Abstract
Objective
To study the effect of apolipoprotein E ε4 status on biomarkers of neurodegeneration (atrophy on magnetic resonance imaging [MRI]), neuronal injury (cerebrospinal fluid [CSF] t-tau), and brain Aβ amyloid load (CSF Aβ1–42) in cognitively normal subjects (CN), amnestic subjects with mild cognitive impairment (aMCI), and patients with Alzheimer disease (AD).
Methods
We included all 399 subjects (109 CN, 192 aMCI, 98 AD) from the Alzheimer's Disease Neuroimaging Initiative study with baseline CSF and MRI scans. Structural Abnormality Index (STAND) scores, which reflect the degree of AD-like anatomic features on MRI, were computed for each subject.
Results
A clear ε4 allele dose effect was seen on CSF Aβ1–42 levels within each clinical group. In addition, the proportion of the variability in Aβ1–42 levels explained by APOE ε4 dose was significantly greater than the proportion of the variability explained by clinical diagnosis. On the other hand, the proportion of the variability in CSF t-tau and MRI atrophy explained by clinical diagnosis was greater than the proportion of the variability explained by APOE ε4 dose; however, this effect was only significant for STAND scores.
Interpretation
Low CSF Aβ1–42 (surrogate for Aβ amyloid load) is more closely linked to the presence of APOE ε4 than to clinical status. In contrast, MRI atrophy (surrogate for neurodegeneration) is closely linked with cognitive impairment, whereas its association with APOE ε4 is weaker. The data in this paper support a model of AD in which CSF Aβ1–42 is the earliest of the 3 biomarkers examined to become abnormal in both APOE carriers and noncarriers.
Apolipoprotein E (APOE) ε4 is the most important known genetic risk factor for typical late onset Alzheimer disease (AD). The lifetime risk of developing AD is increased and the age of onset of the disease is lowered with increasing number of APOE ε4 alleles.1–4 Aβ1–42 and tau levels measured in cerebrospinal fluid (CSF) and atrophy seen on magnetic resonance imaging (MRI) are indicators of important disease-related pathological processes in AD. Low CSF Aβ1–42 reflects deposition of Aβ in plaques.5 High CSF t-tau levels reflect active axonal and neuronal damage.6 Atrophy seen on MRI is the direct result of loss of neurons, synapses, and dendritic arborization.7 In this paper, we use Structural Abnormality Index (STAND) scores as an indicator of severity of an AD-like pattern of atrophy on structural MRI. STAND scores were developed in our lab to condense the severity and location of AD-related atrophy on the 3-dimensional MRI scan into a single number.8
The effect of APOE genotype on neuronal pathology and amyloid load has been studied in autopsy specimens.9–13 Several in vivo CSF Aβ1–42 and t-tau studies,14–17 MRI studies,18–22 and fluorodeoxyglucose-positron emission tomography (PET) imaging studies23–25 have also studied the effect of APOE independently in each of these modalities. The first Alzheimer's Disease Neuroimaging Initiative (ADNI) CSF biomarker study also investigated the effect of APOE on CSF biomarkers, and found that Aβ1–42 concentration is lowest in subjects with 2 APOE ε4 alleles and rises as the number of alleles decreases.26 However, there have not been in vivo studies that have investigated the influence of ε4 allele on the surrogates of Aβ amyloid deposition and neuronal pathology together as measured by CSF and MRI in a cohort of subjects that spans the cognitive spectrum.
The main aim of our paper was to evaluate the effect of APOE genotype on biomarkers of Aβ amyloid load and neuronal pathology by answering these questions: (1) How does APOE genotype effect CSF Aβ1–42 and t-tau levels and atrophy on MRI within each clinical group? (2) How does APOE genotype affect biomarker discrimination between different clinical groups (cognitively normal [CN], amnestic mild cognitive impairment [aMCI], AD)? (3) How much of the variability in the biomarkers is explained by clinical diagnosis versus APOE genotype? and (4) Does the relationship between continuous measures of cognitive performance and the biomarkers differ by APOE genotype?
Subjects and Methods
The data used in this study are from ADNI, a longitudinal multisite observational study of elderly individuals with CN, aMCI, and AD collected from 56 participating institutes.27 Written informed consent was obtained for participation in these studies, as approved by the institutional review board at each of the participating centers. The details of ADNI can be found at http://www.ADNI-info.org
Clinical and Cognitive Assessment
We used Mini Mental State Examination (MMSE)28 and the Clinical Dementia Rating Sum of Boxes (CDR-SB)29 as overall indices of general cognitive performance and global functional status. Baseline clinical diagnosis and cognitive assessments of all 3 clinical groups and clinical/cognitive assessment scores (CDR-SB and MMSE) were considered in this paper. The total sample in this paper consists of 399 subjects (109 CN, 192 aMCI, 98 AD) who had both CSF biomarker data at baseline and usable 1.5T MRI scans (CSF was obtained at baseline in approximately 51% of the ADNI cohort). Two of the 98 AD subjects were subsequently clinically reclassified as having non-AD dementia (formal thought disorder and Dementia with Lewy bodies). Because reclassification occurred after looking forward in their clinical presentation (beyond baseline), and all subjects do not have the same amount of longitudinal follow-up at this time, we considered these 2 subjects as AD for this analysis to be consistent.
