutants. The only established vps mutants uncovered have either excessive fragmentation of vacuoles (Class B) or defects in vacuolar fragmentation (Class D). Among the remaining hhy mutants, our microscopic studies confirmed increased vacuole fragmentation in all but drs2Δ, tpd3Δ, and chc1Δ. Other microscopic studies, however, have reported higher percentages of fragmented vacuoles for those three mutants (Chen et al. 1999; Seeley et al. 2002). Significantly, neither Class A vps mutants (no vacuole morphology or inheritance defects), nor Class C vps mutants (no visible vacuoles) were included in the collection. The lack of Class C mutants is especially interesting since they exhibit the severest pH and osmotic sensitivities within the vps mutant collection (Robinson et al. 1988). Moreover, the Class C genes whose products make up the HOPS complex have been reported to be hypersensitive to gentamicin, another aminoglycoside (Blackburn and Avery 2003; Wagner et al. 2006) and to higher levels of hygromycin B in global multidrug screens (Dudley et al. 2005). Their exclusion from hhy collection suggests that our screen was neither simply picking up the most severe vps mutants nor those most sensitive to aminoglycoside drugs in general. Thus, while none of the 14 hhy mutants exhibit defects in bulk endocytosis or lack of visible vacuoles, ten of the 14 exhibit vacuole morphology defects including a clustering of eight as class B mutants with fragmented vacuoles. Our characterizations suggest that the uncovered mutants share specific defects in vacuole fission/fusion and stress tolerance. All the trafficking pathways to the vacuole involve a final step of membrane fusion at the vacuole; furthermore, the vacuole is a dynamic organelle which undergoes fusion/fission events during the budding stage of the cell cycle and in response to external stress. Thus, the membrane fission/fusion events at the vacuole remain of intense interest (for latest review see Ostrowicz et al. 2008).
As a potential organelle for sequestering hygromycin B from the translation machinery, intact vacuolar function and trafficking may be crucial to cell survival even at low hygromycin B concentrations. However, since only a small fraction of known vacuolar trafficking or function genes were uncovered in our screen, hygromycin B may be directly interfering with a specific step in vacuolar events.Brett et al. (2005) have suggested that pH homeostasis defects may be associated with hygromycin B sensitivity. Our quinacrine staining studies of hhy mutants reveal properly acidified vacuoles, except in hhy1. Hence, any possible defects in pH regulation in hhy mutants may be in other cellular compartments. In mammalian cells, amino-glycoside drugs have been reported to interfere with coatomer formation and secretion (Hu et al. 1999; Hudson and Draper 1997). An attractive model would be that hygromycin B interferes with efficient vacuole fusion/fission events necessary for stress survival and proper cell cycle progression, and that hhy mutants are specifically inefficient at membrane fusion/fission. All hhy mutants showed severe sensitivities to rapamycin—a phenotype associated with defects in Tor1 kinase pathway which regulates cell growth and vacuolar autophagy under various stress and nutrient conditions (Park et al. 2005; Kuranda et al. 2006; reviewed in Levin 2005; Aronova et al. 2007). Additionally, hhy mutants were also hypersensitive to caffeine, a phosphodiesterase inhibitor that has been reported to target yeast TOR Complex1 (TORC1) and TOR Complex2 (TORC2) (Guan et al. 2009; Reinke et al. 2006; Wanke et al. 2008). Most recently, functional interactions have been reported between TORC1, actin, and vesicular trafficking (Aronova et al. 2007), and Tor1-GFP fusion proteins were reported to be concentrated near the vacuolar membrane in live yeast cells (Sturgill et al. 2008). Most relevantly, TPD3 and TORC1 double mutants show synthetic lethality, and Vps34p has been suggested as an upstream regulator of TORC1 (Aronova et al. 2007; Jacinto 2008); both genes were identified as HHY in our screen. The published cell cycle roles of PAF1, TPD3, DHH1, and BUD32 support a connection between cell cycle regulation and vacuole function and morphology (reviewed in Fagarasanu and Rachubinski 2007). It is conceivable that HHY genes may be specifically involved at the interface of TOR kinase pathway and vacuolar events, and hygromycin B may be targeting molecular interactions at that interface.
In this study, a genomic screen for severe hypersensitivity to hygromycin B uncovered 14 hhy mutants all of which were implicated in vacuole trafficking, morphology, and/or stress survival defects by a combination of bioinformatic and experimental approaches. The hhy collection includes two mutants deleted in cell cycle regulatory genes PAF1 and TPD3 with no previously established role in vacuolar events. Additionally, vacuolar fusion/fission defects were enriched in the uncovered collection, and all hhy mutants showed hypersensitivity to caffeine and rapamycin, drug sensitivities associated with Tor kinase pathway defects. Additional studies on hhy strains relative to Tor kinase pathway and vacuole fusion/fission under stress and during cell cycle progression may shed light on the step affected by hygromycin B.
