Abstract
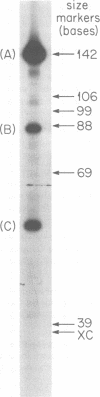
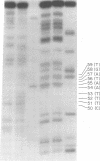
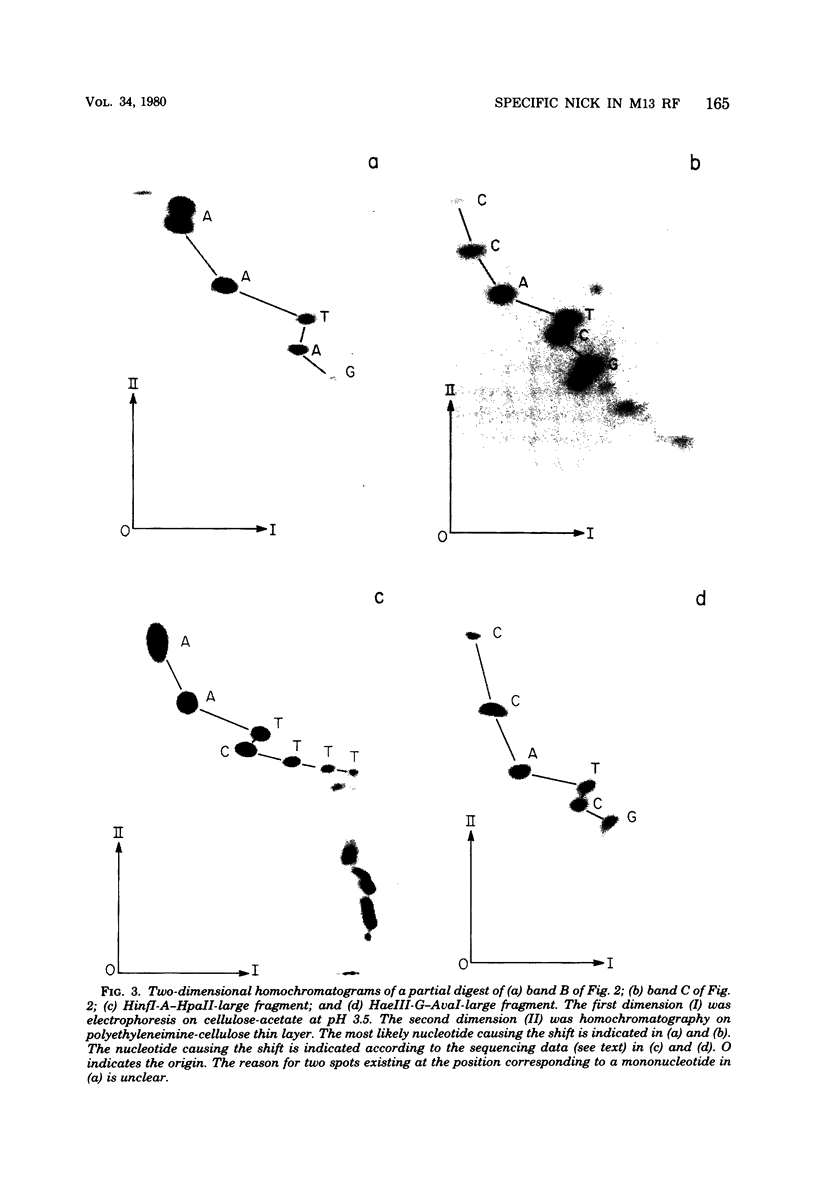
M13 replicative form II (RFII) DNA was prepared from Escherichia coli RS5052 (polAex1) cells in the late stage of infection, and the DNA sequence at the discontinuity was examined. The data presented here suggest that the single discontinuity in the late stage of infection RFII maps at the same position as the gene II protein nicking site on fd RFI which was determined in vitro (Meyer et al., Nature (London) 278:365-367, 1979) and has a 5' terminal nucleotide sequence identical to that at the nick produced by gene II protein in vitro. The discontinuity in the in vivo RFII appears to be a single break in the phosphodiester backbone, leaving a 3' OH terminus. RFII molecules containing a gap, i.e., missing nucleotides at the site of discontinuity, were not detected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. P., Ganesan A. T., Olson A. C., Snyder C. M., Mitra S. Electron microscopic studies of bacteriophage M13 DNA replication. J Virol. 1977 Nov;24(2):673–684. doi: 10.1128/jvi.24.2.673-684.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Chen T. C., Ray D. S. Replication of bacteriophage M13. X. M13 replication in a mutant of Escherichia coli defective in the 5' leads to 3' exonuclease associated with DNA polymerase I. J Mol Biol. 1976 Sep 25;106(3):589–604. doi: 10.1016/0022-2836(76)90253-9. [DOI] [PubMed] [Google Scholar]
- Chen T. C., Ray D. S. Replication of bacteriophage M13. XIV. Differential inhibition of the replication of M13 and M13 miniphage in a mutant of Escherichia coli defective in the 5' leads to 3' exonuclease associated with DNA polymerase I. J Virol. 1978 Dec;28(3):679–685. doi: 10.1128/jvi.28.3.679-685.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Harbers B., Hours C., Denhardt D. T. The mechanism of replication of phiX174 DNA. XII. Non-random location of gaps in nascent phiX174 RF II DNA. J Mol Biol. 1975 Nov 25;99(1):107–123. doi: 10.1016/s0022-2836(75)80162-8. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Scott J. F., Kronberg A. Enzymatic replication of phiX174 duplex circles: continuous synthesis. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):295–302. doi: 10.1101/sqb.1979.043.01.036. [DOI] [PubMed] [Google Scholar]
- Fidanián H. M., Ray D. S. Replication of bacteriophage M13. VII. Requirement of the gene 2 protein for the accumulation of a specific RFII species. J Mol Biol. 1972 Dec 14;72(1):51–63. doi: 10.1016/0022-2836(72)90067-8. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Ravetch J. V., Zinder N. D. DNA replication of bacteriophage f1 in vivo. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):389–399. doi: 10.1101/sqb.1979.043.01.045. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. H., Sinsheimer R. L. Structure of an intermediate in the replication of bacteriophage phiX174 deoxyribonucleic acid: the initiation site for DNA replication. J Mol Biol. 1974 Feb 15;83(1):47–61. doi: 10.1016/0022-2836(74)90423-9. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A simplified method for preparation of mouse satellite DNA. Anal Biochem. 1977 Apr;78(2):561–568. doi: 10.1016/0003-2697(77)90118-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Suggs S. V., Ray D. S. Nucleotide sequence of the origin for bacteriophage M13 DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):379–388. doi: 10.1101/sqb.1979.043.01.044. [DOI] [PubMed] [Google Scholar]
- Suggs S. V., Ray D. S. Replication of bacteriophage M13. XI. Localization of the origin for M13 single-strand synthesis. J Mol Biol. 1977 Feb 15;110(1):147–163. doi: 10.1016/s0022-2836(77)80103-4. [DOI] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Ikeda J. E., Benz E., Marians K. J., Vicuna R., Sugrue S., Zipursky S. L., Hurwitz J. Replication of phiX174 DNA: in vitro synthesis of phiX RFI DNA and circular, single-stranded DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):311–329. doi: 10.1101/sqb.1979.043.01.038. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Jay E., Bahl C. P., Wu R. A reliable mapping method for sequence determination of oligodeoxyribonucleotides by mobility shift analysis. Anal Biochem. 1976 Jul;74(1):73–93. doi: 10.1016/0003-2697(76)90311-0. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]