Abstract
Motile cells regulate their shape and movements largely by remodeling the actin cytoskeleton. Principles of this regulation are becoming clear for simple-shaped steadily crawling cells, such as fish keratocytes. In particular, the shape of the leading edge and sides of the lamellipodium—cell motile appendage—is determined by graded actin distribution at the cell boundary, so that the denser actin network at the front grows, while sparser actin filaments at the sides are stalled by membrane tension. Shaping of the cell rear is less understood. Here we theoretically examine the hypothesis that the cell rear is shaped by the disassembly clock: the front-to-rear lamellipodial width is defined by the time needed for the actin-adhesion network to disassemble to the point at which the membrane tension can crush this network. We demonstrate that the theory predicts the observed cell shapes. Furthermore, turning of the cells can be explained by biases in the actin distribution. We discuss experimental implications of this hypothesis.
1. Introduction
Cell motility depends on a complex dynamic change of cell shape underlined by remodeling of the cytoskeleton coupled to the substrate (Ridley et al 2003). Cell shape also affects cell fate in cell death (Chen et al 1997), development (Nelson 2003) and tumor growth (Vasiliev 2004). Diverse motile behavior of different cell types results in a great variety of shapes (Ridley et al 2003). Molecular components determining these shapes, including the actin–myosin cytoskeleton, cell membrane and adhesions, are well known, but respective molecular mechanisms are not completely clear. The complex questions of how does cell morphology emerge from interactions between these cytoskeletal components and what does cell shape reveal about how a cell moves can be addressed first by studying simple-shaped rapidly crawling cells.
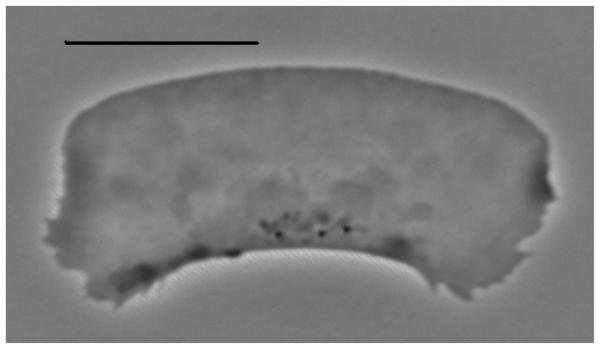
One of these, the half-moon-shaped fish keratocyte (Rafelski and Theriot 2004), is characterized by a flat, fan-shaped lamellipodium at the front and sides and a bulbous cell body in the rear. As they glide on a substrate, keratocytes maintain nearly constant speed and direction over many cell lengths (Euteneuer and Schliwa 1984). The lamellipodium is a broad (tens of microns wide) and flat (about 0.1–0.2 μm thick) motile appendage of the cell (cell body is but a passive cargo mechanically) that consists of a branched network of short actin filaments (Pollard and Borisy 2003) enveloped by the cell membrane. The actin network treadmills: nascent filaments branch off the existent filaments at the leading edge and sides of the lamellipodium and grow forward, advancing the cell boundary until they are capped. This branching and growth is localized to the leading edge/sides, while the actin network disassembles across the lamellipodium (Theriot and Mitchison 1991). The actin network firmly adheres to the substrate (Theriot and Mitchison 1991), so the treadmilling of the actin arrays is translated into the forward translocation of the cell. Importantly, lamellipodial fragments are able to move much like the whole cell while preserving the stereotypical keratocyte shape (Verkhovsky et al 1999) (figure 1). In this paper, we examine the shape of such a fragment.
Figure 1.
Lamellipodial fragment shape. Phase contrast image of polarized locomoting lamellipodial fragment of fish keratocyte cell. Bar, 10 μm. (Image reproduced with permission, courtesy of K Keren and N Ofer (Technion).)