Statistical Analysis
Pair-wise group differences in baseline characteristics and MRI and CSF biomarker measures by APOE genotype and within diagnosis group were tested with a 2-sided Wilcoxon rank sum test or, in the case of gender, a chi-square test. Pair-wise differences in biomarker measures by diagnosis and within APOE genotype were assessed by reporting the area under the receiver operator curves (AUROC) and the corresponding pair-wise Wilcoxon rank sum test p values. The AUROC has the interpretation of the probability of correctly classifying any 2 persons from different clinical groups when the person with the more abnormal biomarker value is assigned to the more abnormal clinical diagnostic category. To test for differences in the proportion of variability in biomarker measures explained by APOE genotype and clinical diagnosis, we generated 95% confidence intervals using bootstrap methods for the difference in R2 between a model with APOE genotype as the only predictor of biomarker and a model with clinical diagnosis as the only predictor of biomarker.
To assess differences in the relationship between cognition and biomarker by APOE genotype, we fit a linear model for each MRI and CSF biomarker with MMSE, APOE genotype, and their interaction as predictors. We allowed the relationship between MMSE and cognition to be nonlinear using restricted cubic splines. We examined the interaction effect to determine if the MMSE and biomarker relationship was different by APOE genotype. To graphically show the differences, we created z scores for each biomarker with mean of 0 and standard deviation of 1 to put all measures on the same scale. The sign of the CSF Aβ1–42 z scores was reversed so that increasing z scores for each biomarker represents the worsening of the biomarker value with disease. We then fit a loss model with MMSE as the predictor of each z score within APOE genotype and plotted the predicted values by MMSE. Because we model the biomarker mean as a smooth function of MMSE using restricted cubic splines, the mean for the biomarker values are estimated at MMSE of 30, MMSE of 29, et cetera. Therefore, these models are not affected by the ceiling effects in MMSE, because there is a sufficient range of MMSE values in the data, as CN, aMCI, and AD subjects are included.
All data manipulation and analysis was performed using SAS version 9.1.3 and R version 2.7.1.
Results
Patient Characteristics
The demographics and clinical summary of CN, aMCI, and AD subjects split by their APOE ε4 status along with the p values are shown in the patient characteristics section of Table 1. As expected, the proportion of ε4 carriers was significantly higher among AD and aMCI than CN. Among aMCI and AD subjects, APOE ε4 carriers tended to be younger than noncarriers, which is consistent with the fact that APOE ε4 allele is associated with earlier onset of the disease. The ages of ε4 carriers and noncarriers were not different among CN subjects. There were no significant differences in the MMSE and CDR-SB among ε4 carriers and noncarriers within each clinical group. MMSE and CDR-SB scaled appropriately by clinical group with CN (least abnormal), and AD (most abnormal) at 2 extremes and aMCI in the middle of the spectrum.
TABLE 1.
Patient Characteristics at the Time of the MRI Scan by Diagnosis and APOE Genotype
| Characteristics | CN | aMCI | AD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ε4 Noncarriers | ε4 Carriers | p | ε4 Noncarriers | ε4 Carriers | p | ε4 Noncarriers | ε4 Carriers | p | |
| Number of subjects | 82 | 27 | 89 | 103 | 29 | 69 | |||
| Women, No. (%) | 43 (52) | 9 (33) | 0.13 | 24 (27) | 40 (39) | 0.11 | 13 (45) | 28 (41) | 0.87 |
| Age, yr | 74 (71, 78) | 77 (72, 78) | 0.34 | 76 (72, 82) | 74 (69, 78) | 0.02 | 79 (72, 82) | 75 (70, 80) | 0.09 |
| Education, yr | 16 (14, 18) | 16 (14, 18) | 0.99 | 16 (14, 18) | 16 (14, 18) | 0.45 | 16 (14, 18) | 16 (12, 17) | 0.12 |
| CDR-SB | 0 (0, 0) | 0 (0, 0) | 0.19 | 2 (1, 2) | 2 (1, 2) | 0.76 | 4 (3, 4) | 4 (4, 5) | 0.21 |
| MMSE | 29 (29, 30) | 29 (28, 30) | 0.41 | 27 (25, 28) | 27 (25, 28) | 0.71 | 24 (22, 25) | 24 (22, 25) | 0.88 |
| Baseline MRI and CSF measurements | |||||||||
| Aβ1-42, pg/mL | 230 (189, 260) | 142 (124, 190) | <0.001 | 171 (139, 246) | 137 (115, 154) | <0.001 | 152 (137, 212) | 131 (111, 149) | <0.001 |
| t-Tau, pg/mL | 60 (45, 80) | 73 (54, 95) | 0.13 | 73 (55, 102) | 101 (78, 146) | <0.001 | 110 (73, 187) | 115 (87, 142) | 0.70 |
| STAND score | –0.9 (–1.4, –0.3) | –1.0 (–1.6, –0.7) | 0.11 | –0.3 (–0.9, 0.4) | 0.1 (–0.4, 0.7) | 0.02 | 0.8 (–0.3, 1.4) | 0.8 (0.3, 1.4) | 0.37 |
Continuous measures reported as median (interquartile range), p Values are based on Wilcoxon rank sum test except in the case of gender, where p values are based on chi-square tests. The proportion of APOE ε4 carriers is lower among CN than aMCI (p < 0.001), CN than AD (p < 0.001), and aMCI than AD subjects (p < 0.009).
