Abstract
Virtually all of the 560 human proteases are stored as inactive proenyzmes and are strictly regulated. We report the identification and characterization of small molecules that directly activate the apoptotic proenzymes, procaspases-3 and -6. Surprisingly, these compounds induce autoproteolytic activation by stabilizing a conformation that is both more active and more susceptible to intermolecular proteolysis. These procaspase activators bypass the normal upstream proapoptotic signaling cascades and induce rapid apoptosis in a variety of cell lines. Systematic biochemical and biophysical analyses identified a cluster of mutations in procaspase-3 that resist small molecule activation both in vitro and in cells. Compounds that induce gain-of-function are rare and the activators reported here will enable direct control of the homodimeric executioner caspases in apoptosis and in cellular differentiation.
Activation of proteases trigger a myriad of fate-determining biological events, such as apoptosis and blood clotting, both inside and outside of the cell (1). Proteases are generally stored as inactive proenzymes that are usually activated by upstream proteases or by themselves. These activation events may sometimes involve binding a protein partner or, in rare instances, interaction with a natural small molecule (2) or peptide (3). In the case of autoproteolysis, the proenzyme must achieve not only an active state, but also one in which the sites of proteolysis are exposed. In situ activation of specific proproteases with synthetic small molecules would uncover new molecular principles in zymogen activation and facilitate direct control of these important processes in biology.
The executioner procaspases, consisting of procaspases-3, -6 and -7, represent excellent initial candidates for discovery of small molecule protease activators. Caspases are a family of homodimeric cysteine proteases responsible for many of the fate-determining processes in cell biology including apoptosis, innate immune signaling, early stages of stem cell differentiation, and cellular remodeling (4–6). As with most proteases, caspases are synthesized as inactive procaspases, or zymogens, and are activated by upstream proteolysis or autoproteolysis. Previous studies have shown that the mature active caspases are intrinsically dynamic (7, 8) and sample both an “on-state” and an “off-state” that structurally resembles the zymogen-like conformation (9, 10). Small molecules have been found to trap these two forms of the mature enzyme (11, 12). We reasoned that, if the procaspases existed in a similar dynamic equilibrium of off- and on-states, it might be possible to find small molecules that promote autoproteolytic activation via stabilization of an on-state conformation. Executioner procaspases are particularly good targets as they are susceptible to rapid activation by both upstream proteases and self-proteolysis. Thus, any activation would be accentuated in trans by autocatalytic activation. Moreover, these particular caspases are essential for executing the final processes of apoptosis, and specific activation by a small molecule would elicit robust and precise phenotypic responses within cells.
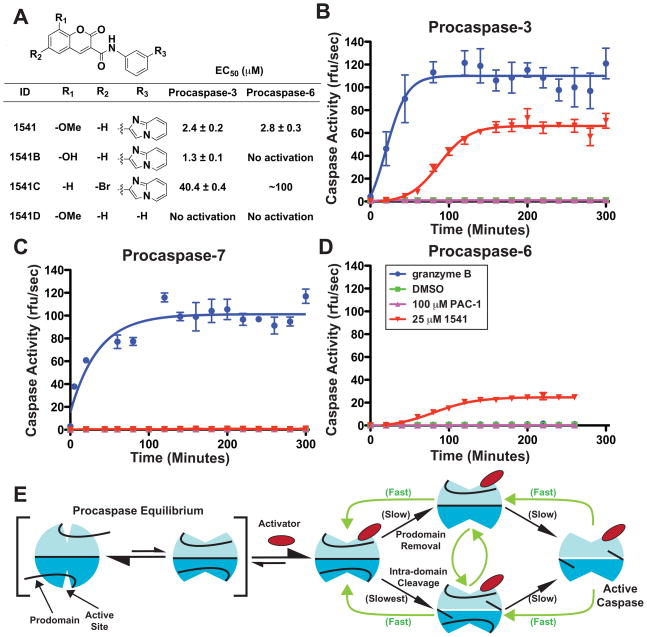
High-throughput screening was employed to identify compounds that could promote autoproteolytic activation of procaspase-3 at physiological concentrations (see Supporting Online Material for HTS design). A dozen compounds out of 62,000 promoted >20-fold activation of procaspase-3 (Fig. S1), and were re-synthesized to validate their chemical composition. To warrant further analysis, dynamic light scattering was used to select compounds with solubilities greater than 100 μM, and a β-lactamase inhibition assay was performed to discard promiscuous aggregators (13). The most robust procaspase activator fulfilling these criteria was compound 1541, a substituted phenyl-imidazopyridine-methoxy coumarin with an EC50 for activation of 2.4 μM (Fig. 1A). Mass spectrometry revealed the compound did not covalently label either the mature or proenzyme forms of caspase-3, nor was 1541 modified by enzyme hydrolysis.