A simple geometric principle—the graded radial extension (GRE) model (Lee et al 1993)—explains how the shape of the leading edge and sides is regulated (figure 2). The GRE model is based on experimental observations that local extension of the cell boundary is perpendicular to the cell's edge. To maintain a steady shape, the magnitude of extension must be graded, from a maximum at the center of the leading edge to zero at the sides (figure 2). More specifically, if the local rate of extension is equal to v, while the steady cell speed is equal to V, then the local normal direction to the cell boundary has to be tilted by the angle θ relative to the direction of movement, so that cos θ = v/V. Indeed, if the boundary extends by the increment vΔt in the locally normal direction, this extension translates into the forward advancement by (v/ cos θ)Δt, which has to be equal to VΔt in order to maintain the steady shape and speed. According to this simple trigonometric formula, this graded protrusion rate defines the cell's shape.
Figure 2.
Model of the lamellipodial shape. Top: the lamellipodial boundary consists of the curved leading and rear edges and straight sides parallel to the movement direction. The leading edge shape is determined by the GRE model (see the text). The rear edge shape is determined by the front–rear distance Y set by the disassembly clock. C, S and R denote the front center, front side and rear side of the lamellipodium. Bottom: the growing actin barbed end density, B, as the function of the arc length along the leading/side edge, s. At the rear side, the density is equal to zero. The front side is located where the actin density is low enough so that the filaments are stalled.
The question of how the cell regulates this protrusion rate along its boundary was answered in Grimm et al (2003) and Lacayo et al (2007). The answer is based on the observation that the density of the actin filaments along the leading edge is graded—maximal at the center and minimal at the sides (figure 2). According to the model proposed in Keren et al (2008), polymerizing actin filaments push the cell membrane from within, generating tension which rapidly equilibrates across the lipid bilayer (Raucher and Sheetz 2000). At the center of the leading edge, where actin filament density is high, the membrane resistance per filament is small, allowing filaments to grow rapidly and generate protrusion. As filament density gradually decreases toward the cell sides, the load force per filament due to membrane tension increases. As a result, local protrusion rates decrease smoothly from the center toward the sides of the leading edge, causing the leading edge to become curved as observed (figure 2). Actin polymerization is stalled for the cell at the sides, which therefore neither protrude nor retract.
How the rear of the cell is shaped is less clear. The observation that cell area is constant (Keren et al 2008) led to the model of the actin treadmill within an inextensible membrane bag (Keren et al 2008), according to which the cell area is simply determined by the total constant surface area of plasma membrane stretched around the lamellipodial actin network and tensed by the filaments pushing at the front and sides. At the rear of the cell, where the actin network disassembles, this membrane tension crushes the actin network weakened by the disassembly and moves actin debris forwards, thereby advancing the cell rear.
In this paper, we complement this model with the hypothesis that the force necessary to crush the actin network at the rear is proportional to the local network density. Essentially then, the disassembly of the network throughout the lamellipodium sets the clock that allows the network retraction after a certain time interval from the network assembly. We demonstrate that this hypothesis explains the observed shapes of the motile lamellipodial fragments.
2. Mathematical model of the cell shape
The leading edge shape can be explained by the model suggested in Grimm et al (2003), Lacayo et al (2007) and Keren et al (2008). Along the leading edge, the growing actin filaments compete for resources (the molecular identity of which is unknown, but Arp2/3 and/or VASP may be involved) to branch out nascent filaments, while existing filaments get capped and lag behind the protruding edge. At the rear corners of the cell, the density of actin filaments is damped, perhaps by large adhesion complexes there that appear to compete with actin polymerization processes for some molecular resources (the identity of which is, again, unknown; VASP is a likely candidate (Lacayo et al 2007)). Filaments at the center of the leading edge may out-compete filaments at the sides because they are not inhibited by the adhesions at the sides, so the actin density is peaked at the center. Mathematical analysis of these processes yielded an inverted parabolic actin filament distribution (figure 2), also observed experimentally:
| (1) |
Here s is the arc length coordinate along the leading edge and sides, so that s = 0 at the center of the leading edge (C in figure 2), s = S at the side of the leading edge (S in figure 2), and s = L at the rear corner of the lamellipodium (R in figure 2), where the density of the growing filaments pushing the boundary is assumed to be equal to zero. The total number of the growing barbed ends along the leading edge and sides is B̄.