MRI = magnetic resonance imaging; APOE = apolipoprotein E; CN = cognitively normal; aMCI = amnestic mild cognitive impairment; AD = Alzheimer disease; CDR-SB = Clinical Dementia Rating Sum of Boxes; MMSE = Mini Mental State Examination; CSF = cerebrospinal fluid; STAND = Structural Abnormality Index.
Effect of APOE ε4 Status on Baseline Biomarkers within Each Clinical Group
MRI and CSF biomarker summary statistics along with p values for differences by APOE genotype are presented in the biomarker measurement section of Table 1. Consistent with the recent report by Shaw et al,30 within each clinical group, APOE ε4 carriers had lower CSF Aβ1–42 than noncarriers ( p < 0.001). Among AD subjects, STAND and t-tau levels did not differ by APOE ε4 status. Among aMCI, both STAND and t-tau were higher (more abnormal) among APOE ε4 carriers. Among CN, STAND and t-tau were not significantly different between ε4 carriers and noncarriers.
Box plots of biomarker distributions by number of APOE ε4 alleles within each clinical group are shown in Figure 1. There was a correlation between number of ε4 alleles and Aβ1–42 among aMCI subjects (ρ = −0.42; p < 0.001) and among AD patients (ρ = −0.50; p < 0.001). In pair-wise comparisons, those with 2 ε4 alleles had significantly lower Aβ1–42 than those with just 1 among aMCI subjects (p = 0.003) and among AD patients (p < 0.001). In contrast, we found no evidence of an APOE ε4 dose effect on either t-tau or STAND among AD patients. On direct pair-wise comparisons, aMCI ε4 homozygotes did not have higher STAND or t-tau values than ε4 heterozygotes (p > 0.70 for both). Because the numbers of CN ε4 homozygotes (n = 2) was small, ε4 heterozygotes and homozygotes were combined together as ε4 carriers for increased power in analyses examining clinical discrimination by biomarkers within APOE genotype groups and also for plotting the biomarker z score curves versus MMSE by APOE genotype groups.
FIGURE 1.
Box plots of Aβ1–42, log (t-tau) and Structural Abnormality Index (STAND) score distributions by apolipoprotein E ε4 dose effect within each clinical group. Larger STAND and cerebrospinal fluid tau values are more abnormal, whereas lower Aβ1–42 values are more abnormal. CN = normal cognition; aMCI = amnestic mild cognitive impairment; AD = Alzheimer disease.
Biomarker-Based Clinical Group Discrimination within ε4 Carriers and Noncarriers
The AUROC and p values for the pair-wise clinical group discrimination within each of the APOE genotype groups are presented in Table 2. STAND score was significant in separating all the clinical group pairs both within carriers and within noncarriers. Within both ε4 carriers and noncarriers, t-tau was significant in separating all clinical group pairs except aMCI versus AD among ε4 carriers. Within ε4 carriers, CSF Aβ1–42 was not significant in separating different clinical group pairs except CN versus AD (p = 0.03); however, among noncarriers, CSF Aβ1–42 was significant in differentiating CN versus aMCI and CN versus AD, but not aMCI versus AD.
TABLE 2.
Clinical Discrimination within APOE Genotype Groups
| Biomarkers | CN vs aMCI | CN vs AD | aMCI vs AD | |||
|---|---|---|---|---|---|---|
| AUROC | p | AUROC | p | AUROC | p | |
| Aβ1-42 | ||||||
| ε4 noncarriers | 0.67 | <0.001 | 0.75 | <0.001 | 0.57 | 0.28 |
| ε4 carriers | 0.58 | 0.20 | 0.64 | 0.03 | 0.56 | 0.19 |
| t-Tau | ||||||
| ε4 noncarriers | 0.63 | 0.003 | 0.78 | <0.001 | 0.68 | 0.003 |
| ε4 carriers | 0.71 | <0.001 | 0.75 | <0.001 | 0.55 | 0.25 |
| STAND score | ||||||
| ε4 noncarriers | 0.68 | <0.001 | 0.84 | <0.001 | 0.70 | 0.001 |
| ε4 carriers | 0.84 | <0.001 | 0.95 | <0.001 | 0.69 | <0.001 |
p Values are based on Wilcoxon rank sum test.
APOE = apolipoprotein E; CN = cognitively normal; aMCI = amnestic mild cognitive impairment; AD = Alzheimer disease; AUROC = area under the receiver operator curve; STAND = Structural Abnormality Index.