Fig. 1.
Specificity of small molecule activators for executioner procaspases. (A) Chemical analogs of 1541 and EC50 values of procaspases-3 and -6 activation. Small molecule activators require the imidazopyridine moiety and prefer a heteroatom substituent at the 8-position on the coumarin ring (39)(40). Substitution of the 8-methoxy with an 8-hydroxy (1541B) improves the EC50 and specificity for activation of procaspase-3 over procaspase-6. Addition of a 6-bromo (1541C) reduces potency to both procaspases. Complete removal of the imidazopyridine substituent (1541D) ablates activation of procaspases-3 and -6. (B–D) Time course of executioner procaspase activation by 1541 facilitates full self-cleavage of procaspases-3 and -6. Procaspases-3 (B), -7 (C) and -6 (D) were incubated at 100 nM with 1 nM granzyme B (blue), DMSO (green on baseline) 100 μM PAC-1 (magenta) or 25 μM 1541 (red) at 37 °C and assayed for activity by addition of fluorogenic peptide substrates Ac-DEVD-AFC (caspases-3 and -7) or Ac-VEID-AFC (caspase-6) every 20 minutes (41). (E) Proposed model for small molecule-assisted procaspase self-activation. We hypothesize that procaspases are in a dynamic equilibrium between an off-state (left) and on-state (right), similar to mature caspases. Unlike mature caspases, the population favors the off-state conformation. Upon binding a small molecule activator, the equilibrium shifts to an on-state (center). This complex slowly undergoes autoproteolytic activation (black arrows) that accelerates with increasing production of mature caspase (green arrows). This model also accounts for activation at low concentrations of small molecule where the on-state is preferred and one active site is available in the dimer for processing, and, at high concentrations, where both sites are saturated and can lead to inhibition.
We compared the rate of 1541 activation and specificity for the executioner procaspases-3, -6 and -7 that share 40–50% sequence identity. Granzyme B, a natural proteolytic activator of procaspase-3, rapidly and fully activates the zymogen within 90 minutes. In contrast, after an initial slow phase that lasts ~30 minutes, 1541 induces accelerated activation of caspase-3 to a level that is finally ~70% that of granzyme B within 2.5 hours (Fig. 1B). Granzyme B and 1541 both induce full proteolytic processing of procaspase-3, as evidenced by production of the large (17 kDa) and small (12 kDa) subunits of caspase-3 (Fig. S2). Procaspase-7 is robustly activated and fully processed by granzyme B, but not by 1541 (Fig. 1C). 1541 does induce ~10% self cleavage of its prodomain (residues 1-23) (Fig. S2), but prodomain removal by itself does not elicit caspase-7 activation (14). Procaspase-6 is resistant to granzyme B (15, 16) but, 1541 promotes activation and complete self-processing similar to procaspase-3 (Fig. 2D) (17). Furthermore, 1541 does not activate or induce self-cleavage of inflammatory procaspase-1 (Fig. S3) (18). Thus, 1541 is a highly specific and robust activator of executioner procaspases-3 and -6, and does not activate procaspases-1 or -7. We also tested PAC-1, a compound previously reported to induce a slight activation of procaspase-3 (19–21), but found no detectable increase in activity or self-proteolysis among the executioner procaspases (Figs. 1B–D). Others have reported a similar inability to activate procaspase-3 with PAC-1 (22).
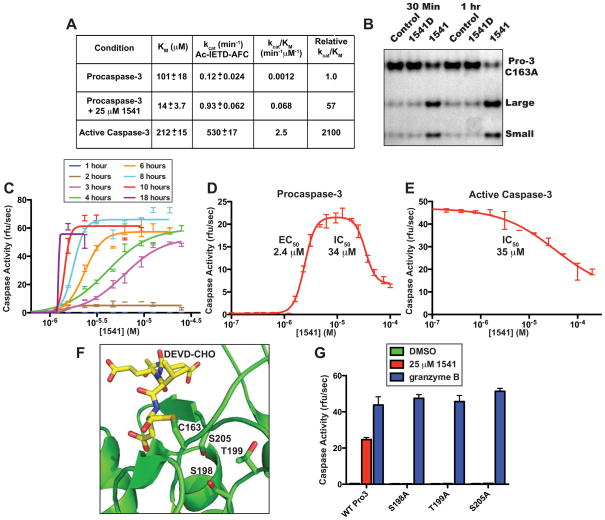
Fig. 2.