Next, according to the model (Keren et al 2008), an actin filament grows with rate V = V0(1 − (f/fS)w) against the force f. Here fS is the stall force at which the filament stops growing, w is the parameter characterizing the shape of the force–velocity relation and V0 is the free polymerization rate. Assuming that the constant membrane tension (force per unit length of the edge) resists actin growth and that the membrane force is shared equally by local actin arrays, the force per filament is inversely proportional to the actin density, f = T/B. Substituting these formulae into (1), we derive the local extension rate of the leading edge:
| (2) |
Then, the shape of the leading edge is determined by the GRE formula (figure 2):
| (3) |
where the cell speed is defined as
| (4) |
The length of the leading edge, 2S, is determined by the condition that the force per filament at the side of the leading edge is equal to the stall force:
| (5) |
At the sides of the lamellipodium (segment SR in figure 2), the actin density is so low that the force per filament is greater than the stall force, and we assume that at such forces the filaments do not grow. Then, according to the GRE model, the sides are straight and parallel to the direction of movement (figures 1 and 2). The recent data indicates that stalling for the actin network most likely means either buckling of individual filaments (Chaudhuri et al 2007) or stopping of the growth, but not depolymerization (Parekh et al 2005). What exactly is the mechanism of the stall, in fact, does not matter, as long as the stall force per filament is constant on average (Schaus and Borisy 2008).
Equations (1)-(5) are the same as those in the model of Keren et al (2008). The novel part of the cell shape model is based on three assumptions. First, we assume that the F-actin network in the lamellipodium disassembles at the constant rate γ. For any material point in the lamellipodium, this means that the filament density decreases with time exponentially, ~ exp[−γ t]. Because the cell moves steadily with speed V, this means that the filament density decreases exponentially with distance y to the rear from the leading edge, ~ exp[−γy/V]. Thus, if the front-to-rear width of the lamellipodium at the point with arc length coordinate s at the leading edge is Y (s) (figure 2), then the local filament density at the rear is B(s) exp[−γ Y (s)/V.
Second, we assume that at the rear edge the membrane tension is constant and equal to that at the front and sides. Third, the actin network is crushed by the membrane tension when the force per filament exceeds the constant breaking force, fB (see section 6). Together, these three assumptions lead to the equation B(s) exp[−γ Y (s)/V] fB = T, which leads to the expression for the lamellipodial front-to-rear width (figure 2):
| (6) |
Effectively, the disassembly rate sets the clock, which measures out the time, 1/γ, slightly altered by the logarithmic force-dependent factor, after which the actin network collapses. This time multiplied by the cell speed determines the lamellipodial front-to-rear width.
Equations (1)-(6), together with the assumptions that the lamellipodial area, A, is constant, determine the lamellipodial shape. The constant model parameters are area A, total number of growing actin filaments maintained by the cell B̄, free polymerization rate V0, stall force fS, breaking force fB, disassembly rate γ and force–velocity exponent w. In fact, as the model's analysis shows, the effective number of parameters is greatly reduced if √A is used as the unit of length, and the combination B̄ fS/ √A is used as the unit of force. Then, just three non-dimensional parameters—α = fB/ fS (ratio of the breaking to stall forces per filament), β = γ √A/V0 (ratio of the time in which the cell crawls one body length to the actin disassembly time) and w—determine the model behavior and cell shape.
A few aspects of the model are worth a brief discussion. First, according to equation (1), the density of the growing filaments at the rear corner of the cell (R in figure 2) is equal to zero. However, very near this corner at the rear edge, the density of the weakened disassembling actin network is equal to the ratio T/ fB. This is not a contradiction, other than the assumption that the narrow band of the growing pushing filaments is infinitely narrow, and the sides are precisely straight. In the model, the length of the sides is determined by the disassembly of the actin network in the vicinity of the rear corners of the cell. Note also that the actin density is observed to increase drastically near the rear (Svitkina et al 1997): however, we hypothesize that this increase is due to actin debris that do not contribute to the force balance at the rear edge.