Acknowledgments
We are grateful to Greg Payne for the gift of the deletion strain library, parent vectors, and for continuous support and guidance. We thank Judy Brusslan for technical advice and helpful discussions, and the laboratories of J. Betz, J. Broach, S. Emr, and D. Wolf for generous gifts of plasmids. This work was supported by NIH grant #R15GM1085794-01 to EG. DEM and CRA-S were supported by NIH-RISE and NIH-MARC undergraduate training grants, respectively. DEM was partially supported by the cited NIH-R15 grant. DKO was supported by a Beckman Scholars award.
References
- Ali R, Brett CL, Mukherjee S, Rao R. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J Biol Chem. 2004;279(6):4498–4506. doi: 10.1074/jbc.M307446200. [DOI] [PubMed] [Google Scholar]
- Aronova S, Wedaman K, Anderson S, Yates J, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18(8):2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83(23):9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107(4):1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergkessel M, Reese JC. An essential role for the Saccharomyces cerevisiae DEAD-box helicase DHH1 in G1/S DNA-damage checkpoint recovery. Genetics. 2004;167(1):21–33. doi: 10.1534/genetics.167.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn AS, Avery SV. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob Agents Chemother. 2003;47(2):676–681. doi: 10.1128/AAC.47.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(7):2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744(3):438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO: Term Finder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16(3):1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, James DE. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 2001;20(13):3380–3388. doi: 10.1093/emboj/20.13.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62(1):230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabañas MJ, Vázquez D, Modolell J. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem Biophys Res Commun. 1978;83(3):991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19(2):1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-Type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147(6):1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy MJ, Cyert MS. Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol Biol Cell. 2000;11(7):2429–2443. doi: 10.1091/mbc.11.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404(1–2):211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11(1):305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:2005.0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchin S, Ruzzene M, Peggion C, Sartori G, Carignani G, Marin O, Brustolon F, Lopreiato R, Pinna LA. Phosphorylation and activation of the atypical kinase p53-related protein kinase (PRPK) by Akt/PKB. Cell Mol Life Sci. 2007;64(19):2680–2689. doi: 10.1007/s00018-007-7179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Rachubinski RA. Orchestrating organelle inheritance in Saccharomyces cerevisiae. Curr Opin Microbiol. 2007;10(6):528–538. doi: 10.1016/j.mib.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12(8):2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5(4):317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K, Wakabayashi H, Ivanova T, Castillon GA, Piccolis M, Abe F, Loewith R, Funato K, Wenk MR, Riezman H. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20(7):2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Barmark G, Larsson J, Mure´n E, Ronne H. Functional genomics of monensin sensitivity in yeast: implications for post-Golgi traffic and vacuolar H+-ATPase function. Mol Genet Genomics. 2008;280(3):233–248. doi: 10.1007/s00438-008-0359-9. [DOI] [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10(12):6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St. Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S, St Onge RP, Giaever G, Nislow C. Yeast chemical genomics and drug discovery: an update. Trends Pharmacol Sci. 2008;29(10):499–504. doi: 10.1016/j.tips.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, DeWald DB, Emr SD. Protein transport to the yeast vacuole. Curr Opin Cell Biol. 1995;7(4):544–551. doi: 10.1016/0955-0674(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Hu T, Kao CY, Hudson RT, Chen A, Draper RK. Inhibition of secretion by 1, 3-Cyclohexanebis(methylamine), a dibasic compound that interferes with coatomer function. Mol Biol Cell. 1999;10(4):921–933. doi: 10.1091/mbc.10.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell. 2002;13(9):3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6(15):1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Hudson RT, Draper RK. Interaction of coatomer with aminoglycoside antibiotics: evidence that coatomer has at least two dilysine binding sites. Mol Biol Cell. 1997;8(10):1901–1910. doi: 10.1091/mbc.8.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ. SAC1 Encodes a Regulated Lipid Phosphoinositide Phosphatase, Defects in Which Can Be Suppressed by the Homologous Inp52p and Inp53p Phosphatases. J Biol Chem. 2000;275(2):801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- Jacinto E. What controls TOR? IUBMB Life. 2008;60(8):483–496. doi: 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bankaitis VA, Emr SD. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987;48(5):875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Jones EW. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW, Webb GC, Hiller MA. Molecular biology of the yeast Saccharomyces cerevisiae. vol 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 363–469. [Google Scholar]
- Ka M, Park Y, Kim J. The DEAD-box RNA helicase, Dhh1, functions in mating by regulating Ste12 translation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2008;367(3):680–686. doi: 10.1016/j.bbrc.2007.12.169. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3(12):893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162(3):413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren R, Rainis L, Kleinberger T. The scaffolding A/Tpd3 subunit and high phosphatase activity are dispensable for Cdc55 function in the Saccharomyces cerevisiae spindle checkpoint and in cytokinesis. J Biol Chem. 2004;279(47):48598–48606. doi: 10.1074/jbc.M409359200. [DOI] [PubMed] [Google Scholar]