In vitro characterization of apoptotic procaspase small molecule activators. (A) Initial rates of activation of procaspase-3 in presence of 1541B, as measured by Ac-IETD-AFC compared to unstimulated zymogen and mature caspase-3. Procaspase-3 (100 nM) was incubated with 1541B (0.78 to 50 μM) and initial rates of activity were measured over ~15 min by addition of Ac-IETD-AFC (1.2 to 300 μM) at 37 °C. Presence of 1541 improves the kcat/KM of procaspase-3 by 57-fold and stimulates autocatalytic processing. (B) Silver-stained SDS-PAGE gel of inactive C163A procaspase-3 (250 nM) incubated with granzyme B (0.5 nM) with and without 1541 (25 μM) shows increased cleavage susceptibility by granzyme B with 1541 but not with the inactive analog 1541D. (C) Procaspase-3 was incubated at 100 nM with 1541 (0.1 to 100 μM) to assess rate of self-activation. Kinetic activity of the mixtures was determined every hour for 18 hours by incubation with 20 μM Ac-DEVD-AFC. Once procaspase-3 is activated by 1541, the zymogen rapidly self-activates. The effective concentration of 1541 is approximately 1.5 μM with no activation below this concentration despite extended incubation periods. (D) Procaspase-3 at 100 nM was incubated with 1541 (100 nM to 100 μM). After a 4-hour incubation at 37 °C, the samples were assayed for caspase activity by addition of Ac-DEVD-AFC. Low concentrations of 1541 induce activity of procaspase-3 with an EC50 of 2.4 μM, and, at high concentrations, inhibit with an IC50 of 34.0 μM. (E) 1541 exerts a similar inhibitory effect on active caspase-3 (IC50 = 35 μM), as seen for procaspase-3. Mature caspase-3 was incubated at 25 nM in various concentrations of 1541 for 10 minutes and assayed with Ac-DEVD-AFC. (F) Structural model of the active site of procaspase-3 showing the residues targeted for mutation. The location of the DEVD substrate is shown and modeled from caspase-3 in complex with Ac-DEVD-CHO (PDB 2DKO). (G) Identification of mutations in procaspase-3 that confer resistance to activation by 1541. Wild type and mutant (S198A, T199A and S205A) procaspases were incubated for 5 hours with 25 μM 1541 (red), 1 nM granzyme B (blue) or DMSO (green on baseline) at 37 °C and sampled for activation via kinetic assays initiated by addition of substrate Ac-DEVD-AFC. The procaspase-3 variants S198A, T199A and S205A displayed wild type activity when activated by granzyme B, but were unable to self-activate in the presence of 1541.
To further validate activity and specificity, several 1541 analogs with modifications on the coumarin ring were tested (Fig. 1A, and Fig. S4). Substitution of the 8-methoxy with an 8-hydroxy (1541B) improves the EC50 by about 2-fold for activation of procaspase-3, but dramatically reduces activity toward procaspase-6. A 6-bromo substitution (1541C) reduces potency by ~15-fold for procaspase-3 and ~35-fold for procaspase-6. Deletion of the imidazopyridine substituent meta to the coumarin on the phenyl ring (1541D) eliminates activation of procaspases-3 and -6 (Fig. 1A). Thus, relatively modest modifications to 1541 can alter efficacy and selectivity and suggest 1541 has high specificity to the site of interaction.
A series of in vitro biochemical and kinetic analyses were performed to assess the activation mechanism of procaspase-3 by 1541 and test our initial hypothesis of autoproteolysis through stabilization of an on-state conformation (Fig. 1E). Mature caspase-3 has a catalytic efficiency (kcat/KM) of 2.5 min−1μM−1 for hydrolyzing a fluorogenic Ac-IETD-AFC reporter substrate that mimics the activating cleavage site (residues 172-175) between the large and small subunits (Fig. 2A). Procaspase-3 is substantially less active (2100-fold) with a corresponding kcat/KM of 0.0012 min−1μM−1. We titrated procaspase-3 with compound 1541B (from 0 to 50 μM) and measured the kinetic parameters for Ac-IETD-AFC by Michaelis-Menten analysis within 15 min, where no detectable processing or self-activation of procaspase-3 occurred that would contribute to substrate recognition and cleavage. As 1541B is increased, a dose-dependent 7-fold decrease is observed in KM (101 μM to 14 μM) and an 8-fold increase in kcat (0.12 to 0.93 min−1) (Fig. 2A). Compound 1541B, therefore, induces an immediate kinetic state where the catalytic efficiency is increased 57-fold (kcat/KM of 0.068 min−1μM−1) over unstimulated procaspase-3. Importantly, the KM value for procaspase-3 improves to 14 μM in the presence of 1541B, compared to 212 μM and 101 μM for the mature enzyme and unstimulated proenzyme, respectively. These data strongly imply that the initial catalytic activity of 1541B-stimulated procaspase-3 results from a different on-state conformation than that of mature caspase-3, and represents the first intermediate of the activator-bound procaspase-3 (Fig. 1E). For hydrolyzing amide substrates, KM is a reasonable estimate of the KD (23, 24). Thus, the 1541B-activated procaspase-3 binds Ac-IETD-AFC about 15-fold tighter than the mature enzyme albeit with a kcat value substantially lower than the mature enzyme (Fig. 1B) (25). Small amounts of mature caspase-3 that are generated in the presence of 1541 eventually overtake the activation process and account for the subsequent rapid increase in activity.