Second, adhesion to the substrate, which is essential for translating the actin treadmill into forward translocation of the cell, can also be an important part of the disassembly clock. Indeed, it appears that the adhesion of the keratocytes is graded—stronger at the front and weaker at the rear (Lee and Jacobson 1997, Anderson and Cross 2000). One attractive hypothesis (Lee and Jacobson 1997) explaining this fact is that rapid assembly of integrin, talin, vinculin and other adhesion molecules takes place at the front followed by slower ageing of the adhesion complexes. This hypothesis is similar to the mathematical model (Choi et al 2008) predicting that the adhesions assemble into a maximal density at a very small, submicron distance behind the leading edge and then disassemble exponentially. In fact, very recent data (Digman et al 2009) suggests that the mature adhesions uncouple from the substrate and disappear together with the actin filaments to which they attach. Thus, the adhesions could disassemble with the same rate as the actin network, and their density could follow the same pattern ~ exp[−γ y/V] as that of actin: because of the cell translocation, the adhesions weaken by the time the cell rear catches up with them. So, it could be that the membrane tension has to be balanced by the force of removing these adhesions. The mathematical formalism of the model would not change in this case.
Third, molecular processes of the actin network breaking are largely unknown. A force of the order of 100 pN can break one filament in two (Tsuda et al 1996) by torsion, but a smaller force in the range of tens of pN can break a filament by bending. Actin crosslinks (Delatour et al 2008) and actin-Arp2/3 bonds (Fujiwara et al 2002) detach rapidly (under a second) when a force of ~10 pN is applied. A nN-range force is sufficient to ‘peel’ the adhesions of hundreds of filaments off the substrate (Ra et al 1999), which translates into ~10 pN per filament to break its adhesion. Finally, perhaps the most relevant data is the observation and modeling of the fracture of actin gel (growing around a plastic bead) is reminiscent of the fracture of a brittle elastic material (van der Gucht et al 2005). Energy needed to break a filament or an inter-filament crosslink was estimated as ~10kBT in this study, which corresponds to the effective breaking force of 4–40 pN per filament (10kBT ≈ 40 pN × nm divided by effective bond breaking deformation ~1–10 nm). Thus, we assume in the model that the network breaking means either filaments' breaking, or crosslink or adhesion detaching or, most likely, a combination of all these processes, so that the resulting breaking force per filament is ~10 pN. The breaking force per filament does not have to be exactly constant, as far as the total such force is proportional to the average filament number. Finally, let us note that the exact processes responsible for the localization of the polymerization to the leading edge and delocalization of the depolymerization are not known (Pollard and Borisy 2003). Localization of signaling molecules such as Rho-family GTPases, PIP2 and SH3 adapter proteins (activating nucleation-promoting factors including WASp, N-WASP and Scar/WAVE, making them available to activate in turn Arp2/3 complex) to the curved plasma membrane at the leading edge is the key to the first part of this riddle. The second part depends on delocalization of ADF/cofilins accelerating F-actin disassembly and slow ageing of the filaments.
3. Predicted shape of the lamellipodium
We used equations (1)-(6) non-dimensionalized as described above to compute the lamellipodial shape using the following numerical iteration procedure: (1) choose some initial half arc length of the leading edge S and cell speed V ; (2) use formulae (5) and (6) to compute the membrane tension T and length of the sides, and then the parameter L; (3) use formulae (2) and (4) to compute corrected parameter V and function V (s); (4) compute the shape and the arc length of the protruding part of the leading edge; (5) use formulae (1) and (3) to compute the shape of the leading edge and the x coordinates of the sides ±Xside; (6) use formulae (6) to compute function Y (s); (7) compute the area using the formula ; (8) if the computed area is smaller than the given parameter A, increase the half arc length of the leading edge S, else decrease it; and (9) stop when the error of the area is smaller than 1%.
