Abstract
Wg/Wnt signals specify cell fates in both invertebrate and vertebrate embryos and maintain stem-cell populations in many adult tissues. Deregulation of the Wnt pathway can transform cells to a proliferative fate, leading to cancer. We have discovered that two Drosophila proteins that are crucial for cytokinesis have a second, largely independent, role in restricting activity of the Wnt pathway. The fly homolog of RacGAP1, Tumbleweed (Tum)/RacGAP50C, and its binding partner, the kinesin-like protein Pavarotti (Pav), negatively regulate Wnt activity in fly embryos and in cultured mammalian cells. Unlike many known regulators of the Wnt pathway, these molecules do not affect stabilization of Arm/β-catenin (βcat), the principal effector molecule in Wnt signal transduction. Rather, they appear to act downstream of βcat stabilization to control target-gene transcription. Both Tum and Pav accumulate in the nuclei of interphase cells, a location that is spatially distinct from their cleavage-furrow localization during cytokinesis. We show that this nuclear localization is essential for their role in Wnt regulation. Thus, we have identified two modulators of the Wnt pathway that have shared functions in cell division, which hints at a possible link between cytokinesis and Wnt activity during tumorigenesis.
Keywords: Tum, RacGAP, Pav, Wg, Wnt, Drosophila, Embryo, Signal transduction
Introduction
The Wnt family of growth factors are highly conserved throughout the animal kingdom (for a review, see Logan and Nusse, 2004). In vertebrates, Wnt proteins promote a variety of cell-fate decisions during embryogenesis, whereas excess Wnt-pathway activity in adult tissues is associated with certain cancers (for a review, see Polakis, 2000). The Drosophila Wnt protein, encoded by wingless (wg), also directs embryonic cell-fate decisions, and excess Wg activity in flies produces characteristic cell-fate transformations that are easily detected (for a review, see Bejsovec, 2006). The cell-surface components and intracellular components of the Wnt pathway are highly conserved between vertebrates and Drosophila, and many were first identified through mutational analysis of Drosophila embryonic epidermal patterning.
At the end of fly embryogenesis, epidermal cells secrete a cuticle layer decorated with stereotyped pattern elements. On the ventral surface, segmentally repeating belts of small hooks, called denticles, are interspersed with naked cuticle. wg, expressed in a stripe of cells in each segment, directs naked-cuticle specification: wg loss of function produces a lawn of denticles, whereas wg gain of function generates excess naked cuticle at the expense of denticles (Hays et al., 1997; Noordermeer et al., 1992) and ectopic naked cuticle in other patterning mutants consistently correlates with hyperactivity of the Wg pathway (Bejsovec and Wieschaus, 1993; Peifer et al., 1994; Siegfried et al., 1992). Indeed, mutations in the fly homolog of the human tumor suppressor Apc were identified based on their naked-cuticle phenotype (McCartney et al., 1999). Similar genetic screens have yielded mutations in other negative regulators of Wg/Wnt signaling, such as the transcription factor SoxN (Chao et al., 2007) and the fly RacGAP1 homolog, tumbleweed (tum), also known as RacGAP50C (Jones and Bejsovec, 2005).
Our current understanding of the Wnt pathway hinges on regulation of β-catenin (βcat), which is called Armadillo (Arm) in the fly (reviewed by Bejsovec, 2006; Jones and Bejsovec, 2003; Peifer and Polakis, 2000). In the absence of Wnt signaling, βcat levels are kept low by a set of proteins known collectively as the destruction complex. These proteins, which include glycogen synthase kinase 3β (GSK), called Zeste-white3 (Zw3) in the fly, and the scaffolding proteins Axin and Apc, target Arm/βcat for destruction via ubiquitylation. When Wg/Wnt binds to its receptor complex, which consists of Frizzled and Arrow (LRP5 and LRP6 in vertebrates), this inactivates the destruction complex and allows βcat to accumulate. In the simplest view, βcat accumulation drives its interaction with the HMG-box transcription factors Tcf or Lef in the nucleus. Tcf can bind DNA in the absence of βcat, repressing Wnt target genes in conjunction with the transcriptional co-repressor Groucho (Gro/TLE1) (Cavallo et al., 1998). When βcat binds to Tcf, it displaces Gro and recruits other proteins such as Legless (Lgs/BCL-9) and Pygopus (Pygo) to form an activation complex that promotes expression of Wnt target genes (Brunner et al., 1997; Daniels and Weis, 2005; Kramps et al., 2002; van de Wetering et al., 1997).
Our discovery that Tum has a role in negatively regulating activity of the Wg pathway (Jones and Bejsovec, 2005) was surprising because Tum is known to be essential for positioning the contractile ring during cytokinesis. RNA interference of Tum, or of the human homolog, MgcRacGAP with which Tum shares 34% identity, leads to binucleated and multinucleated cells in culture (Minoshima et al., 2003; Mishima et al., 2002; Somers and Saint, 2003). Likewise, loss-of-function tum mutations in Drosophila embryos lead to binucleate cells at late stages of embryogenesis, when the maternal contribution of Tum is depleted (Jones and Bejsovec, 2005; Zavortink et al., 2005). Tum/MgcRacGAP binds to the kinesin-like protein Pav/mitotic kinesin-like protein 1 (MKLP-1) to form an evolutionarily conserved complex, centralspindlin, which bundles microtubules (Mishima et al., 2002) and localizes to the central spindle during division (Hirose et al., 2001; Somers and Saint, 2003). During all stages of the cell cycle, Tum/MgcRacGAP and Pav/MKLP-1 colocalize, and physical association between the two proteins is detected (Somers and Saint, 2003; Zhao and Fang, 2005). Like Drosophila tum mutants, pav mutants accumulate binucleate cells because of failure in cytokinesis at stage 11 (Adams et al., 1998). Thus, for both Tum and Pav, perduring maternal gene product is sufficient for normal cell division through all but the last postblastoderm division in fly embryos. Because there is no further cell division during normal embryogenesis, the late-stage Drosophila embryo provides us with a system in which to monitor the effects of Tum and Pav on Wg-mediated events without interference from their pleiotropic effects on cytokinesis. Indeed, we have found conditions under which the tum mutant pattern defect can be rescued without rescue of the binucleate-cell phenotype, indicating that these functions of Tum are separable (Jones and Bejsovec, 2005). Here, we report that Tum and Pav act in the nucleus to exert control over the Wg pathway in Drosophila and the Wnt pathway in mammalian cells in culture.
Results
Tum and its binding partner Pav modulate Wg-mediated embryonic patterning
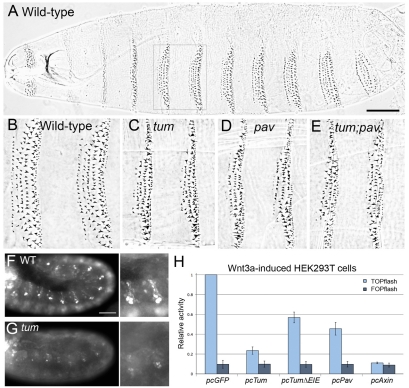
Mutations disrupting Drosophila tum were isolated in a genetic screen for modifiers of wg mutant phenotypes (Jones and Bejsovec, 2005). tum mutants show excess specification of naked cuticle (Fig. 1C) when compared with the wild-type epidermal pattern (Fig. 1A,B), consistent with a negative regulatory role for Tum in Wg signaling. To understand how Tum functions in Wnt repression, we performed a yeast two-hybrid screen for Tum-interacting proteins. Among the interactors, we recovered several isolates of pavarotti cDNAs as well as cDNAs encoding other cytokinesis-related proteins, such as Anillin (Gregory et al., 2008). We expected to recover pav because its yeast two-hybrid interaction with Tum has been well characterized and the Pav-binding domain within the N-terminal region of Tum has been described (Somers and Saint, 2003). What we did not expect was that pav, like tum, also modulated Wg signaling. Loss-of-function mutations in the fly pav gene are known to result in embryonic lethality with cell division defects in late stage embryos (Adams et al., 1998). We found that pav mutant embryos also had epidermal pattern defects similar to those seen in tumAR2 mutant embryos (Fig. 1C,D). This phenotype was unchanged when the pavB200 mutation was placed in trans with a deficiency removing pav (Table 1). Thus, Pav, like Tum, is required for negative regulation of Wg-pathway activity. To test for additive effects, we generated tumAR2; pavB200 double-mutant embryos, but observed no significant difference from the tum and pav single-mutant phenotypes (Fig. 1E; Table 1). These data suggest that Tum and Pav work together, possibly in a complex, and that both are required to properly regulate patterning in embryos.