Variability in the Biomarkers Explained by Clinical Diagnosis versus APOE Genotype
R2 values examining the proportion of the variability in each biomarker value that is explained by clinical diagnosis versus APOE genotype are shown in Table 3. The proportion of the variability in CSF Aβ1–42 levels explained by the APOE genotype (R2 = 0.28) was greater than the proportion of the variability in CSF Aβ1–42 that was explained by clinical diagnosis (R2 = 0.17). The point estimate of the difference in the proportion of the variability in CSF Aβ1–42 explained by the APOE versus clinical diagnosis is 0.11, that is, 11%, and is significant because the 95% confidence interval (CI) does not include zero. There was some evidence that the proportion of the variability in CSF t-tau explained by clinical diagnosis (R2 = 0.15) was slightly higher than the proportion of the variability explained by APOE genotype (R2 = 0.08), but the difference in R2 was not significant, because the 95% CI included zero. On the other hand, the proportion of the variability in STAND scores explained by the clinical diagnosis (R2 = 0.27) was significantly higher than the proportion of the variability explained by APOE genotype (R2 = 0.06), with the point estimate and 95% CI for the difference in R2 being 0.21 (0.14 – 0.29).
TABLE 3.
Summary of R2 Values Examining the Proportion of the Variability in Each Biomarker Value That Is Explained by Dx versus APOE Genotype
| Biomarkers | Clinical Diagnosis R2 | APOE ε4 Dose R2 | Dx vs APOE ε4 Dose Difference in R2 |
|---|---|---|---|
| Aβ1-42a | 0.17 | 0.28 | –0.11 (–0.19,–0.02) |
| log (t-tau) | 0.15 | 0.08 | 0.07 (–0.01,0.13) |
| STAND score | 0.27 | 0.06 | 0.21 (0.14,0.29) |
Differences between the proportion of the variability explained by Dx versus APOE genotype with 95% bootstrap confidence interval around the point estimate of R2 is also shown.
For example, 17% of the variability in the cerebrospinal fluid (CSF) Aβ1-42 is explained by clinical diagnosis versus 28% by the APOE ε4 dose. The point estimate of the difference in the proportion of the variability in CSF Aβ1-42 that is explained by the APOE versus clinical diagnosis is 11%, and is significant because the 95% confidence interval does not include zero.
Dx = clinical diagnosis; APOE =apolipoprotein E; STAND = Structural Abnormality Index.
Effect of APOE ε4 Status on the Relationship between Cognitive Performance and Biomarkers
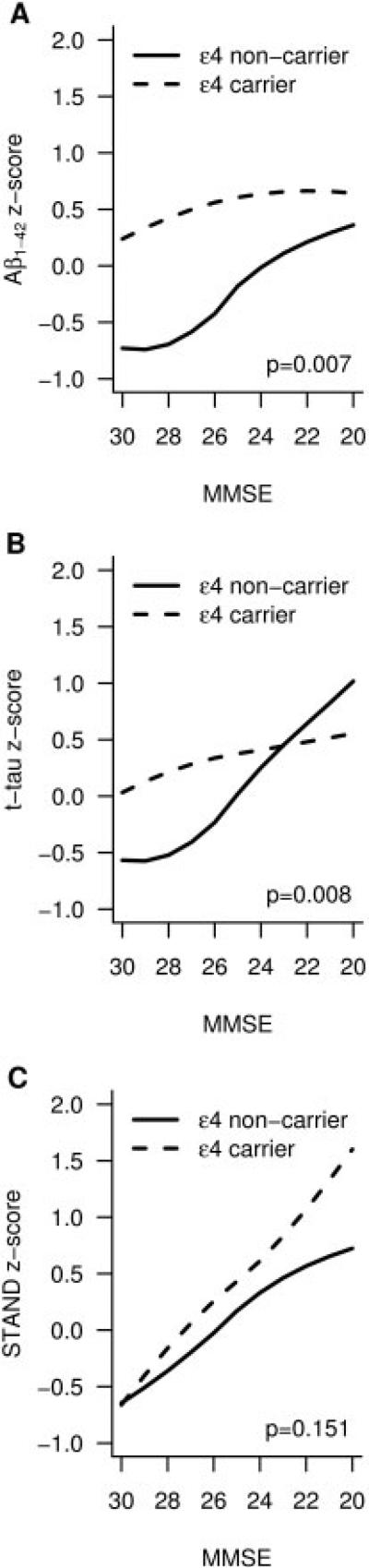
Biomarker z scores are plotted as a function of MMSE in ε4 carriers as well as noncarriers across the normal to AD cognitive continuum in Figure 2. The plots with the underlying data are shown as a Supplementary Figure 2. The curves relating biomarker values as a function of MMSE differed between APOE ε4 carriers and noncarriers for CSF Aβ1–42 (p = 0.007) and CSF t-tau (p = 0.008) levels, but did not differ for MRI atrophy (p = 0.151). Further testing found no relationships between MMSE and CSF Aβ1–42 (p = 0.16) nor MMSE and CSF t-tau (p = 0.24) among ε4 carriers, that is, the slope of the fit is not different from zero.
FIGURE 2.
Smoothed biomarker z score curves plotted as a function of cognitive performance (Mini Mental State Examination [MMSE]) across the Alzheimer disease continuum in apolipoprotein E ε4 carriers and noncarriers. STAND = Structural Abnormality Index.