This iteration procedure converges rapidly, indicating local stability of the model cell shape. More elaborate investigation of the stability, in which the model was made dynamic (the leading edge evolved from an arbitrary shape in small time steps according to the GRE model, while the actin density inside the lamellipodium was decreasing exponentially, and the rear followed the level curve of the critical breaking density) also confirmed the stability of the cell shape (data not shown).
The numerical solution produced shapes shown in figure 3 and their dependence on three non-dimensional model parameters. The general feature of the predicted shape—lamellipodium looking like a bent rectangle—was reproduced by the model. The force–velocity exponent w was estimated to be between 1 and 10 (Prass et al 2006, Keren et al 2008). Figure 3(a) illustrates that the exact value of this parameter does not affect the cell shape significantly. Figure 3(b) shows that the cell shape is not very sensitive to the ratio of the breaking to stall forces per filament either, though when this ratio is closer to 1 (it has to be greater than 1, otherwise the filaments break before they are stalled, which is very unlikely), the rear edge becomes convex up, rather than concave up. Finally, figure 3(c) demonstrates that the cell shape is sensitive to parameter β, so the rate of disassembly of the actin network, as well as the cell area and speed are important factors determining the cell shape. The parameters tried give the aspect ratio of the shape (see below) close to 2, which is the average observed value (Keren et al 2008). The model makes the following prediction testable in the future: the radius of curvature of the rear edge of the lamellipodial fragment is greater than or equal to that of the leading edge.
Figure 3.
The lamellipodial shapes predicted by the model. For all shapes, the total constant lamellipodial area A is the unit of area, and the unit of length is equal to . (a) Two shapes characterized by the exponents of the force–velocity relation w = 2, 6; α = 3, β = 2.5 (the leading and rear edges are flatter for higher values of w). (b) Three shapes characterized by the breaking/stall force ratios α = 2, 3, 4; β = 2.5, w = 6 (the higher the ratio, the greater the front-to-rear width of the lamellipodium). (c) Three shapes characterized by the non-dimensionalized disassembly rates β = 1.5, 2.5, 3.5; α = 3, w = 6 (the greater the disassembly rate, the smaller the front-to-rear width of the lamellipodium).
4. Approximate analytical results for the lamellipodial shape
Figure 3 demonstrates that in the wide range of parameters (in fact, the only requirement is that w ⪢ 1, which is the case) the lamellipodium has roughly a rectangular shape. Also, for the realistic parameter values, V ≈ V0 (see also Keren et al 2008 for relevant discussions). This suggests the approximate equation for the aspect ratio of the lamellipodium. Indeed, because Bside fS = T and , where Y is the approximate front-to-rear width of the lamellipodium, then Y = (V0/γ) ln[fB/fS]. If X is the side-to-side lamellipodial length, then X = A/Y, and we predict the following aspect ratio of the lamellipodium:
| (7) |
This formula explains why the lamellipodial shape is not very sensitive to the ratio of the breaking to stall forces per filament: this ratio affects the shape through the slowly varying logarithmic factor. Equation (7) predicts that the aspect ratio increases with the membrane area (which agrees with the observations (Keren et al 2008)), with the rate of actin disassembly and decreases with cell speed. The last prediction does not agree with the data in Keren et al (2008), but it is possible that the actin disassembly rate is greater in faster moving cells.
5. Asymmetric actin density causes cell turning
The actin distribution along the leading edge can vary in time and space (Keren et al 2008), or it can be altered by external or intracellular signals. Therefore it is interesting to see how the shape and movement of the cell change if the actin filament density becomes skewed (figure 4). We investigated the consequences of the actin distribution along the leading edge and sides of the form
| (8) |
such that the maximum of the distribution is shifted to the left by the distance , where ε is a small non-dimensional parameter. In this case, the condition defining location of the cell sides remains B = Bside. Because the actin distribution is skewed to the left, the arc length coordinates of the front side corners also shift to the left, s ≈ ±S − δ (figure 4), so the length of the left side, l1, becomes shorter than that of the right side, l2 (figure 4).
Figure 4.