Fig. 1.
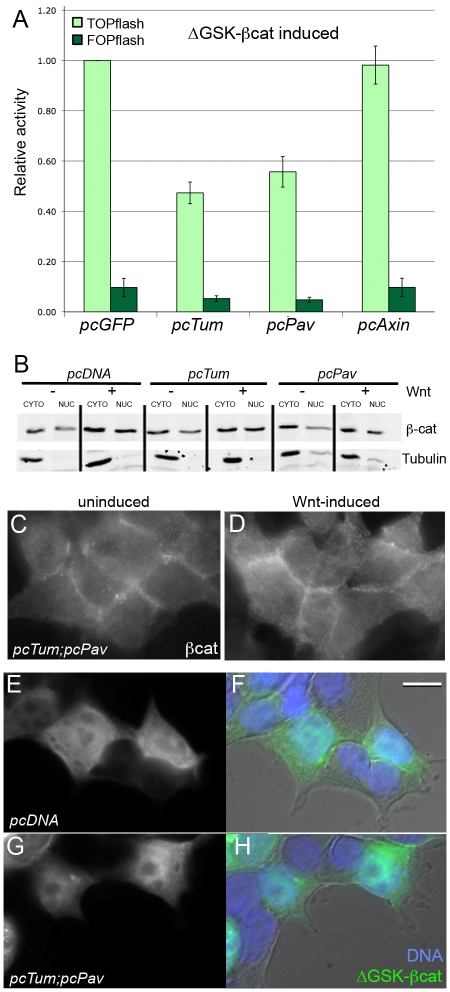
Tum and Pav control epidermal patterning in the Drosophila embryo and repress Wnt-induced TOPflash activity in HEK293T cells. (A) Wild-type embryos secrete a cuticle pattern with expanses of naked cuticle separating ventral denticle belts. (B) Higher magnification of ventral abdominal segments 2 and 3 indicated by box in A. Anterior is to the left in these and all subsequent panels. (C,D) Representative phenotypes of (C) tumAR2 and (D) pavB200 homozygous mutants show a reduced number of denticle rows in each belt compared with wild-type (B). (E) The most severe phenotypes observed among embryos segregating from the tum/+;pav/+ self-cross are not substantially different from either tum or pav single mutant phenotypes (see Table 1). Scale bar in A: 100 μm (A), 50 μm (for B-E). (F) rhomboid-lacZ marker is expressed in segmental stripes in the late epidermis. (G) tum mutant embryos show repression of this rho-lacZ expression. A GFP-marked balancer was used to distinguish homozygous mutants from their wild-type siblings (n>100 embryos at or later than stage 11). Note that rhomboid expression in the head (left in F and G) was detected at similar levels in both tum mutants and their wild-type siblings, providing an internal control for staining intensity. Scale bar: 50 μm (F,G); 25 μm (for zooms). (H) Tum represses TOPflash activation to 23% of pcGFP controls (P=0.001), when HEK293T cells are induced by Wnt-conditioned medium. Mutation of the GAP domain (pcTumΔEIE) does not eliminate the ability of Tum to repress TOPflash (P=0.008). Pav transfection represses activity to 45% of pcGFP controls (P=0.007). Axin, a known negative regulator of Wnt signaling, represses TOPflash activity to 11% of pcGFP (P<0.001). FOPflash values were not significantly altered by any of these treatments. Results are means ± s.d.
Table 1.
tum and pav loss-of-function phenotypes
Expression of engrailed (en) in cells posterior to the wg-expressing cells is a commonly used molecular marker for Wg signaling, and we have previously shown that neither wg nor en gene expression is altered significantly in tum mutant embryos (Jones and Bejsovec, 2005). Since maternal Tum permits normal cell division in zygotic mutants before stage 11, we reasoned that it could provide sufficient regulatory function during stages 8-10, the Wg-dependent phase of en gene regulation (Bejsovec and Martinez Arias, 1991; Heemskerk et al., 1991). We therefore examined the epidermal expression of rhomboid (rho), which is repressed by Wg signaling at stage 11 in each embryonic segment (Gritzan et al., 1999; Sanson et al., 1999). Using a rho-lacZ enhancer trap, we found that rho expression in the head region (Fig. 1F,G, left panels) was not affected by Wg activity, providing an internal control for staining intensity. By contrast, rho expression was reduced in the epidermis of tum homozygous mutants compared with wild-type embryos at the same stage (Fig. 1F,G). The reduction of epidermal rho is consistent with excessive Wg signaling in these cells, indicating that Tum function is required to restrict activity of the Wg pathway.
Tum and Pav repress Wnt signaling in a heterologous system
Remarkably, the fly Tum protein produces a similar negative regulation of Wnt signaling in mammalian cells in culture. We used the TOPflash assay system, which employs a luciferase reporter under the control of Tcf-optimal binding sites, to measure Wnt response compared with the basal transcription of FOPflash, a construct where the Tcf consensus sequences are mutated. This system has been used extensively to monitor the effects of fly components on the mammalian Wnt pathway (van de Wetering et al., 1997; Cavallo et al., 1998). The fly Axin, a negative regulator, produces strong repression of Wnt-induced TOPflash when overexpressed in human embryonic kidney (HEK293T) cells (Fig. 1H). Similarly, ectopic Tum was able to repress Wnt-induced reporter activity in HEK293T cells compared with control cells expressing green fluorescent protein (GFP) (Fig. 1H). Overexpression of Tum does not disrupt cell division or cell health, because we did not observe any binucleate cells nor any decrease in cell proliferation or increase in cell death in Tum-transfected cells compared with GFP-transfected cells (not shown). To test the possibility that Tum acts as a general repressor of transcription, we substituted the TOPflash reporter with pGL4.13 (Promega), which contains the SV40 early enhancer/promoter region upstream of the luciferase (luc2) reporter gene. This construct is not Wnt responsive and we observed no repression of luc2 expression in cells transfected with Tum compared with the empty vector (pcDNA3.1) control (data not shown). These data are consistent with our previous observations that tum mutants do not show generally deregulated gene expression patterns (Jones and Bejsovec, 2005).
The GAP activity of Tum is not essential for its Wnt repression function. ΔEIE is a deletion that removes three essential amino acids from the GAP domain: the mutant protein can still bind to Rac but cannot catalyze GTPase function (Sotillos and Campuzano, 2000). TumΔEIE was able to repress TOPflash activity, although to a lesser extent than full-length Tum (Fig. 1H). This is consistent with our previous result that TumΔEIE rescues pattern defects in tum mutant fly embryos (Jones and Bejsovec, 2005). By contrast, TumΔEIE was unable to rescue the cytokinesis defect of tum mutant embryos (Zavortink et al., 2005), showing that cytokinesis requires an intact GAP domain and further indicating that the Wnt repressor function of Tum is genetically distinct from its role in positioning the contractile ring during cell division.
In the initial analysis of Tum-interacting proteins found in our yeast two-hybrid screen, we tested each in the mammalian TOPflash system to determine whether any showed an ability to modulate Wnt response. We found that Pav, like Tum, has the ability to repress TOPflash induction in HEK293T cells (Fig. 1H), whereas other Tum-interactors in the cytoskeletal class, such as Anillin, do not (not shown). As with Tum, overexpression of Pav did not disrupt cell division in HEK293T cells. Repression by Drosophila Tum and Pav was observed whether cells were induced by treatment with Wnt-conditioned medium (Fig. 1H) or by cotransfection with a construct overexpressing wild-type βcat (not shown). Thus the negative regulatory functions of Tum and Pav are highly conserved, because the fly proteins are clearly able to control mammalian components in the Wnt signaling pathway. This degree of conservation has been observed for other transcriptional regulators of the Wnt pathway, such as Tcf, Groucho and SoxN (van de Wetering et al., 1997; Cavallo et al., 1998; Chao et al., 2007).