As activation of caspase zymogens requires cleavage of the junction between the large and small subunits, we tested whether these compounds could also enhance the proteolytic susceptibility of the proenzyme itself. We generated an inactive procaspase-3 by mutating the catalytic cysteine, C163A, and then tested whether 1541 enhances C163A processing by granzyme B. Incubation with granzyme B alone resulted in ~10% cleavage of C163A after 30 minutes and 20% after 60 minutes. Significantly, addition of 1541 substantially increased the rate of processing to 40% after 30 minutes, and 80% after 60 minutes (Fig. 2B). Co-incubation with a non-activating analog 1541D (Fig. 1A) did not enhance proteolysis by granzyme B. Thus, these activating compounds increase both the catalytic efficiency of unprocessed procaspase-3 (Fig. 2A) and its susceptibility to proteolysis (Fig. 2B). The ability to induce autoproteolytic activation was then evaluated by titrating 1541 (0.1 to 100 μM) with wild type procaspase-3 and measuring activation as a function of time (Fig. 2C). As the incubation times increase, the EC50 values of the activation curves shift to lower values and the slopes dramatically increase, which are indicative of a highly cooperative activation process consistent with the feedback activation proposed in Fig. 1E (26).
We next investigated why the maximal autoproteolytic activation of procaspase-3 was only about 70% of that induced by granzyme B (Fig. 1B) even though procaspase-3 was fully processed (Fig. S2). Titration of procaspase-3 with 1541 (from 0.1 to 100 μM) assayed after 4 hours shows a biphasic dose-response (Fig. 2D). At low concentrations, the enzyme is activated (EC50 of 2.4 μM) whereas, at high concentrations, it is partially inhibited (IC50 of 34 μM). The proenzyme becomes fully processed over the time course of the dose response study (EC50 of ~2μM; Fig. S6). To understand the basis for this inhibition, we directly tested 1541 on mature caspase-3. Indeed, caspase-3 is inhibited by 1541 with a virtually identical IC50 of 35 μM (Fig. 2E). A similar biphasic dose-response curve was observed for procaspase-6 (EC50 of 2.8 μM and IC50 of 38 μM, data not shown). Michaelis-Menten analyses revealed that 1541 acts as a competitive or mixed non-competitive inhibitor of caspases-3 and -6 (Fig. S7). Overall, these results are consistent with a mechanism whereby, at low concentration, a single 1541 molecule binds near one active site of the dimer as an inhibitor and stabilizes an on-state conformation that promotes self-cleavage using the unoccupied subunit (Fig. 1E). At very high concentrations, 1541 can inhibit both active sites.
Based on kinetic inhibition data of caspase-3 by 1541 (Fig. S4) and conservation with caspase-6, we employed alanine-scanning mutagenesis on residues near the active site to probe for resistance mutations (Fig. 2F). Three procaspase-3 mutants (S198A, T199A and S205A) rendered the proenzyme insensitive to 1541 activation. Each of the three variants were efficiently activated by granzyme B showing these were fully functional enzymes (Fig. 2G). These mutational studies indicate specific residues near the active site that alter binding of 1541 and/or susceptibility to autoproteolysis of wild type procaspase-3.