Cell turning. Top: computed shape of the cell turning to the left. The cell sides become radial arcs; the right side of the cell slightly lags behind the left side. The total constant lamellipodial area A is the unit of area, and the unit of length is equal to . Bottom: the F-actin distribution skewed to the left causing the lamellipodial deformation and left turning shown at the top. The stalling condition at the front sides and zero actin density condition at the rear sides determine the arc lengths of the sides and leading edge. Explanations of the notations are in the text.
According to the disassembly clock hypothesis, the times that pass from the actin assembly at the front sides of the cell to disassembly of these actin arrays at the rear corners of the lamellipodium have to be equal, because the actin densities at the front sides are equal. If the cell continues to move steadily along the straight path, this is impossible: l1/V ≠ l2/V. The only steady state solution in this case corresponds to the cell moving along a curvy path (curving to the left with radius of curvature R) with angular velocity ω = V/R. Then, because the left and right sides move with linear velocities ωR and ω(R + l), respectively, the times of disassembly along the left and right sides are equal if . (Here l is the lamellipodial length, see figure 4.) Thus, l1(R + l) = l2R and the radius of curvature of the cell path is given by the formula
| (9) |
To find this radius and the cell shape, we consider for simplicity the case in which the cell aspect ratio is approximately equal to 2, and . In this symmetric (non-dimensional) case, . The shift of the arc length coordinates of the front side corners also to the left, s ≈ ±S − δ, can be found using the condition , which in the case of the asymmetric distribution (8) has the form . Approximate solution of this equation is , so . According to equation (9), the radius of curvature of the path along which the cell moves and the angular speed of the cell are
| (10) |
where χ is the shift of the maximum of the actin distribution from the center.
The shape of the cell in this case can be found by simply taking into account that the linear advancement rate along the curvy path of the point at the leading edge with coordinate s is approximately . Then, according to the GRE model, , and
| (11) |
The front-to-rear width of the lamellipodium can be found using the formula
| (12) |
Using equations (2), (4), (8), (11) and (12), formula (10) for the radius of the left side of the cell (figure 4), and the iteration procedure described above, we obtained the asymmetric shape of the turning cell shown in figure 4. The right side of the cell turning to the left lags behind the left side, and the left side is shorter than the right one. The most advanced point of the leading edge is shifted to the left. One interesting possibility that has to be explored in the future is that the turning movement and the skewing of the F-actin distribution are involved in a feedback, making the turning behavior more stable and predictable.
6. Discussion
Here we explored mathematically the hypothesis that the actin network breaks when the force per filament exceeds a certain threshold. Assuming that the membrane tension at the lamellipodial rear is the source of the breaking force and that the actin network disassembles across the lamellipodium with a constant rate, we demonstrated that this hypothesis predicts the observed characteristic bent rectangular shapes of the keratocyte lamellipodia. Effectively, the disassembly rate sets the clock, which measures out the time after which the actin network collapses. This time multiplied by the cell speed determines the lamellipodial front-to-rear width. The model makes the testable prediction: the radius of curvature of the rear edge of the lamellipodial fragment is greater than or equal to that of the leading edge. Furthermore, we demonstrated that, if the F-actin distribution at the leading edge becomes skewed, asymmetric, then the cell, according to the model, will turn in the direction of the maximum of the F-actin density. This prediction can be tested in the future, because the F-actin density fluctuates significantly both in space and in time due to natural stochasticity of the branching and capping processes (Keren et al 2008). Perhaps more importantly, this prediction is relevant to significant recent interest in mechanisms of cell changing direction of migration in response to environmental signals through remodeling of the actin cytoskeleton (Sidani et al 2007, Wessels et al 2007).