- Kuranda K, Leberre V, Sokol S, Palamarczyk G, Franc¸ois J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol. 2006;61(5):1147–1166. doi: 10.1111/j.1365-2958.2006.05300.x. [DOI] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69(2):262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793(4):650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi R, Riezman H. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J Biol Chem. 2001;276(8):6016–6022. doi: 10.1074/jbc.M008735200. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Bright NA, Pryor PR. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem Soc Trans. 2007;35(Pt 5):1088–1091. doi: 10.1042/BST0351088. [DOI] [PubMed] [Google Scholar]
- Martínez-Muñoz GA, Kane P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem. 2008;283(29):20309–20319. doi: 10.1074/jbc.M710470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaha SM, Champney WS. Hygromycin B inhibition of protein synthesis and ribosome biogenesis in Escherichia coli. Antimicrob Agents Chemother. 2007;51(2):591–596. doi: 10.1128/AAC.01116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ. Autophagy and vacuole homeostasis: a case for self-degradation? Autophagy. 2007;3(5):417–421. doi: 10.4161/auto.4441. [DOI] [PubMed] [Google Scholar]
- Moehle CM, Aynardi MW, Kolodny MR, Park FJ, Jones EW. Protease B of Saccharomyces cerevisiae: Isolation and regulation of the PRB1 structural gene. Genetics. 1986;115:255–263. doi: 10.1093/genetics/115.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniaux N, Nemos C, Schmied BM, Chauhan SC, Deb S, Morikane K, Choudhury A, Vanlith M, Sutherlin M, Sikela JM, Hollings-worth MA, Btra SK. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25(23):3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- Moriya H, Isono K. Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast. 1999;15(6):481–496. doi: 10.1002/(SICI)1097-0061(199904)15:6<481::AID-YEA391>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Kallay L, Brett CL, Rao R. Mutational analysis of the intramembranous H10 loop of yeast Nhx1 reveals a critical role in ion homoeostasis and vesicle trafficking. Biochem J. 2006;398(1):97–105. doi: 10.1042/BJ20060388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127(2):373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12(7):2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269(14):10185–10188. [PubMed] [Google Scholar]
- Ostrowicz CW, Meiringer CTA, Ungermann C. Yeast vacuole fusion: a model system for eukaryotic endomembrane dynamics. Autophagy. 2008;4(1):5–19. doi: 10.4161/auto.5054. [DOI] [PubMed] [Google Scholar]
- Park I, Erbay E, Nuzzi P, Chen J. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp Cell Res. 2005;309(1):211–219. doi: 10.1016/j.yexcr.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost R, Ho C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126(3):611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Insights from yeast endosomes. Curr Opin Cell Biol. 2002;14(4):454–462. doi: 10.1016/s0955-0674(02)00352-6. [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SE, Washburn TM, Chang M, Jaehning JA. The yeast pafl-rNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot Cell. 2002;1(5):830–842. doi: 10.1128/EC.1.5.830-842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3(12):1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996;7(11):166716–166777. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 2006;281(42):31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- Ripmaster TL, Vaughn GP, Woolford JL. DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(12):7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8(11):4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K-H, Sheehan A, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402(6760):413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47(6):1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260(5104):88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Seeley ES, Kato M, Margolis N, Wickner W, Eitzen G. Genomic analysis of homotypic vacuole fusion. Mol Biol Cell. 2002;13(3):782–794. doi: 10.1091/mbc.01-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell JL, Kahn RA. Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc Natl Acad Sci USA. 1988;85(13):4620–4624. doi: 10.1073/pnas.85.13.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16(2):669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17(3):1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771(3):353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7(10):1819–1830. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MK, Frost C, Oyadomari K, Pinho M, Sao D, Chima-Okereke O, Gharakhanian E. A novel immunodetection screen for vacuolar defects identifies a unique allele of VPS35 in S. cerevisiae. Mol Cell Biochem. 2008;311(1–2):121–136. doi: 10.1007/s11010-008-9703-y. [DOI] [PubMed] [Google Scholar]
- Thumm M. Structure and function of the yeast vacuole and its role in autophagy. Microsc Res Tech. 2000;51(6):563–572. doi: 10.1002/1097-0029(20001215)51:6<563::AID-JEMT6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Toikkanen J, Gatti E, Takei K, Saloheimo M, Olkkonen VM, Söderlund H, De Camilli P, Keranen S. Yeast protein translocation complex: isolation of two genes SEB1 and SEB2 encoding proteins homologous to the Sec61 beta subunit. Yeast. 1996;12(5):425–438. doi: 10.1002/(SICI)1097-0061(199604)12:5%3C425::AID-YEA924%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(11):4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267(26):18665–18670. [PubMed] [Google Scholar]
- Wagner MC, Molnar EE, Molitoris BA, Goebl MG. Loss of the homotypic fusion and vacuole protein sorting or Golgi-associated retrograde protein vesicle tethering complexes results in gentamicin sensitivity in the yeast Saccharomyces cerevisiae. Antimicrob Agents Chemother. 2006;50(2):587–595. doi: 10.1128/AAC.50.2.587-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(2):620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69(1):277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Bacallao R, Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J Cell Biol. 1987;105(4):1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J Cell Biol. 2002;158(4):761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]