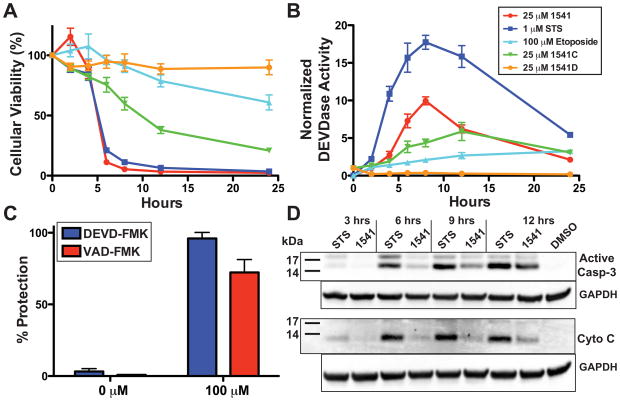
We next tested the ability of 1541 to induce apoptosis in a p53-deficient breast cancer cell line, BT549. Cells were treated with 1541 (25 μM), as well as with traditional inducers, such as staurosporine (STS) (a non-specific kinase inhibitor (27)) and etoposide (a topoisomerase II inhibitor (28–30)). Cells were probed every two hours for cellular viability and executioner caspase activity (DEVDase). After 4 hours, 1541 and STS induce comparable and near complete apoptosis of BT549 cells (Fig. 3A). Etoposide induced much slower apoptosis due to dependency on p53 for activity. The decrease in cellular viability in presence of STS and 1541 is accompanied by a reciprocal increase in DEVDase activity, characteristic of caspase-3, which peaks after 8 hours (Fig. 3B). Interestingly, DEVDase activity is significantly higher with STS than 1541. Western blots confirmed production of active caspases-3, -6 and -7 and the hallmark apoptotic cleavage of poly(ADP-ribose) polymerase (PARP); however, more extensive cleavage is observed with STS (Fig. S8). Procaspase-7 is also cleaved in cells treated with 1541, which likely reflects the ability of mature caspases-3 and/or -6 to activate procaspase-7 as previously reported (14) (Fig. S8).
Fig. 3.
Induction of apoptosis in a p53-deficient breast cancer cell line by 1541. (A) Time course of cell viability in BT549 cells. 2,000 BT549 cells were incubated with either 25 μM 1541 (red), 25 μM 1541C (green), 25 μM 1541D (orange), 1 μM STS (blue), 100 μM etoposide (light blue), or DMSO (final 0.5%) and assayed for cell viability at 2, 4, 6, 8, 12 and 24 hours. Analogs 1541C and D induce limited to no cellular death, respectively, with correlation to the in vitro effects on procaspase-3 self-activation. (B) Time course of DEVDase activity in BT549 cells. All concentrations and variables are conserved with panel 3A. 1541 induces less DEVDase activity in comparison to STS, but results in similar rates of apoptosis. (C) Protection against 1541-induced apoptosis with caspase inhibitors. 2,500 cells from BT549 were incubated overnight with the caspase-3/-7 inhibitor Biotin-DEVD-FMK (blue) or the general caspase inhibitor Biotin-VAD-FMK (red) from 0.78 to 100 μM. Cells were exchanged into new media containing 25 μM 1541 and respective concentration of peptide inhibitor. After 24 hours of incubation at 37 °C with 1541, the cells were assayed for cell viability. Caspase inhibitors prevented cellular apoptosis induced by 1541. (D) Cytoplasmic extracts of BT549 cells reveal limited cytochrome C release from mitochondria during 1541-induced apoptosis. One million BT549 cells were incubated with 25 μM 1541, 1 μM STS, or DMSO (0.5% final) and harvested every 3 hours. Cytoplasmic supernatants were subjected to Western blot analysis for presence of active caspase-3 or cytochrome C. Compared to STS, 1541 induced far less release of cytochrome C during apoptosis.
Two other cell lines were tested: MDA-MB361, a p53-proficient breast cancer cell line; and HEK293, a transformed human embryonic kidney cell line (Fig. S9). A similar pattern of cell viability and DEVDase activity was observed in MDA-MB361, as for BT549 cells. In HEK293 cells, 1541 was the most potent apoptotic inducer, compared to STS and etoposide, and generated the highest level of DEVDase activity. These data suggest that 1541 is as potent an inducer of cell death as STS and functions independent of p53. The fact that STS and 1541 induce comparable apoptotic rates, despite the higher caspase activity associated with STS, suggests that activation of only a fraction of the pool of executioner procaspases in these cells is required to induce cell death. Compounds 1541C and D, respectively, induce limited to no apoptosis or DEVDase activity in these three cell lines (Figs. 3A, S9). Significantly, the cellular results for these 1541 analogs correlate with their in vitro activity against procaspase-3 (Fig. 1A). Preincubation of cells with irreversible cell-permeable caspase inhibitors (VAD-FMK or DEVD-FMK) completely blocked apoptosis with 1541 indicating the caspase activity is responsible for the observed apoptosis (Fig. 3C).