The model we explored here has a number of limitations, the main one of which is the absence of a myosin-induced contractile stress at the rear of the lamellipodium that causes a centripetal flow of the actin network (Schaub et al 2007). Myosin does not seem to be crucial for movement: keratocytes continue to move after it is inhibited, albeit slower and with less regular shapes (Keren et al 2008). However, myosin that is swept to the rear as the cell moves forward helps to contract the actin network weakened by depolymerization, probably contributing to pulling the cell body forward (Svitkina et al 1997), as well as to F-actin disassembly accelerating the actin treadmill, and to pulling inward the cell sides, containing their spread. Most importantly, myosin-induced forces and movements seem to contribute significantly to the cell turning behavior (Barnhart 2009). The model will have to be tested by correlating time series for the angular velocity, cell shape asymmetry and F-actin distribution asymmetry in the case of the myosin-inhibited cells. Then, the same correlations have to be re-examined in addition to time series of the myosin distribution, and the model will have to be expanded to include the myosin effects. Another limitation stems from recent observations that the disassembly rate varies across the lamellipodium (Schaub et al 2007). Also, we have yet to investigate what effect could the membrane-associated proteins, as well as the adhesion complexes at the ventral surface, have on the tension distribution across the plasma membrane. The model conclusions would remain valid as far as the membrane tension equilibrates rapidly (on the 10 s scale or faster) and does not vary more than an order of magnitude spatially.
An alternative possibility for the rear edge shaping mechanism is that the transverse actin bundle that lies along the rear is under tension. Then, the radius of curvature, r, of the rear edge would be determined by the Laplace law: r = F/T, where F is the tensile force in the bundle and T is the membrane tension (force per unit length) (Bar-Ziv et al 1999, Kozlov and Mogilner 2007). The tensile force, very likely, depends on the myosin-generated contraction. Testing this possibility is not going to be easy, because this tensile force also depends on the strength of the large adhesions in the rear corners of the lamellipodium (Lacayo et al 2007)where the transverse actin bundle terminates. This strength, in turn, could depend on the myosin-generated contraction (Balaban et al 2001). It is also possible that a combination of the disassembly clock mechanism and actin–myosin bundle tension mechanism are responsible for the rear edge shape.
Last, but not least, the cell body may not be entirely passive, ‘riding’ on the treadmilling actin network (Anderson et al 1996). Further research on the mechanisms of the cell body mechanical coupling to the lamellipodium and comparison of the lamellipodial fragments lacking the cell body and whole cells will be needed to evaluate this factor for the cell shaping. Finally, it is worth noting that many of the more complex and slowly moving motile cells, such as fibroblasts, have long tails at the rear that seem to be stuck to the surface because of the large mature focal adhesions (Lewis et al 1982). Special mechanisms including directed endo/exocytosis and regulation of the dynamic graded adhesion are deployed by the cell to retract those tails (Ridley et al 2003). In such cells, it is unclear whether the membrane area and tension are the factors limiting motility, so the relation of our model to such tails is an open question. Furthermore, in three-dimensional motility through an extracellular matrix, recent data (Doyle et al 2009) indicates that the cell is less restrained by the membrane at the rear and relies more on the actin–myosin contraction to pull up the rear. In the future, modifying the model to remove these limitations and indeterminacies by comparison with quantitative data, and then testing whether the model is applicable to more complex motile cells will contribute to understanding of the molecular mechanisms of the cell migration.
Acknowledgments
We thank K Keren and J Theriot for fruitful discussions and K Keren and N Ofer for the image included in figure 1. This work was supported by NIH grant NIGMS U54 GM64346 and by NSF grant DMS-0315782 to AM.