Tum and Pav act synergistically to regulate Wg/Wnt signaling
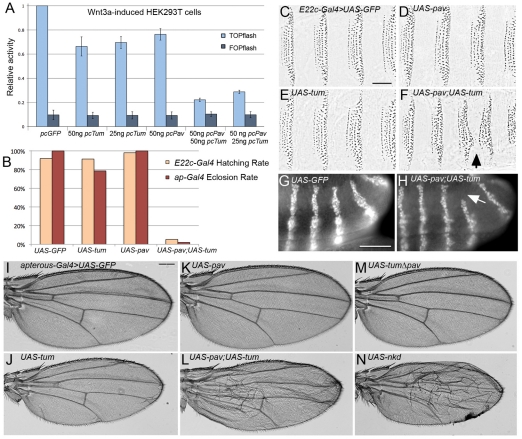
We titrated the Tum and Pav expression constructs in HEK293T cells to determine the threshold concentration at which each begins to repress TOPflash with statistical significance. Cotransfection of Tum and Pav at these low concentrations produced a more substantial repression than either molecule singly produced (Fig. 2A). This enhancement was not due to increased stability of Pav in the presence of Tum; we found no difference in steady state Pav protein levels in the presence or absence of ectopic Tum (not shown). Overexpression of either Tum or Pav alone in Drosophila embryos had little effect on viability or on epidermal patterning (Fig. 2B-E). However, driving Tum and Pav together in the developing Drosophila embryonic epidermis increased embryonic lethality (Fig. 2B), and caused reduction of the segmental naked cuticle expanse (Fig. 2F, arrow). This pattern disruption is diagnostic of decreased Wg signaling activity (Bejsovec and Wieschaus, 1993; Moline et al., 1999). Indeed, the penetrance of full denticle-belt fusions was increased from 22.5% (n=223) to 81.1% (n=132) when Tum and Pav were overexpressed in embryos heterozygous for a wg-null allele. Disruption of the wg target gene en was also detected in this sensitized background (Fig. 2G,H). These data show that the effects of Tum and Pav are sensitive to the dosage of Wg activity, and suggest that the lack of effect when each transgene is expressed singly in vivo is due to limited endogenous levels of the other binding partner.
Fig. 2.
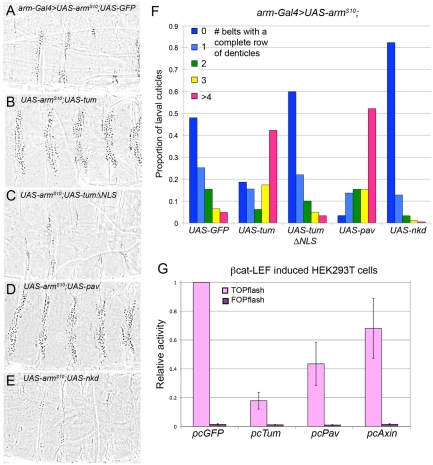
Tum and Pav act more strongly when coexpressed. (A) Minimal doses required for statistically significant repression are 25 ng Tum (P=0.01) and 50 ng Pav (P=0.02). Tum and Pav together at these doses repress TOPflash activation to 29% of vector alone controls (P=0.0002). This repression is also significantly different when compared with repression by Tum alone (P=0.005) or Pav alone (P=0.005). Results are means ± s.d. (B) Overexpression of Tum or Pav in the embryonic epidermis, with E22c-Gal4, or in the dorsal domain of wing imaginal discs, with apterous-Gal4, has no effect on embryonic hatching rate or adult eclosion rate respectively. Co-expressing Tum and Pav dramatically decreases both rates (n>200 fertilized eggs for each hatching experiment, n>100 balancer siblings from each ap-Gal4/CyO cross). E22c-Gal4-driven UAS-GFP (C), UAS-pav (D) or UAS-tum (E) does not affect epidermal patterning. Co-expression of both transgenes (F) variably reduces the naked cuticle expanse (arrow) to cause denticle belt fusions (22.5%, n=223). Penetrance of this segmentation defect is increased when embryos are heterozygous for the wgCX4-null allele (81.1%, n=132). (G) No disruption of the striped en expression is detected when UAS-GFP was driven in the wg heterozygous background (n>100). (H) In the sensitized wg heterozygous background, defects in the segmental expression of en (arrow) can be detected when UAS-pav and UAS-tum are coexpressed with E22c-Gal4 (29.8%, n=161). Without the sensitizing wg heterozygosity, virtually no defects in en expression can be detected when UAS-pav and UAS-tum are coexpressed (1.2%, n=160), indicating that cuticle pattern is a more sensitive assay for Wg activity. ap-Gal4-driven UAS-GFP does not affect wild-type wing patterning (I), whereas ap-Gal4-driven UAS-tum (J) consistently reduces the overall size of the wing. This phenotype is not shared by UAS-pav (K). ap-Gal4 driving both transgenes simultaneously killed most animals before or during pupation. (L) The few escapers (n=3) consistently show wings that are reduced in size and blistered. (M) ap-Gal4-driven UAS-tumΔpav does not reduce wing size. (N) ap-Gal4-driven UAS-nkd produces wing pattern defects similar to those observed with Tum-Pav coexpression (L). Scale bars: 50 μm (C-H); 100 μm (I-N).
We were unable to examine loss-of-function tum and pav phenotypes in later stages of Drosophila development: because both are essential for cell division, somatic clones of either mutant were not detected. Knockdown of tum expression with a double-stranded RNA construct severely disrupts cell division in the wing imaginal disc (Somers and Saint, 2003), deforming the wing profoundly so that patterning cannot be assessed. However, overexpression of Tum or Pav did not disrupt cell division in the wing, and we found that their activity in imaginal discs paralleled their effects on embryogenesis. We used the apterous (ap)-Gal4/CyO line to express high levels of UAS-transgene expression in the dorsal portion of the wing disc, and compared the rate of adult eclosion between Gal4-expressing progeny and their CyO siblings. Crosses with the single UAS-tum or UAS-pav transgenes showed no difference between progeny classes, but crosses with the doubly transgenic UAS-pav;UAS-tum line showed a profound decrease in eclosion rate (Fig. 2B). Overexpression of Tum in the wing consistently produced a decrease in the overall size of the wing (Fig. 2I,J). This effect was not observed with Pav overexpression (Fig. 2K), but did depend on the ability of Tum to interact with Pav because a mutant form of Tum that removes the Pav-binding domain (Zavortink et al., 2005) did not alter wing size (Fig. 2M). We are currently investigating the source of this size reduction. Despite the dramatic decrease in adult eclosion when Tum and Pav were co-expressed, a few flies did eclose. All three showed the same, more severe pattern disruption, which included blistering of the wing (Fig. 2L). This phenotype is strikingly similar to that observed when we overexpressed naked cuticle (nkd) (Fig. 2N), a known negative Wg regulator (Zeng et al., 2000) which interacts genetically with tum (Jones and Bejsovec, 2005). Thus we believe that at least some of the wing defect caused by misexpression of Tum and Pav might involve Wg-dependent pattern disruption.