The breadth of 1541 to induce apoptosis was then assessed in a variety of cancerous and transformed cell lines including BT549, MDA-MB361, HEK293, HeLa, and HCC1954. Cells were incubated with increasing concentrations of 1541 and assayed for cell death after 24 hours (Fig. S10). All cell lines underwent apoptosis with EC50 values ranging between 4 and 9 μM, similar to those measured in vitro for procaspases-3 and -6 (EC50 2–3 μM). Compound 1541B, which is comparably active toward procaspase-3, but inactive against procaspase-6, exhibited virtually the same EC50 for inducing apoptosis (Fig. S10). These results imply that direct activation of procaspase-3 alone is sufficient to induce rapid apoptosis.
We assessed the effects of 1541-induced apoptosis on apoptotic hallmarks and signaling pathways. Typical inducers of the intrinsic apoptosis pathway, such as STS, operate indirectly to activate executioner procaspases by releasing mitochondrial cytochrome C into the cytoplasm that leads to activation of procaspase-9, which targets executioner procaspases. Direct activation of procaspase-3 by 1541 should be independent of cytochrome C in the cytoplasm. BT549 cells were incubated with either 1541 or STS, harvested every 3 hours, and cytoplasmic extracts probed by Western blot to establish levels of activated caspase-3 and cytochrome C (top and bottom panel of Fig. 3D, respectively). Indeed, STS induced much greater and more rapid mitochondrial cytochrome C release than did 1541. As expected, small amounts of cytochrome C appeared in the cytosol at later time points with 1541, since positive feedback loops from activated caspase-3 are known to eventually produce mitochondrial damage and subsequent cytochrome C release (31).
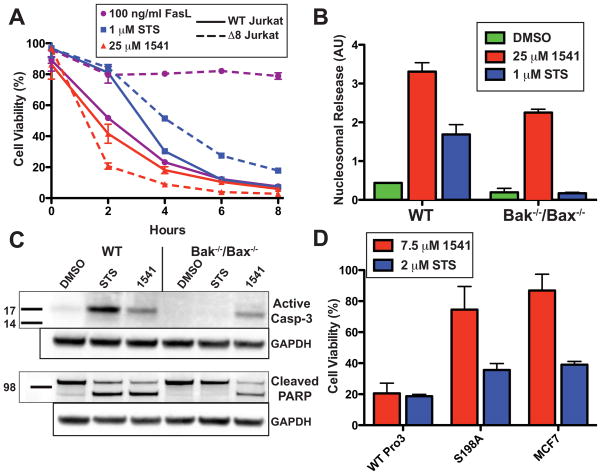
We next evaluated if 1541 induces apoptosis directly through the executioner caspases or if the compound requires an intact extrinsic or intrinsic pathway. To test the requirement for the extrinsic cell death pathway, wild type (A3) and caspase-8 deficient human Jurkat cells (I9.2) (32) were treated with 1541, STS and FasL, a specific inducer of the extrinsic pathway (Fig. 4A). Compound 1541 produced rapid and comparable rates of apoptosis in both wild type and caspase-8 deficient Jurkat cells. As expected, FasL was active in wild type Jurkats, but not in caspase-8 deficient cells. STS induction was comparable to 1541 in wild type Jurkats, and somewhat slower in caspase-8 deficient cells suggesting that the extrinsic pathway augments some of its activity. Thus, cells with genetic lesions within either the intrinsic (p53) or extrinsic (caspase-8) pathways are resistant to traditional intrinsic or extrinsic inducers, respectively, but both are susceptible to cell death by 1541.
Fig. 4.
Validation of executioner procaspases as cellular target of 1541. (A) Wild type (A3) (solid line) or caspase-8 deficient (I9.2) (dotted line) Jurkat cells (100,000 cells) were incubated with 25 μM 1541 (red), 1 μM STS (blue) or 100 ng/ml FasL (purple) and assayed for cellular viability every 2 hours. 1541 induces cell death independent of the extrinsic pathway. (B) 5,000 cells derived from either wild type or Bak−/−/Bax−/− DKO MEFs were incubated with either 1 μM STS (blue), 25 μM 1541 (red) or DMSO (green) (final 0.5%) at 37 °C. A histone sandwich ELISA (Roche) was performed on the lysates to determine the extent of apoptosis after 12 hours. Induction of cell death by 1541, in the absence of Bak and Bax, supports direct activation of the executioner procaspases. (C) For Western blot analysis, one million cells of both wild type and Bak−/−/Bax−/− DKO MEFs were plated and incubated with either 1 μM STS, 25 μM 1541 or DMSO (final 0.5%) at 37 °C for 12 hours. Western blots of whole-cell lysates were probed for the presence of active caspase-3 and cleavage of PARP. (D) Transfection of MCF-7 cells with wild type procaspase-3 and 1541-resistant S198A procaspase-3 supports ablation of 1541-induced procaspase-3 activation in cells. 6,000 cells from stably transfected MCF-7 cell lines were incubated for 24 hours with either 7.5 μM 1541 (red), 2 μM STS (blue) or DMSO (final concentration 0.5%) and assayed for cell viability. S198A procaspase-3 MCF-7 cells resisted apoptosis similarly to the parental MCF-7 cell line, while wild type procaspase-3 transfection of MCF-7 cells had increased apoptotic susceptibility to 1541.