References
- Anderson KI, Wang YL, Small JV. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J. Cell Biol. 1996;134:1209–18. doi: 10.1083/jcb.134.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KI, Cross R. Contact dynamics during keratocyte motility. Curr. Biol. 2000;10:253–60. doi: 10.1016/s0960-9822(00)00357-2. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Barnhart E. 2009. personal communication (Stanford)
- Bar-Ziv R, Tlusty T, Moses E, Safran SA, Bershadsky A. Pearling in cells: a clue to understanding cell shape. Proc. Natl Acad. Sci. USA. 1999;96:10140–5. doi: 10.1073/pnas.96.18.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–8. doi: 10.1038/nature05459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–50. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatour V, Helfer E, Didry D, Le KH, Gaucher JF, Carlier MF, Romet-Lemonne G. Arp2/3 controls the motile behavior of N-WASP-functionalized GUVs and modulates N-WASP surface distribution by mediating transient links with actin filaments. Biophys. J. 2008;94:4890–905. doi: 10.1529/biophysj.107.118653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Wiseman PW, Choi C, Horwitz AR, Gratton E. Stoichiometry of molecular complexes at adhesions in living cells. Proc. Natl Acad. Sci. USA. 2009;106:2170–5. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184:481–90. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Schliwa M. Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature. 1984;310:58–61. doi: 10.1038/310058a0. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Suetsugu S, Uemura S, Takenawa T, Ishiwata S. Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem. Biophys. Res. Commun. 2002;293:1550–5. doi: 10.1016/S0006-291X(02)00421-7. [DOI] [PubMed] [Google Scholar]
- Grimm HP, Verkhovsky AB, Mogilner A, Meister JJ. Analysis of actin dynamics at the leading edge of crawling cells: implications for the shape of keratocyte lamellipodia. Eur. Biophys. J. 2003;32:563–77. doi: 10.1007/s00249-003-0300-4. [DOI] [PubMed] [Google Scholar]
- Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453:475–80. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, Mogilner A. Model of polarization and bistability of cell fragments. Biophys. J. 2007;93:3811–9. doi: 10.1529/biophysj.107.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, Mogilner A, Theriot JA. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993;362:167–71. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- Lee J, Jacobson K. The composition and dynamics of cell-substratum adhesions in locomoting fish keratocytes. J. Cell Sci. 1997;110:2833–44. doi: 10.1242/jcs.110.22.2833. [DOI] [PubMed] [Google Scholar]
- Lewis L, Verna JM, Levinstone D, Sher S, Marek L, Bell E. The relationship of fibroblast translocations to cell morphology and stress fibre density. J. Cell Sci. 1982;53:21–36. doi: 10.1242/jcs.53.1.21. [DOI] [PubMed] [Google Scholar]
- Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–74. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nat. Cell Biol. 2005;7:1219–23. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 2006;174:767–72. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra HJ, Picart C, Feng H, Sweeney HL, Discher DE. Muscle cell peeling from micropatterned collagen: direct probing of focal and molecular properties of matrix adhesion. J. Cell Sci. 1999;112:1425–36. doi: 10.1242/jcs.112.10.1425. [DOI] [PubMed] [Google Scholar]
- Rafelski SM, Theriot JA. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 2004;73:209–39. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- Raucher D, Sheetz MP. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 2000;148:127–36. doi: 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Schaub S, Bohnet S, Laurent VM, Meister JJ, Verkhovsky AB. Comparative maps of motion and assembly of filamentous actin and myosin II in migrating cells. Mol. Biol. Cell. 2007;18:3723–32. doi: 10.1091/mbc.E06-09-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaus TE, Borisy GG. Performance of a population of independent filaments in lamellipodial protrusion. Biophys. J. 2008;95:1393–411. doi: 10.1529/biophysj.107.125005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, Wang W, Kuhl S, El-Sibai M, Backer JM, Eddy R, Soll D, Condeelis J. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 2007;179:777–91. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin–myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–31. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin–actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc. Natl Acad. Sci. USA. 1996;93:12937–42. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Gucht J, Paluch E, Plastino J, Sykes C. Stress release drives symmetry breaking for actin-based movement. Proc. Natl Acad. Sci. USA. 2005;102:7847–52. doi: 10.1073/pnas.0502121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliev JM. Cytoskeletal mechanisms responsible for invasive migration of neoplastic cells. Int. J. Dev. Biol. 2004;48:425–39. doi: 10.1387/ijdb.041806jv. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Self-polarization and directional motility of cytoplasm. Curr. Biol. 1999;9:11–20. doi: 10.1016/s0960-9822(99)80042-6. [DOI] [PubMed] [Google Scholar]
- Wessels D, Lusche DF, Kuhl S, Heid P, Soll DR. PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J. Cell Sci. 2007;120:2517–31. doi: 10.1242/jcs.010876. [DOI] [PubMed] [Google Scholar]