Discussion
We investigated the effect of APOE ε4 status on brain amyloid load (measured by CSF Aβ1–42 levels), neuronal injury (measured by CSF t-tau), and neurodegeneration (measured by atrophy on MRI) across the cognitive continuum. The major findings regarding the effect of APOE genotype on biomarkers were: (1) CSF Aβ1–42 is closely linked to APOE genotype, but is less strongly associated with cognitive impairment; (2) in contrast, MRI atrophy is closely linked with cognitive impairment, whereas its association with APOE ε4 is weaker; and (3) of all the biomarkers, MRI retains the strongest relationship with cognitive impairment in the later stages. The other main conclusion from this paper was support for a model where the biomarker for Aβ amyloid deposition (CSF Aβ1–42) is the earliest of the 3 biomarkers examined to become abnormal.
We regard imaging and CSF biomarkers as in vivo indicators of specific pathologies in AD. Low CSF Aβ1–42 is a marker of Aβ amyloid plaque load, and CSF Aβ1–42 levels correlate inversely with total Aβ load in the brain.5,31 In this study, we found that Aβ amyloid deposition was significantly greater among ε4 carriers within each clinical group, which is consistent with earlier CSF14,15,32 and PET amyloid imaging33 studies. Increased CSF t-tau is a marker of neuronal injury, which correlates well with neurofibrillary tangle (NFT) stage and NFT load.5,34 Our results indicate that t-tau does not significantly differ by APOE genotype among CN or AD, which is in agreement with a majority of CSF t-tau studies.14,32 Atrophy on structural MRI is a biomarker of neurodegeneration, and it too correlates with Braak NFT stage and quantitative NFT burden.35–40 However, the most proximate histological correlate of MRI volume loss is loss of synapses and neurons.7,41 Our finding of no association of neurodegeneration (as measured by MRI) and APOE genotype among CN or AD subjects is also consistent with some earlier MRI studies.18,19,42–44
Observed Relationships between APOE, Biomarkers, and Baseline Clinical Status
CSF Aβ1–42 is low in APOE ε4 carriers in all clinical groups, and therefore our data support the hypothesis that the primary pathological effect of APOE ε4 is to increase Aβ amyloid plaque formation by any of several potential mechanisms, including reducing the efficiency of Aβ clearance.45 A plausible model of the development of AD posits that amyloid deposition occurs early in the process but by itself does not directly cause clinical symptoms.46–48 Impaired cognitive performance is largely driven by neurodegeneration, which may be mediated by tau pathology. Based on this evidence, it has been hypothesized that AD pathological cascade is a 2-stage process where amyloidosis and neuronal pathology (tauopathy, neuronal injury, and neurodegeneration) are largely sequential rather than simultaneous processes.47,49
Our data show that MRI correlates more closely with cognitive status than with APOE genotype. Also, there is some evidence that t-tau correlates better with cognitive status than with APOE genotype. Thus, whereas we see significant differences between the CSF Aβ1–42 levels of ε4 carriers and noncarriers in all clinical groups, t-tau and MRI values do not differ significantly between ε4 carriers and noncarriers among CN or AD subjects. In patients with clinically diagnosed AD, the influence of APOE genotype on cognitive decline appears most consistently present in milder patients, and less evident or absent when patients with more advanced cognitive decline are examined.50 This is not to say that APOE ε4 is unrelated to indicators of neuronal pathology. When all subjects are combined, APOE ε4 clearly increases the odds that any individual will be more impaired clinically, and have higher t-tau and a higher STAND score. APOE ε4 is not deterministic, in the sense that there are many ε4 carriers who are not demented and many ε4 noncarriers who are demented. In contrast, subjects with highly abnormal STAND values are almost invariably demented, and those with normal STAND are almost invariably cognitively normal regardless of APOE genotype.
There was evidence of lower median age in aMCI ε4 carriers when compared with ε4 noncarriers, which suggests that ε4 carriers might have slightly more cognitive reserve (brain reserve, ie, less age-related atrophy and brain resiliency) when compared with noncarriers. This possibly explains why STAND was worse in aMCI ε4 carriers when compared with ε4 noncarriers, that is, more atrophy in younger subjects brought them to the same cognitive level of less atrophy in older subjects. This along with evidence that MRI atrophy does not differ by APOE ε4 status in CN and AD subjects strengthens the argument that MRI as a marker of the actual stage of neurodegeneration is more closely related to the present clinical status.
Effect of APOE on the Biomarkers across the Alzheimer's Disease Continuum
EFFECT OF APOE ON CSF Aβ1–42
Age of clinical AD onset is lowered by 5 to 10 years in ε4 carriers relative to noncarriers.1,51,52 This is supported in our data by the fact that among both AD and aMCI subjects ε4 carriers are younger than noncarriers; that is, carriers reach the same clinical disease stage at a younger age. Our data show that CSF Aβ1–42 is lower in ε4 carrier CN subjects relative to noncarriers, and does not differ noticeably between AD/aMCI ε4 carriers and CN ε4 carriers. This can be interpreted to indicate that CSF Aβ1–42 has reached a nadir while APOE 4 carrier subjects are still cognitively normal, whereas Aβ1–42 falls progressively in ε4 noncarriers from CN to aMCI to AD.