Reduction of tum and pav activity suppresses a weak arm mutant phenotype
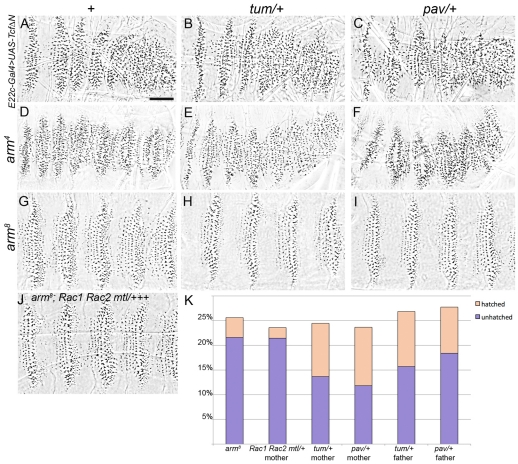
We next explored interactions of Tum and Pav with known components of the Wg signaling pathway. Epistasis analysis with Tcf mutations is complicated because this transcription factor can act as either activator or repressor, depending on availability of Arm (Cavallo et al., 1998). Therefore we used TcfΔN, which provides constitutive repressor function when overexpressed in the embryo (Cavallo et al., 1998). The elimination of naked cuticle observed in these E22c-Gal4>UAS-TcfΔN embryos was unchanged in either the tum or the pav mutant background (Fig. 3A-C), indicating that they do not act by promoting the repressor activity of Tcf. This result contrasts with mutations in another negative Wg/Wnt regulator, SoxNeuro, which acts at the transcriptional level and reduces the severity of E22c-Gal4>UAS-TcfΔN cuticle defects (Chao et al., 2007).
Fig. 3.
arm and Tcf loss-of-function phenotypes are epistatic to tum and pav. Driving TcfΔN with E22c-Gal4 represses Wg target genes and produces a ‘lawn of denticles’ phenotype (A) that is similar to the severe loss-of-function arm4 mutation (D). In crosses expected to generate 25% tum homozygous mutants (B) or pav mutants (C), no embryos show any alteration from the typical E22c-Gal4>UAS-TcfΔN mutant pattern (n>100 fertilized eggs for each), nor any change in the arm4 severe phenotype (E,F, respectively) (n>50 arm mutants for each). (G) The weak arm8 mutation retains sufficient Arm activity to generate proper denticle diversity but shows reduced naked cuticle separating the belts. Naked cuticle is consistently restored in crosses where the mothers are heterozygous for tum (H) or pav (I). (J) Mothers heterozygous for three Rac mutations do not produce altered arm8 mutant progeny. (K) Heterozygosity for tum or pav, but not Rac1 Rac2 mtl, increases the hatching rate of arm8 mutants, expected to represent 25% of the eggs produced in each cross (n>300 for each cross). In these crosses, only 50% of the arm8 embryos were expected to be heterozygous for the triple Rac mutant, the tum or the pav mutation. Scale bar: 50 μm.
Tum and Pav also do not appear to act in the absence of functional Arm/βcat. Severe arm mutations that eliminate zygotic arm gene product, such as arm4 (also known as armYD35), produce drastic pattern disruptions (Peifer and Wieschaus, 1990). The severe arm4 mutant phenotype (Fig. 3D) was not altered in either the tum or the pav mutant background (Fig. 3E-F). Weak arm mutations, such as arm8 (also known as armH8.6), provide enough Arm activity to generate denticle diversity in each segment, but lack the wide expanse of naked cuticle separating the denticle belts (Fig. 3G) and most fail to hatch (Fig. 3K). In this sensitized background, simply reducing the dose of tum or pav by half restored areas of naked cuticle between the belts to almost wild-type levels (Fig. 3H,I, compare with Fig. 1A), and many more arm8 mutants were able to hatch into the first instar stage (Fig. 3K). This effect largely results from reduced zygotic dose of Tum or Pav, rather than reduced maternal product, because we observed similar effects with heterozygous fathers (Fig. 3K). Since some functional Arm is required to observe the mutant effects of tum and pav, and these mutations have no effect on a constitutive-repressor form of Tcf, it is most likely that Tum and Pav act upstream of or at the level of Arm-dependent transcriptional activation. Recently, vertebrate Rac1 has been implicated in chaperoning βcat into the nucleus (Wu et al., 2008) and/or promoting Wnt target expression (Buongiorno et al., 2008). We wondered whether the ability of Tum to bind Rac might be relevant to the dosage effects we observed. We reduced the dose of Rac by introducing a triple-mutant chromosome bearing mutations in three Rac family members: Rac1, Rac2 and mtl (Hakeda-Suzuki et al., 2002). Heterozygosity for this triple-mutant chromosome did not alter arm8 mutant patterning (Fig. 3J), nor its rate of hatching (Fig. 3K). This is consistent with our previous work showing that manipulation of Rac activity did not mimic tum loss-of-function or gain-of-function phenotypes (Jones and Bejsovec, 2005).
Tum and Pav act downstream of βcat stabilization and nuclear translocation
In previous work we showed that Arm levels were elevated in tum mutant embryos, suggesting that Tum might promote Arm degradation. To test this hypothesis, we used a form of βcat that evades the destruction complex. In normal βcat, Ser33, Ser37, Ser45 and Thr41 are phosphorylated by GSK-3β and casein kinase1α (Ikeda et al., 1998; Kikuchi, 1999; Liu et al., 2002), creating a binding site for βTrCP, a component of the ubiquitin ligase pathway (Kitagawa et al., 1999; Liu et al., 2002). When all four residues are mutated to alanine, the mutant βcat (ΔGSK-βcat) cannot be targeted for degradation (Barth et al., 1999). Transfection of a construct encoding ΔGSK-βcat produced strong induction of TOPflash activity in control HEK293T cells transfected with empty pcDNA3.1 vector (not shown) or pcGFP (Fig. 4A). We were surprised to find that both Tum and Pav were able to antagonize this ΔGSK-βcat-induced reporter expression (Fig. 4A). By contrast, Axin overexpression failed to repress ΔGSK-βcat-induced reporter expression (Fig. 4A), as expected, because Axin provides a scaffold for the GSK-βcat interaction (Salahshor and Woodgett, 2005). Since Tum and Pav can repress ΔGSK-βcat-induced Wnt signaling, we conclude that their action cannot be explained exclusively through destruction complex modulation, and that our initial observations of elevated Arm accumulation (Jones and Bejsovec, 2005) might reflect a secondary consequence of pathway activation in tum mutants. Rather, Tum and Pav appear to act downstream of βcat stabilization. Furthermore, this result rules out other possible cytoplasmic and cell surface mechanisms for modulating the Wnt pathway, such as indirect effects on receptor recycling.
Fig. 4.
Pav and Tum do not alter β-cat stability or nuclear translocation. (A) TOPflash activity induced by the constitutively-active ΔGSK-βcat mutant is repressed by Tum (P=0.003) and Pav (P=0.009). Axin, a destruction complex component, was not able to repress ΔGSK-βcat (P>0.76). Results are means ± s.d. (B) Immunoblots show that βcat levels in the nuclear fraction of HEK293T cells increased in response to Wnt stimulation, as shown in the pcDNA3.1 empty vector control lanes. This pattern of Wnt-induced nuclear βcat accumulation is not altered in HEK293T cells transfected with either Tum or Pav. β-tubulin was used as a loading and fractionation control. (C) Uninduced and (D) Wnt-induced HEK293T cells that overexpress Tum and Pav show similar patterns of endogenous βcat distribution, with higher overall levels in the Wnt-induced cells. HEK293T cells transfected with ΔGSK-GFP-βcat and stained for GFP show high uniform βcat levels in the transfected cells (E), which have normal morphology compared with neighboring untransfected cells (F, brightfield image merged with fluorescent images). (G) Co-transfection with Tum and Pav does not alter the distribution of ΔGSK-GFP-βcat compared with the empty vector control, nor does it alter cell morphology (H). Scale bar: 10 μm (C-H).
We next asked whether Tum and Pav might regulate βcat levels in the cytoplasm versus the nucleus. Such a role has been postulated for Apc and Axin: in addition to their scaffolding role in the destruction complex, they elevate βcat levels in the cytoplasm by sequestering the protein (Krieghoff et al., 2006; Seo and Jho, 2007). We subjected HEK293T cell lysates to fractionation and assessed levels of βcat in the cytoplasm and in the nucleus. Ectopic Tum or Pav expression did not alter the level of βcat detected in the nuclear fraction of induced versus uninduced cells, compared with control cells (Fig. 4B). These results were corroborated by examining βcat protein distribution in situ. Transfection with Tum and Pav did not alter βcat localization in uninduced versus Wnt-induced cells (Fig. 4C,D), nor did it alter subcellular localization of the stabilized ΔGSK-βcat (Fig. E-H). Therefore, Tum and Pav do not sequester βcat in the cytoplasm, but rather must act downstream of nuclear βcat translocation to block activation of TOPflash reporter expression.