To further probe if 1541 targets are downstream of the mitochondria within the intrinsic pathway, we compared induction of apoptosis in wild type murine embryonic fibroblasts (MEF), and in MEFs that lack the pro-apoptotic proteins Bak and Bax (33–35). Deletion of Bak and Bax prevents release of mitochondrial cytochrome C and blocks the apoptotic response of intrinsic apoptotic inducers STS and etoposide. Wild type (MEFs) and Bak−/−/Bax−/− double knockout (DKO MEFs) were incubated with STS or 1541, and assayed for apoptosis using a histone ELISA that measures solubilization of histones caused by DNA fragmentation, a hallmark of apoptosis (36). As expected, wild type MEFs were susceptible to apoptosis by either 1541 or STS (Fig. 4B). However, DKO MEFs retained sensitivity to 1541, but were completely resistant to STS. Western blots probed for active caspase-3 and PARP confirmed cleavage of these markers in 1541-induced DKO MEF lysates, but not in STS-treated cells (Fig. 4C). Thus, 1541 can bypass the lesion of Bak and Bax consistent with direct activation of procaspase-3.
To further test if the compounds are acting specifically through procaspase-3 in cells, we tested the ability of 1541 to induce apoptosis in human MCF-7 cells that lack a functional caspase-3 gene (37, 38). Indeed, MCF-7 cells were far less susceptible to 1541-induced apoptosis, which could be restored upon stable transfection with wild type procaspase-3 (Fig. 4D). Cells transfected with the S198A procaspase-3 mutant, previously determined in vitro to resist 1541-induced activation, also largely resisted apoptosis in cells. This mutant did exhibit some apoptosis after a 24-hour incubation that may arise from leaky activation of S198A procaspase-3. Overall, these data provide strong support that procaspase-3 is the primary intracellular target of 1541 and a major mediator of 1541-induced apoptosis in vivo. To begin to explore a therapeutic window for procaspase activators, we tested the effects of 1541B on a cancerous B cell line (DB) in comparison to immortalized normal B cells. We found the cancerous cell line to be more sensitive to apoptosis induced by 1541B (Fig. S11). Further experiments will be needed to explore these compounds as novel and selective chemotherapeutic agents that bypass typical proapoptotic lesions.
In conclusion, our studies reveal a unique mechanism for activation of homodimeric executioner procaspases by synthetic small molecules. Remarkably, the compounds both reorganize the active site for catalysis and render their internal cleavage sites more accessible to proteolysis, which may reflect aspects of the natural zymogen activation process. These compounds could either stabilize the on-state of procaspase-3, or destabilize the off-state and enhance susceptibility to proteolysis through a more flexible on-state conformation. The small molecule analogs exhibit selectivity towards procaspase-3 in vitro and in cells. Selective small molecule activators of procaspases will have great utility for in-depth studies of caspase-dependent apoptotic and non-apoptotic processes, such as stem cell differentiation (6) and cellular remodeling (5). Finally, our results, together with the recently characterized allosteric activation of the monomeric Vibrio cholerae RTX cysteine protease by a natural small molecule metabolite (2), suggest it may be possible to discover synthetic small molecule activators for other proenzymes to facilitate functional and mechanistic studies and further enhance their utility in biology and disease.
Supplementary Material
Acknowledgments
We thank J. Williams, B. Wolff and the UCSF Small Molecule Discovery Center for assistance in the HTS of procaspase-3 activators, Drs. S. Mahrus, H. Yoshihara, A. Renslo, M. Burlingame and J. Sadowsky for critical suggestions, Drs. Jack Taunton and Zev Gartner for critical reading of the manuscript, J. Gao for procaspase-1, J. Gray for cell lines, and C. Craik and D. Hostetter for granzyme B. This research was supported by grants from the National Institutes of Health (R21 N5057022, RO1 CA136779), National Cancer Institute Postdoctoral Fellowship (F32 CA119641-03) (to D.W.W.), UC Cancer Research Committee Fellowship (to D.C.G.) and the Hartwell Foundation.