The observed effect of APOE ε4 is to cause a plateau in the CSF Aβ1–42 levels early in the clinical disease progression, such that worsening MMSE is not accompanied by worsening CSF Aβ1–42. In contrast, in ε4 noncarriers the relationship between CSF Aβ1–42 and MMSE remains roughly linear into lower levels of MMSE performance. Both these relationships can be observed in Figure 2. We do acknowledge that the assumption here that APOE ε4 carriers who are currently cognitively normal had normal CSF Aβ1–42 at an earlier time in life cannot be proven by our data. However, a recent nonselected all-age autopsy series53 convincingly demonstrates that APOE ε4 does shift the onset of Aβ accumulation to an earlier age relative to noncarriers, with the greatest difference in the plaque load as a function of APOE genotype occurring in the 50-to 59-year age group.
EFFECT OF APOE ON CSF T-TAU
There was no cross-sectional difference in t-tau between aMCI and AD in ε4 carriers presumably with more advanced disease, but t-tau does differ between aMCI and AD in ε4 noncarriers (see Table 2). These data can be interpreted to mean that t-tau increases may have plateaued by the aMCI stage in the more advanced ε4 aMCI carriers, but not in the less advanced ε4 noncarriers. This argument is strengthened by Figure 2B.
EFFECT OF APOE ON MRI ATROPHY
There were cross-sectional differences on MRI between aMCI and AD in both ε4 carriers and noncarriers (see Table 2), and the variability in STAND scores is largely driven by cognitive status and less by APOE genotype (see Table 3). These data can be interpreted to indicate that, unlike t-tau, brain atrophy does not plateau by the aMCI stage even in the more advanced ε4 carriers, and hence MRI retains its close relationship with clinical status later into the clinical disease progression than t-tau. The evidence for this can also be seen in Figure 2, where the relationship between MMSE and STAND scores remains linear across the cognitive spectrum in both ε4 carriers and noncarriers.
TEMPORAL ORDERING OF BIOMARKERS
Although biomarker assessments were obtained only at baseline in this study, we found evidence for a temporally ordered sequencing of CSF Aβ1–42, CSF t-tau, and MRI. The specific findings in this study support the comprehensive model of AD proposed earlier.47,54 The main observed effect of APOE genotype was to shift the entire AD biomarker cascade toward younger age, which results in an earlier onset of AD in ε4 carriers.
An important point is that the aMCI group is heterogeneous. Based on prior studies, some of these individuals simply have poor memory performance and will never progress to dementia, whereas others will go on to develop clinical AD. Some (particularly ε4 noncarriers) likely have substrates for cognitive impairment other than AD, for example, vascular disease or Lewy body disease. Many likely have a mixture of pathologies including but not confined to AD.55,56
There are some limitations to the study. First, the ADNI cohort is not a population-based cohort. The recruitment mechanisms were those used for clinical trials in AD, and included memory clinics, patient registries, public media campaigns, and other forms of public advertisements. Consequently inferences about the diagnostic sensitivity and specificity of biomarkers in the general population cannot be drawn from ADNI data. However, biologically based conclusions concerning the effect of APOE genotype on AD biomarkers are valid.
Acknowledgments
This work was supported by NIH National Institute on Aging grant AG11378; a Robert H. Smith Family Foundation Research Fellowship; the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation, U.S.A.; and Opus building NIH grant C06 RR018898.
The Foundation for the National Institutes of Health (www.fnih.org) coordinates the private sector participation of the $60 million ADNI public–private partnership that was begun by the National Institute on Aging and supported by the NIH. To date, more than $27 million has been provided to the Foundation for NIH by Abbott, AstraZeneca AB, Bayer Schering PharmaAG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly and Co., Merck & Co. Inc., Novartis AG, Pfizer Inc., F. Hoffmann-LaRoche, Schering-Plough, Synarc Inc., and Wyeth, as well as nonprofit partners the Alzheimer's Association and the Institute for the Study of Aging.
Footnotes
Additional supporting information may be found in the online version of this article.
Statistical analysis was conducted by Heather J. Wiste, BA and Stephen D. Weigand, MS.