Tum and Pav act within the nucleus to repress Wnt target genes
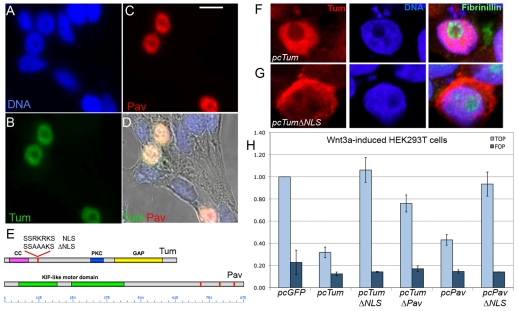
Although Tum and Pav act in the cytoplasm during cytokinesis, both molecules are known to accumulate in nuclei in the Drosophila embryonic epidermis (Guerin and Kramer, 2009; Minestrini et al., 2003; Minestrini et al., 2002) and in postmitotic Drosophila neurons (Goldstein et al., 2005), as well as during interphase in cultured insect cells (Somers and Saint, 2003). Likewise, their vertebrate homologs, MgcRacGAP and MKLP-1, colocalize in the interphase nuclei of HeLa cells (Hirose et al., 2001; Mishima et al., 2002). We verified that Tum and Pav proteins accumulated in the nuclei of interphase HEK293T cells (Fig. 5A-D). Antibody staining for Tum and Pav often appeared as a ring within the nucleus, because of their exclusion from the nucleolus, as shown by costaining for the nucleolar protein fibrinillin (Fig. 5F,G). Pav contains several nuclear localization sequences (NLSs). When crucial charged amino acids within three of these NLSs are replaced with alanine, the protein was excluded from the nucleus but was able to rescue cell division defects in pav mutants (Minestrini et al., 2002). We found that this PavΔNLS protein was significantly compromised in its ability to repress TOPflash reporter activity, compared with full-length Pav (Fig. 5H). We also found that Tum behaved in the same manner. Tum contains a predicted unconventional NLS (Fig. 5E). To determine whether this predicted NLS was functional, we mutated three core basic amino acids to alanine. TumΔNLS was expressed at levels comparable to wild-type Tum, but was excluded from nuclei (Fig. 5F,G). As with PavΔNLS, TumΔNLS showed a significantly reduced capacity to repress TOPflash reporter activity (Fig. 5H). Tum also requires its association with Pav for its full repressive effect: deletion of the Pav-binding domain diminished the repressive capacity of Tum (Fig. 5H), even though TumΔPav was able to accumulate in nuclei (Fig. 6E).
Fig. 5.
Pav and Tum act in the nucleus to repress Wnt pathway activity. DAPI staining of DNA (A) visualizes the nuclei in cells cotransfected with Tum (B) and Pav (C), where they colocalize (overlap is white) in HEK293T cells (D). Untransfected neighboring cells stain only with DAPI (blue). Scale bar: 10 μm. (E) Schematic diagrams of Tum and Pav proteins, with position of nuclear-localization sequences highlighted in red, and mutational changes in putative NLS of Tum shown (top). The coiled-coil domain (CC, pink box) includes the Pav-binding domain (Somers and Saint, 2003), which is deleted in the pcTumΔPav construct (Zavortink et al., 2005). HEK293T cells transfected with wild-type Tum (F) or TumΔNLS (G) stained with anti-Tum antibody (red), DAPI (blue nuclei), and anti-Fibrinillin (green), a marker for the nucleolus. (H) Tum or Pav repression of Wnt-induced TOPflash, compared with FOPflash, is severely compromised when the NLS is mutated (P<0.0001 for each comparison). Cells transfected with TumΔPav also show significantly reduced repression compared with cells expressing full-length Tum (P<0.0001). Results are means ± s.d. Mutant transgenes were expressed at levels equivalent to or greater than the full-length Tum or Pav, as assayed by immunoblot (data not shown).
Fig. 6.
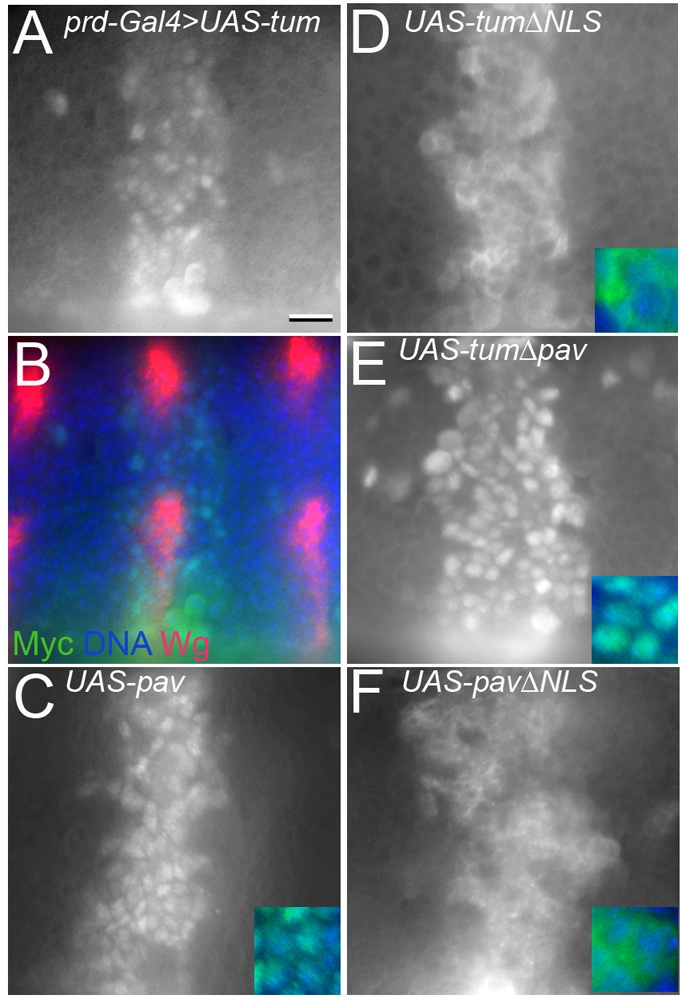
Wg signaling does not affect localization of Tum or Pav. (A) Anti-Myc antibody staining detects nuclear transgenic Tum, driven with prd-Gal4 in odd-numbered segments. This expression domain (green) encompasses the wg-expressing stripes (red) in alternate segments of the same embryo (B). (C-F) Embryos with same orientation, position and developmental stage. (C) prd-Gal4>UAS-pavGFP shows no change in nuclear Pav within the prd expression domain. (D) prd-Gal4>UAS-tumΔNLS is excluded from the nucleus. (E) prd-Gal4>UAS-tumΔpav localizes to the nucleus. (F) prd-Gal4>UAS-pavΔNLS is excluded from the nucleus. Insets show merge of transgenic protein (green) and DAPI (blue). Scale bar: 10 μm (A-F), 5 μm (insets).
The subcellular localizations of Tum and Pav were not altered by Wg signaling in fly embryos. We used prd-Gal4 to drive wild-type or mutant forms of Tum or Pav in the embryonic epidermis, in broad domains centered over the odd-numbered wingless expression stripes (Fig. 6). At mid-stage 10, a dorsolateral portion of the wg stripe shuts off (Bejsovec and Martinez Arias, 1991; Gonzalez et al., 1991). In stage 11 embryos, the dorsolateral portion of prd-driven Tum or Pav showed no difference from regions where wg was still expressed (Fig. 6A-C). Likewise, the mutant ΔNLS forms of Tum and Pav, which clearly failed to localize to the nucleus in the embryonic epidermis (Fig. 6D,F respectively), did not show any redistribution in response to Wg.