References and Notes
- 1.Neurath H, Walsh KA. Proc Natl Acad Sci USA. 1976;73:3825. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Science. 2008;322:265. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stack MS, Pizzo SV. J Biol Chem. 1993;268:18924. [PubMed] [Google Scholar]
- 4.Earnshaw WC, Martins LM, Kaufmann SH. Annu Rev Biochem. 1999;68:383. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 5.Yi CH, Yuan J. Dev Cell. 2009;16:21. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdul-Ghani M, Megeney LA. Cell Stem Cell. 2008;2:515. doi: 10.1016/j.stem.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Boatright KM, Salvesen GS. Curr Opin Cell Biol. 2003;15:725. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Shi YG. Mol Cell. 2002;9:459. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 9.Wei Y, et al. Chem Biol. 2000;7:423. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 10.Gao J, Sidhu SS, Wells JA. Proc Natl Acad Sci USA. 2009;106:3071. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Proc Natl Acad Sci USA. 2004;101:12461. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheer JM, Romanowski MJ, Wells JA. Proc Natl Acad Sci USA. 2006;103:7595. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng BY, et al. J Med Chem. 2007;50:2385. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 14.Van de Craen M, Declercq W, Van den brande I, Fiers W, Vandenabeele P. Cell Death Differ. 1999;6:1117. doi: 10.1038/sj.cdd.4400589. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri T, et al. Proc Natl Acad Sci USA. 1996;93:7464. [Google Scholar]
- 16.Srinivasula SM, et al. J Biol Chem. 1996;271:27099. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- 17.Activation of procaspase-6 with 1541 achieves a specific activity that is >90% that of purified fully active caspase-6.
- 18.The DMSO negative controls show no activation or self-processing over the time course for any of the procaspases.
- 19.Putt KS, et al. Nat Chem Biol. 2006;2:543. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 20.Recent evidence suggests that the ~2-fold activation seen for PAC-1 is caused through chelation of an inhibitory zinc ion present in some assay buffers. Please see reference 21.
- 21.Peterson QP, et al. J Mol Biol. 2009;388:144. doi: 10.1016/j.jmb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denault JB, et al. Nat Chem Biol. 2007;3:519. doi: 10.1038/nchembio0907-519. author reply 520. [DOI] [PubMed] [Google Scholar]
- 23.Asboth B, Stokum E, Khan IU, Polgar L. Biochemistry. 1985;24:606. doi: 10.1021/bi00324a010. [DOI] [PubMed] [Google Scholar]
- 24.Hedstrom L. Chem Rev. 2002;102:4501. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 25.The kinetic parameters of procaspase-3 with 1541 after 4 hours are virtually identical to mature caspase-3 indicating both forms are kinetically similar. The EC50 for this transition from procaspase-3 to active capase-3 based on Michaelis-Menton analysis is 1.5 μM (Fig. S5) and closely matches the EC50 of 1.3 μM based on activity measurements with the preferred substrate Ac-DEVD-AFC (Fig. 1A).
- 26.Rominger CM, et al. Arch Biochem Biophys. 2007;464:130. doi: 10.1016/j.abb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Bertrand R, Solary E, O’Connor P, Kohn KW, Pommier Y. Exp Cell Res. 1994;211:314. doi: 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 28.Kaufmann SH. Cancer Res. 1989;49:5870. [PubMed] [Google Scholar]
- 29.Walker PR, et al. Cancer Res. 1991;51:1078. [PubMed] [Google Scholar]
- 30.Kaufmann SH, Earnshaw WC. Exp Cell Res. 2000;256:42. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 31.Lakhani SA, et al. Science. 2006;311:847. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juo P, Kuo CJ, Yuan J, Blenis J. Curr Biol. 1998;8:1001. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- 33.Wei MC, et al. Science. 2001;292:727. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei MC, et al. Genes Dev. 2000;14:2060. [PMC free article] [PubMed] [Google Scholar]
- 35.Executioner caspase-3 is 86% identical between human and mouse.
- 36.The histone ELISA was used instead of the simpler viability assay measuring ATP levels because the mitochondria, and hence ATP levels, are preserved in the DKO MEFs.
- 37.Jänicke RU, Sprengart ML, Wati MR, Porter AG. J Biol Chem. 1998;273:9357. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 38.Blanc C, et al. Cancer Res. 2000;60:4386. [PubMed] [Google Scholar]
- 39.Compound 1541 contains no free amines or hydroxyls to perturb buffer pH or ionic strength. Therefore, 1541 is unlikely to involve the “safety catch” region (residues 179-181) of procaspase-3 previously proposed to promote susceptibility towards autoactivation via environmental pH reduction.
- 40.Roy S, et al. Proc Natl Acad Sci USA. 2001;98:6132. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Error bars on all figures represent the standard deviation of n=3 to 5 replicates.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.