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in analysis or the writing of this report. A complete listing of ADNI investigators is available at www.loni.ucla.edu\ADNI\Collaboration\ADNI_Manuscript_Citations.pdf
References
- 1.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 4.Mayeux R, Stern Y, Ottman R, et al. The apolipoprotein epsilon 4 allele in patients with Alzheimer's disease. Ann Neurol. 1993;34:752–754. doi: 10.1002/ana.410340527. [DOI] [PubMed] [Google Scholar]
- 5.Tapiola T, Alafuzoff I, Herukka SK, et al. Cerebrospinal fluid {beta}-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch Neurol. 2009;66:382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 6.Blennow K, Wallin A, Agren H, et al. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol Chem Neuropathol. 1995;26:231–245. doi: 10.1007/BF02815140. [DOI] [PubMed] [Google Scholar]
- 7.Bobinski M, de Leon MJ, Wegiel J, et al. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer's disease. Neuroscience. 2000;95:721–725. doi: 10.1016/s0306-4522(99)00476-5. [DOI] [PubMed] [Google Scholar]
- 8.Vemuri P, Gunter JL, Senjem ML, et al. Alzheimer's disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Isla T, West HL, Rebeck GW, et al. Clinical and pathological correlates of apolipoprotein E epsilon 4 in Alzheimer's disease. Ann Neurol. 1996;39:62–70. doi: 10.1002/ana.410390110. [DOI] [PubMed] [Google Scholar]
- 10.Hyman BT, Gomez-Isla T, West H, et al. Clinical and neuropatho-logical correlates of apolipoprotein E genotype in Alzheimer's disease. Window on molecular epidemiology. Ann N Y Acad Sci. 1996;777:158–165. doi: 10.1111/j.1749-6632.1996.tb34414.x. [DOI] [PubMed] [Google Scholar]
- 11.Mukaetova-Ladinska EB, Harrington CR, Roth M, Wischik CM. Presence of the apolipoprotein E type epsilon 4 allele is not associated with neurofibrillary pathology or biochemical changes to tau protein. Dement Geriatr Cogn Disord. 1997;8:288–295. doi: 10.1159/000106646. [DOI] [PubMed] [Google Scholar]
- 12.Hartman RE, Laurer H, Longhi L, et al. Apolipoprotein E4 influences amyloid deposition but not cell loss after traumatic brain injury in a mouse model of Alzheimer's disease. J Neurosci. 2002;22:10083–10087. doi: 10.1523/JNEUROSCI.22-23-10083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy Z, Esiri MM, Jobst KA, et al. Influence of the apolipoprotein E genotype on amyloid deposition and neurofibrillary tangle formation in Alzheimer's disease. Neuroscience. 1995;69:757–761. doi: 10.1016/0306-4522(95)00331-c. [DOI] [PubMed] [Google Scholar]
- 14.Galasko D, Chang L, Motter R, et al. High cerebrospinal fluid tau and low amyloid beta42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch Neurol. 1998;55:937–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- 15.Tapiola T, Pirttila T, Mehta PD, et al. Relationship between apoE genotype and CSF beta-amyloid (1-42) and tau in patients with probable and definite Alzheimer's disease. Neurobiol Aging. 2000;21:735–740. doi: 10.1016/s0197-4580(00)00164-0. [DOI] [PubMed] [Google Scholar]
- 16.Ganzer S, Arlt S, Schoder V, et al. CSF-tau, CSF-Abeta1–42, ApoE-genotype and clinical parameters in the diagnosis of Alzheimer's disease: combination of CSF-tau and MMSE yields highest sensitivity and specificity. J Neural Transm. 2003;110:1149–1160. doi: 10.1007/s00702-003-0017-7. [DOI] [PubMed] [Google Scholar]
- 17.Engelborghs S, Sleegers K, Cras P, et al. No association of CSF biomarkers with APOEepsilon4, plaque and tangle burden in definite Alzheimer's disease. Brain. 2007;130:2320–2326. doi: 10.1093/brain/awm136. [DOI] [PubMed] [Google Scholar]
- 18.Jack CR, Jr, Petersen RC, Xu YC, et al. Hippocampal atrophy and apolipoprotein E genotype are independently associated with Alzheimer's disease. Ann Neurol. 1998;43:303–310. doi: 10.1002/ana.410430307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barber R, Gholkar A, Scheltens P, et al. Apolipoprotein E epsilon4 allele, temporal lobe atrophy, and white matter lesions in late-life dementias. Arch Neurol. 1999;56:961–965. doi: 10.1001/archneur.56.8.961. [DOI] [PubMed] [Google Scholar]
- 20.Moffat SD, Szekely CA, Zonderman AB, et al. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurology. 2000;55:134–136. doi: 10.1212/wnl.55.1.134. [DOI] [PubMed] [Google Scholar]
- 21.Fleisher A, Grundman M, Jack CR, Jr, et al. Sex, apolipoprotein E epsilon 4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62:953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- 22.Schuff N, Woerner N, Boreta L, et al. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drzezga A, Riemenschneider M, Strassner B, et al. Cerebral glucose metabolism in patients with AD and different APOE genotypes. Neurology. 2005;64:102–107. doi: 10.1212/01.WNL.0000148478.39691.D3. [DOI] [PubMed] [Google Scholar]
- 24.Mielke R, Zerres K, Uhlhaas S, et al. Apolipoprotein E polymorphism influences the cerebral metabolic pattern in Alzheimer's disease. Neurosci Lett. 1998;254:49–52. doi: 10.1016/s0304-3940(98)00673-9. [DOI] [PubMed] [Google Scholar]
- 25.Reiman EM, Chen K, Alexander GE, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci U S A. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's Disease Neuroimaging Initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack CR, Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 29.Hughes CP, Berg L, Danziger WL, et al. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 30.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Characterization of CSF Aβ1-42, Tau And P-Tau 181P concentrations at baseline In Alzheimer's Disease Neuroimaging Initiative (ADNI) study cohorts. Alzheimers Dement. 2008;4:545–546. [Google Scholar]