The nuclear localization of Tum and Pav is relevant for their activity in fly embryos. E22c-Gal4-driven expression of wild-type UAS-tum and UAS-pav transgenes rescued epidermal pattern defects of the tum and pav mutants, respectively (Table 1). However, neither ΔNLS form was able to rescue mutant pattern defects (Table 1). They were also unable to block ectopic activation of the Wg pathway in the embryo. Expressing the UAS-armS10 transgene, which lacks the Zw3/GSK phosphorylation sites (Pai et al., 1997), transforms most of the ventral denticle-secreting epidermal cells to a naked-cuticle cell fate. Coexpression of a UAS-GFP transgene did not significantly diminish this effect (Fig. 7A,F), but coexpression of either Tum or Pav partially blocked this cell-fate transformation and allowed some denticle production (Fig. 7B,D,F). By contrast, coexpression of TumΔNLS (Fig. 7C,F) or PavΔNLS (not shown) did not block the armS10-induced cell-fate transformation. Thus, the ability of Tum and Pav to translocate into the nucleus is crucial for their downregulation of the Wg/Wnt pathway. Co-expression of Nkd, which is thought to inhibit the Wnt pathway upstream of Arm stabilization (Waldrop et al., 2006), was also unable to block the armS10-induced cell-fate transformation, as expected (Fig. 7E,F).
Fig. 7.
Tum and Pav act downstream of βcat stabilization and βcat-Tcf-Lef complex formation. (A-E) arm-Gal4 driving constitutively stabilized armS10 transforms denticle-secreting regions of the epidermis into ectopic naked cuticle. Coexpression of a second transgene, such as UAS-GFP shown here, does not diminish the effect by competing for Gal4. (B) Coexpression with UAS-tum rescued denticle formation in most denticle belts. (C) This rescue was not observed with UAS-tumΔNLS coexpression. (D) Coexpression with UAS-pav also rescued denticle formation. (E) No rescue was observed with UAS-nkd coexpression. (F) Chart quantifying the severity of phenotypes shown in A-E produced by arm-Gal4-driven armS10. Extent of ectopic naked cuticle production is expressed as the number of ventral belts that show at least one complete row of denticles. n>100 larval cuticles were scored for each genotype. (G) Tum and Pav repress TOPflash activity induced by a βcat-Lef fusion construct. Tum represses activation to 18% of pcGFP control (P<0.0001), Pav represses activation to 43% of pcGFP control (P=0.0002), whereas Axin shows a small but statistically significant repression (P=0.025). Results are means ± s.d.
We conclude that the nuclear localization of both Tum and Pav is relevant to their roles in controlling activity of the Wnt pathway. This subcellular localization raised the possibility that Tum and Pav act within the nucleus to block the interaction between βcat and the Tcf-Lef transcription factor. If so, they should be unable to affect Wnt activity induced by a chimeric protein that fuses the 87 amino acid activation domain of βcat with the Lef-1 protein (Hsu et al., 1998). However, we found that cotransfection with either Tum or Pav repressed βcat-Lef-induced reporter expression very robustly (Fig. 7G). In this assay, Axin showed a modest ability to repress βcat-Lef-induced TOPflash (Fig. 7G), perhaps because of its proposed secondary role in mediating nuclear export of βcat (Cong and Varmus, 2004; Wiechens et al., 2004). Taken together, these observations suggest that Tum and Pav act within the nucleus to antagonize Wnt-stimulated gene expression at a stage between formation of the βcat-Tcf complex and formation of an active transcriptional complex at Wnt target-gene promoters.
Discussion
Here, we present evidence that Tum and its binding partner, Pav, are required for negative regulation of Tcf-mediated transcription. In retrospect, it seems likely that their pleiotropic effects on cytokinesis have obscured their role in regulation of the Wnt pathway until now. For example, neither of these genes was identified in genomics-based screens for Wg pathway components in cultured cells (DasGupta et al., 2005). Reduction of Tum or Pav function cannot be easily assessed in cell culture because tum- or pav-deficient cells cease to divide. The fly embryo, which uses maternally loaded Tum and Pav until midway through development, provided a fortuitous and genetically tractable model that allowed the detection of their involvement in Wnt modulation. This raises the question of how many pathway components have been overlooked because they have other essential cellular functions.
The roles of Tum and Pav in cytokinesis are genetically separable from their roles in Wnt signaling. The GAP activity of Tum is required for cytokinesis: TumΔEIE was unable to support normal cell division (Somers and Saint, 2003; Zavortink et al., 2005). However, we found that TumΔEIE had a significant capacity to repress TOPflash activity in cultured cells (this work) and to rescue pattern defects in tum mutant fly embryos (Jones and Bejsovec, 2005). Similarly, the Pav-binding domain of Tum is essential for cytokinesis (Zavortink et al., 2005), but we found that TumΔPav retained a modest ability to repress Wg/Wnt signaling in cultured cells, indicating that some of the Tum regulatory function is independent of Pav. Conversely, nuclear localization of Pav is irrelevant to its role in cytokinesis (Minestrini et al., 2003), whereas we found that nuclear localization of both Tum and Pav was required for rescuing pattern defects in embryos and for repressing TOPflash activity in cultured cells. Thus the Tum-Pav interaction is important for both cytokinesis and full Wnt regulation, but the cellular activity of the complex differs both spatially and enzymatically in the two processes.
Most molecules that negatively regulate the Wnt pathway act through the ubiquitylation-based pathway that controls βcat stability. However, Tum and Pav were able to repress TOPflash induced by the constitutively active ΔGSK-βcat and to diminish pattern disruption caused by the analogous mutant form of Arm in fly embryos. This observation places Tum and Pav activity downstream of the well-characterized control exerted by the destruction complex. Tum and Pav did not prevent βcat from entering the nucleus, but did prevent a βcat-Lef fusion protein from successfully activating gene expression. Therefore we favor a model where Tum and Pav have a constitutive role that impedes binding of the βcat-Tcf complex to DNA or that dampens transcriptional activation by the DNA-bound complex. Curiously, the Tum homolog in humans MgcRacGAP has been connected with transcriptional regulation via HIF-1α, with which it interacts physically (Lyberopoulou et al., 2007). MgcRacGAP has also been associated with nuclear translocation and activation of STAT transcription factors (Kawashima et al., 2009). Tum itself was shown to regulate activity of the EGF pathway in the Drosophila wing (Sotillos and Campuzano, 2000). These observations suggest that Tum/MgcRacGAP has a broader role within the nucleus, interacting with transcription factors in other cellular pathways.
Since Drosophila Tum and Pav are able to repress the Wnt response in human cells, this raises the possibility that MgcRacGAP and MKLP-1, their vertebrate counterparts, might also regulate Wnt signaling. It is tempting to speculate that a direct connection between Wnt regulation and cell division is relevant to oncogenesis, in much the same way that Wnt signaling and cell adhesion are integrally connected through the shared component βcat (Hinck et al., 1994; Pai et al., 1996). Recent work has demonstrated that cell-division defects leading to tetraploidy might ultimately lead to cancer (Fujiwara et al., 2005; Ganem et al., 2007). Multinucleate cells are more likely to experience genetic instability, increasing the risk of transformation to malignancy. Thus, any genetic or epigenetic change that reduces the function of Tum/MgcRacGAP or Pav/MLKP-1 could have a double impact on the cell, compromising the cytokinesis machinery and elevating activity of the Wnt pathway.
Materials and Methods
Drosophila stocks and culture conditions
The tumAR2 mutation was isolated in an EMS mutagenesis (Jones and Bejsovec, 2005). All stocks were obtained from the Bloomington Stock Center, except UAS-tum and UAS-tumΔEIE flies which were a gift from Sonsoles Campuzano (Centro de Biologia Molecular Severo Ochoa, Madrid, Spain), UAS-GFP-Pav+ and UAS-GFP-PavΔNLS flies a gift from David Glover (University of Cambridge, Cambridge, UK) and UAS-nkd flies a gift from Keith Wharton and Matthew Scott (Stanford University, Palo Alto, CA). UAS-tumΔNLS flies were created by subcloning tum sequences from pcTumΔNLS into pUAST and transforming into fly embryos to derive transgenic lines. Flies were cultured, and embryos and hatched larvae were collected and examined, as described previously (Chao et al., 2007). To examine embryonic cuticles, eggs were allowed to develop fully (24 hours at 25°C), dechorionated in bleach, and transferred to a drop of Hoyer's medium, mixed 1:1 with lactic acid (Wieschaus and Nüsslein-Volhard, 1986), on a microscope slide. Mechanical devitellinization was performed by exerting gentle pressure on the coverslip to burst the vitelline membrane. Cuticle preparations were baked at 65°C overnight before viewing on a Zeiss Axioplan microscope with phase optics. Wings were dissected from 2-day-old adult females that had been dehydrated in 95% ethanol overnight. Wings were then mounted in Hoyer's medium:lactic acid and photographed using a 5× objective on a Zeiss Axioplan microscope with brightfield illumination. Images were captured using a SPOT camera (Diagnostic Instruments) and were processed with SPOT imaging software and Adobe Photoshop.
Yeast two-hybrid screen
A 0-4 hour Drosophila embryonic cDNA library in pGAD10 (Clontech) was kindly provided by Robin Wharton (Ohio State University, Columbus, OH). Tum was inserted into pGBT9 (Clontech) using PCR strategies. Library transformation and screening followed standard protocols (Clontech).
DNA constructs and TOPflash assays
All plasmids constructed for testing in HEK293T cells were cloned into the vector pcDNA3.1 (Invitrogen), and verified by DNA sequencing. Expression levels were confirmed by immunoblotting. pcβcat-myc and pcβcat-LEF were gifts from Patrick Casey and Tannishtha Reya, respectively (Duke University, Durham, NC). pEGFP:ΔGSK-βcat was a gift from Angela Barth and James Nelson (Stanford University, Palo Alto, CA).
HEK293T cells were grown in 24-well plates at 0.5 ml cells per well. Cells were Lipofectamine-transfected at approximately 80% confluence, using 0.1 μg TOPflash or FOPflash firefly luciferase reporter plasmid (Upstate), 0.25 ng phRG-TK Renilla luciferase control plasmid (Promega), and 0.2 μg test plasmid. Total amount of DNA was kept constant by adding pcGFP construct or empty pcDNA3.1 vector. Cells were induced using Wnt-conditioned medium recovered from L-cells stably transfected with Wnt-3a (ATCC CRL-2647) or by cotransfecting with either pcβcat-myc, pEGFP:ΔGSK-βcat, or pcβcat-LEF at a concentration of 0.25 μg per 0.5 ml well. Uninduced controls were incubated with media from L-cells (ATCC CRL-2648) or cotransfected with pcGFP, respectively. Expression was monitored using the Dual-Luciferase Reporter Assay System (Promega) on a Veritas luminometer. General cell health was assayed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). All transfections for TOPflash analysis were done in duplicate, and three independent experiments were performed for each analysis. TOPflash or FOPflash values were divided by the Renilla luciferase transfection control value. Average luciferase ratio was calculated for induced and uninduced cells in each treatment category. Relative activity was calculated by dividing the luciferase ratio of induced cells by the luciferase ratio of uninduced cells. Each experiment was normalized to pcGFP or empty pcDNA3.1 vector controls, set to 100% activation. P values were calculated using two-tailed Student's t-tests.
Cell fractionation and antibody staining
Nuclear and cytoplasmic fractions of HEK293T cells were collected following manufacturer's protocol, using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce Biotechnology). Immunoblots with anti-βcat E247 (Abcam) and anti-β-tubulin (Labvision) were visualized using the Odyssey infrared imaging system (Li-Cor), with secondary antibodies from Rockland Immunochemicals.
HEK293T cells were grown on 10 mm coverslips in 24-well plates and transfected as described above. Cells were harvested and fixed for 20 minutes in 4% paraformaldehyde 24-30 hours after transfection, blocked in 20% normal goat serum and 0.1% Tween-20, and incubated in primary antibody overnight at 4°C. Stained cells were mounted in Aquapolymount (Polysciences) for viewing on a Zeiss LSM510 confocal microscope or on a Zeiss Axioplan microscope using epifluorescence. Antibodies and dilutions used included anti-βcat E247 1:250 (Abcam), anti-Tum 1:250 (Somers and Saint, 2003), anti-Flag M2 used to detect Flag-tagged Pav 1:1000 (Sigma), anti-fibrinillin 1:100 (Abcam). DAPI was used at 1:10,000 (Roche). Secondary antibodies (1:500) were purchased from Jackson ImmunoResearch.
Fly embryos were collected overnight at 18°C, and processed as previously described (Dierick and Bejsovec, 1998). The tum mutant chromosome was maintained over a balancer chromosome marked with twist-Gal4 UAS-Green Fluorescent Protein (GFP), so that homozygotes could be identified by their failure to express GFP. Anti-β-galactosidase (Chemicon) was used at 1:500, anti-Wg at 1:500 and anti-En at 1:50 (both from Developmental Studies Hybridoma Bank), anti-GFP (Chemicon) at 1:500, and anti-Myc (Cell Signaling) at 1:400. Cy2 and Cy3 IgG secondary antibodies (Jackson ImmunoResearch) were used to detect fluorescence staining. Stained embryos were mounted in Aquapolymount (Polysciences), viewed on a Zeiss Axioplan microscope using epifluorescence and imaged using SPOT imaging software (Diagnostic Instruments).
Acknowledgments
We wish to thank our colleagues in the Wray, McClay, Casey and Reya labs for sharing reagents, equipment and advice and we thank the Duke Model System Genomics Group for assistance with transgenic fly lines. As always, we are deeply indebted to the dedicated staff at the Bloomington Stock Center and at Flybase. This work was supported by grants from the National Science Foundation and National Institutes of Health to A.B. and from the Australian Research Council and the ANU Institute of Advanced Studies to R.S. Deposited in PMC for release after 12 months.