- 31.Strozyk D, Blennow K, White LR, Launer LJ. CSF Abeta 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology. 2003;60:652–656. doi: 10.1212/01.wnl.0000046581.81650.d0. [DOI] [PubMed] [Google Scholar]
- 32.Sunderland T, Mirza N, Putnam KT, et al. Cerebrospinal fluid beta-amyloid1–42 and tau in control subjects at risk for Alzheimer's disease: the effect of APOE epsilon4 allele. Biol Psychiatry. 2004;56:670–676. doi: 10.1016/j.biopsych.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Reiman EM, Chen K, Liu X, et al. Fibrillar amyloid-{beta} burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buerger K, Ewers M, Pirttila T, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer's disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- 35.Csernansky JG, Hamstra J, Wang L, et al. Correlations between antemortem hippocampal volume and postmortem neuropathology in AD subjects. Alzheimer Dis Assoc Disord. 2004;18:190–195. [PubMed] [Google Scholar]
- 36.Gosche KM, Mortimer JA, Smith CD, et al. Hippocampal volume as an index of Alzheimer neuropathology: findings from the Nun Study. Neurology. 2002;58:1476–1482. doi: 10.1212/wnl.58.10.1476. [DOI] [PubMed] [Google Scholar]
- 37.Jack CR, Jr, Dickson DW, Parisi JE, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002;58:750–757. doi: 10.1212/wnl.58.5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silbert LC, Quinn JF, Moore MM, et al. Changes in premorbid brain volume predict Alzheimer's disease pathology. Neurology. 2003;61:487–492. doi: 10.1212/01.wnl.0000079053.77227.14. [DOI] [PubMed] [Google Scholar]
- 39.Jagust WJ, Zheng L, Harvey DJ, et al. Neuropathological basis of magnetic resonance images in aging and dementia. Ann Neurol. 2008;63:72–80. doi: 10.1002/ana.21296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vemuri P, Whitwell JL, Kantarci K, et al. Antemortem MRI based STructural Abnormality iNDex (STAND)-scores correlate with postmortem Braak neurofibrillary tangle stage. Neuroimage. 2008;42:559–567. doi: 10.1016/j.neuroimage.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarow C, Vinters HV, Ellis WG, et al. Correlates of hippocampal neuron number in Alzheimer's disease and ischemic vascular dementia. Ann Neurol. 2005;57:896–903. doi: 10.1002/ana.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jak AJ, Houston WS, Nagel BJ, et al. Differential cross-sectional and longitudinal impact of APOE genotype on hippocampal volumes in nondemented older adults. Dement Geriatr Cogn Disord. 2007;23:382–389. doi: 10.1159/000101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemaitre H, Crivello F, Dufouil C, et al. No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage. 2005;24:1205–1213. doi: 10.1016/j.neuroimage.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt H, Schmidt R, Fazekas F, et al. Apolipoprotein E e4 allele in the normal elderly: neuropsychologic and brain MRI correlates. Clin Genet. 1996;50:293–299. doi: 10.1111/j.1399-0004.1996.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 45.Mahley RW, Huang Y. Alzheimer disease: multiple causes, multiple effects of apolipoprotein E4, and multiple therapeutic approaches. Ann Neurol. 2009;65:623–625. doi: 10.1002/ana.21736. [DOI] [PubMed] [Google Scholar]
- 46.Jack CR, Jr, Lowe VJ, Senjem ML, et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008;131:665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jack CR, Jr, Lowe VJ, Weigand SD, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: implications for sequence of pathological events in Alzheimer's disease. Brain. 2009;132:1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mormino EC, Kluth JT, Madison CM, et al. Episodic memory loss is related to hippocampal-mediated {beta}-amyloid deposition in elderly subjects. Brain. 2009;132(pt 5):1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingelsson M, Fukumoto H, Newell KL, et al. Early Abeta accumulation and progressive synaptic loss, gliosis, and tangle formation in AD brain. Neurology. 2004;62:925–931. doi: 10.1212/01.wnl.0000115115.98960.37. [DOI] [PubMed] [Google Scholar]
- 50.Growdon JH, Locascio JJ, Corkin S, et al. Apolipoprotein E genotype does not influence rates of cognitive decline in Alzheimer's disease. Neurology. 1996;47:444–448. doi: 10.1212/wnl.47.2.444. [DOI] [PubMed] [Google Scholar]
- 51.Rebeck GW, Perls TT, West HL, et al. Reduced apolipoprotein epsilon 4 allele frequency in the oldest old Alzheimer's patients and cognitively normal individuals. Neurology. 1994;44:1513–1516. doi: 10.1212/wnl.44.8.1513. [DOI] [PubMed] [Google Scholar]
- 52.Ashford JW. APOE genotype effects on Alzheimer's disease onset and epidemiology. J Mol Neurosci. 2004;23:157–165. doi: 10.1385/JMN:23:3:157. [DOI] [PubMed] [Google Scholar]
- 53.Kok E, Haikonen S, Luoto T, et al. Apolipoprotein E-dependent accumulation of Alzheimer disease-related lesions begins in middle age. Ann Neurol. 2009;65:650–657. doi: 10.1002/ana.21696. [DOI] [PubMed] [Google Scholar]
- 54.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Modeling dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider JA, Arvanitakis Z, Bang W, Bennett DA. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69:2197–2204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- 56.Jicha GA, Parisi JE, Dickson DW, et al. Neuropathologic outcome of mild cognitive impairment following progression to clinical dementia. Arch Neurol. 2006;63:674–681. doi: 10.1001/archneur.63.5.674. [DOI] [PubMed] [Google Scholar]