References
- Adams R. R., Tavares A. A., Salzberg A., Bellen H. J., Glover D. M. (1998). pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 15, 1483-1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A. I., Stewart D. B., Nelson W. J. (1999). T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant beta-catenin. Proc. Natl. Acad. Sci. USA 96, 4947-4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A. (2006). Flying at the head of the pack: Wnt biology in Drosophila. Oncogene 25, 7442-7449 [DOI] [PubMed] [Google Scholar]
- Bejsovec A., Martinez Arias A. (1991). Roles of wingless in patterning the larval epidermis of Drosophila. Development 113, 471-485 [DOI] [PubMed] [Google Scholar]
- Bejsovec A., Wieschaus E. (1993). Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development 119, 501-517 [DOI] [PubMed] [Google Scholar]
- Brunner E., Peter O., Schweizer L., Basler K. (1997). pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385, 829-833 [DOI] [PubMed] [Google Scholar]
- Buongiorno P., Pethe V. V., Charames G. S., Esufali S., Bapat B. (2008). Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol. Cancer 7, 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. (1998). Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395, 604-608 [DOI] [PubMed] [Google Scholar]
- Chao A. T., Jones W. M., Bejsovec A. (2007). The HMG-box transcription factor SoxNeuro acts with Tcf to control Wg/Wnt signaling activity. Development 134, 989-997 [DOI] [PubMed] [Google Scholar]
- Cong F., Varmus H. (2004). Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc. Natl. Acad. Sci. USA 101, 2882-2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Weis W. I. (2005). Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12, 364-371 [DOI] [PubMed] [Google Scholar]
- DasGupta R., Kaykas A., Moon R. T., Perrimon N. (2005). Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826-833 [DOI] [PubMed] [Google Scholar]
- Dierick H. A., Bejsovec A. (1998). Functional analysis of Wingless reveals a link between intercellular ligand transport and dorsal-cell-specific signaling. Development 125, 4729-4738 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Bandi M., Nitta M., Ivanova E. V., Bronson R. T., Pellman D. (2005). Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437, 1043-1047 [DOI] [PubMed] [Google Scholar]
- Ganem N. J., Storchova Z., Pellman D. (2007). Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17, 157-162 [DOI] [PubMed] [Google Scholar]
- Goldstein A. Y., Jan Y. N., Luo L. (2005). Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 102, 3834-3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Swales L., Bejsovec A., Skaer H., Martinez Arias A. (1991). Secretion and movement of wingless protein in the epidermis of the Drosophila embryo. Mech. Dev. 35, 43-54 [DOI] [PubMed] [Google Scholar]
- Gregory S. L., Ebrahimi S., Milverton J., Jones W. M., Bejsovec A., Saint R. (2008). Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr. Biol. 18, 25-29 [DOI] [PubMed] [Google Scholar]
- Gritzan U., Hatini V., DiNardo S. (1999). Mutual antagonism between signals secreted by adjacent Wingless and Engrailed cells leads to specification of complementary regions of the Drosophila parasegment. Development 126, 4107-4115 [DOI] [PubMed] [Google Scholar]
- Guerin C. M., Kramer S. G. (2009). RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development 136, 1411-1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S., Ng J., Tzu J., Dietzl G., Sun Y., Harms M., Nardine T., Luo L., Dickson B. J. (2002). Rac function and regulation during Drosophila development. Nature 416, 438-442 [DOI] [PubMed] [Google Scholar]
- Hays R., Gibori G. B., Bejsovec A. (1997). Wingless signaling generates pattern through two distinct mechanisms. Development 124, 3727-3736 [DOI] [PubMed] [Google Scholar]
- Heemskerk J., DiNardo S., Kostriken R., O'Farrell P. H. (1991). Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature 352, 404-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L., Nelson W. J., Papkoff J. (1994). Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J. Cell Biol. 124, 729-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Kawashima T., Iwamoto I., Nosaka T., Kitamura T. (2001). MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol. Chem 276, 5821-5828 [DOI] [PubMed] [Google Scholar]
- Hsu S. C., Galceran J., Grosschedl R. (1998). Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 18, 4807-4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida S., Yamamoto H., Murai H., Koyama S., Kikuchi A. (1998). Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 17, 1371-1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. M., Bejsovec A. (2003). Wingless signaling: an Axin to grind. Curr. Biol. 13, R479-R481 [DOI] [PubMed] [Google Scholar]
- Jones W. M., Bejsovec A. (2005). RacGap50C negatively regulates Wingless pathway activity during Drosophila embryonic development. Genetics 169, 2075-2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Bao Y. C., Minoshima Y., Nomura Y., Hatori T., Hori T., Fukagawa T., Fukada T., Takahashi N., Nosaka T., et al. (2009). A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol. Cell. Biol. 29, 1796-1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A. (1999). Roles of Axin in the Wnt signalling pathway. Cell Signal 11, 777-788 [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K. (1999). An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 18, 2401-2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T., Peter O., Brunner E., Nellen D., Froesch B., Chatterjee S., Murone M., Zullig S., Basler K. (2002). Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109, 47-60 [DOI] [PubMed] [Google Scholar]
- Krieghoff E., Behrens J., Mayr B. (2006). Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J. Cell Sci. 119, 1453-1463 [DOI] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002). Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837-847 [DOI] [PubMed] [Google Scholar]
- Logan C. Y., Nusse R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781-810 [DOI] [PubMed] [Google Scholar]
- Lyberopoulou A., Venieris E., Mylonis I., Chachami G., Pappas I., Simos G., Bonanou S., Georgatsou E. (2007). MgcRacGAP interacts with HIF-1alpha and regulates its transcriptional activity. Cell. Physiol. Biochem. 20, 995-1006 [DOI] [PubMed] [Google Scholar]
- McCartney B. M., Dierick H. A., Kirkpatrick C., Moline M. M., Baas A., Peifer M., Bejsovec A. (1999). Drosophila APC2 is a cytoskeletally-associated protein that regulates Wingless signaling in the embryonic epidermis. J. Cell Biol. 146, 1303-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini G., Mathe E., Glover D. M. (2002). Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J. Cell Sci. 115, 725-736 [DOI] [PubMed] [Google Scholar]
- Minestrini G., Harley A. S., Glover D. M. (2003). Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol. Biol. Cell 14, 4028-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Kawashima T., Hirose K., Tonozuka Y., Kawajiri A., Bao Y. C., Deng X., Tatsuka M., Narumiya S., May W. S., Jr, et al. (2003). Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 4, 549-560 [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41-54 [DOI] [PubMed] [Google Scholar]
- Moline M. M., Southern C., Bejsovec A. (1999). Directionality of Wingless protein transport influences epidermal patterning in the Drosophila embryo. Development 126, 4375-4384 [DOI] [PubMed] [Google Scholar]
- Noordermeer J., Johnston P., Rijsewijk F., Nusse R., Lawrence P. A. (1992). The consequences of ubiquitous expression of the wingless gene in the Drosophila embryo. Development 116, 711-719 [DOI] [PubMed] [Google Scholar]
- Pai L. M., Kirkpatrick C., Blanton J., Oda H., Takeichi M., Peifer M. (1996). Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J. Biol. Chem. 271, 32411-32420 [DOI] [PubMed] [Google Scholar]
- Pai L. M., Orsulic S., Bejsovec A., Peifer M. (1997). Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 124, 2255-2266 [DOI] [PubMed] [Google Scholar]
- Peifer M., Wieschaus E. (1990). The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell 63, 1167-1178 [DOI] [PubMed] [Google Scholar]
- Peifer M., Polakis P. (2000). Wnt signaling in oncogenesis and embryogenesis-a look outside the nucleus. Science 287, 1606-1609 [DOI] [PubMed] [Google Scholar]
- Peifer M., Sweeton D., Casey M., Wieschaus E. (1994). wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development 120, 369-380 [DOI] [PubMed] [Google Scholar]
- Polakis P. (2000). Wnt signaling and cancer. Genes Dev. 14, 1837-1851 [PubMed] [Google Scholar]
- Salahshor S., Woodgett J. R. (2005). The links between axin and carcinogenesis. J. Clin. Pathol. 58, 225-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson B., Alexandre C., Fascetti N., Vincent J. P. (1999). Engrailed and hedgehog make the range of Wingless asymmetric in Drosophila embryos. Cell 98, 207-216 [DOI] [PubMed] [Google Scholar]
- Seo E., Jho E. H. (2007). Axin-independent phosphorylation of APC controls beta-catenin signaling via cytoplasmic retention of beta-catenin. Biochem. Biophys. Res. Commun. 357, 81-86 [DOI] [PubMed] [Google Scholar]
- Siegfried E., Chou T. B., Perrimon N. (1992). wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell 71, 1167-1179 [DOI] [PubMed] [Google Scholar]
- Somers W. G., Saint R. (2003). A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29-39 [DOI] [PubMed] [Google Scholar]
- Sotillos S., Campuzano S. (2000). DRacGap, a novel Drosophila gene inhibits EGFR/Ras signalling in the developing imaginal wing disc. Development 127, 5427-5438 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A., et al. (1997). Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789-799 [DOI] [PubMed] [Google Scholar]
- Waldrop S., Chan C. C., Cagatay T., Zhang S., Rousset R., Mack J., Zeng W., Fish M., Zhang M., Amanai M., et al. (2006). An unconventional nuclear localization motif is crucial for function of the Drosophila Wnt/wingless antagonist Naked cuticle. Genetics 174, 331-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechens N., Heinle K., Englmeier L., Schohl A., Fagotto F. (2004). Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-beta-catenin Pathway. J. Biol. Chem. 279, 5263-5267 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C. (1986). Looking at embryos. In Drosophila, A Practical Approach (ed. Roberts D. B.). Oxford, England: IRL Press; [Google Scholar]
- Wu X., Tu X., Joeng K. S., Hilton M. J., Williams D. A., Long F. (2008). Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell 133, 340-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavortink M., Contreras N., Addy T., Bejsovec A., Saint R. (2005). Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J. Cell Sci. 118, 5381-5392 [DOI] [PubMed] [Google Scholar]
- Zeng W., Wharton K. A., Jr, Mack J. A., Wang K., Gadbaw M., Suyama K., Klein P. S., Scott M. P. (2000). naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403, 789-795 [DOI] [PubMed] [Google Scholar]
- Zhao W. M., Fang G. (2005). MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA 102, 13158-13163 [DOI] [PMC free article] [PubMed] [Google Scholar]