Abstract
The kidney is one of the main targets of drug toxicity, but early detection of renal damage is often difficult. As part of the InnoMed PredTox project, a collaborative effort aimed at assessing the value of combining omics technologies with conventional toxicology methods for improved preclinical safety assessment, we evaluated the performance of a panel of novel kidney biomarkers in preclinical toxicity studies. Rats were treated with a reference nephrotoxin or one of several proprietary compounds that were dropped from drug development in part due to renal toxicity. Animals were dosed at two dose levels for 1, 3, and 14 days. Putative kidney markers, including kidney injury molecule-1 (Kim-1), lipocalin-2 (Lcn2), clusterin, and tissue inhibitor of metalloproteinases-1, were analyzed in kidney and urine using quantitative real-time PCR, ELISA, and immunohistochemistry. Changes in gene/protein expression generally correlated well with renal histopathological alterations and were frequently detected at earlier time points or at lower doses than the traditional clinical parameters blood urea nitrogen and serum creatinine. Urinary Kim-1 and clusterin reflected changes in gene/protein expression and histopathological alterations in the target organ in the absence of functional changes. This confirms clusterin and Kim-1 as early and sensitive, noninvasive markers of renal injury. Although Lcn2 did not appear to be specific for kidney toxicity, its rapid response to inflammation and tissue damage in general may suggest its utility in routine toxicity testing.
Keywords: kidney, drug-induced nephrotoxicity, biomarker, Kim-1, clusterin, lipocalin-2
The discovery and development of new drugs is very costly, and the attrition rate of drug candidates due to late-breaking findings in preclinical toxicity studies is high. To reduce the cost of drug development and speedup the delivery of safe medicines to patients, it is therefore critical to identify and eliminate at the earliest possible stage the compounds with potential safety issues that are unlikely to become successfully marketed pharmaceuticals. Thus, pharmaceutical companies have increasingly incorporated in vivo signal generation studies into early phases of the drug discovery process to establish dose-limiting toxicities, understand the mechanism of toxicity, and to better manage and/or screen out safety liabilities before advancing drug candidates into formal development (Kramer et al., 2007).
Molecular profiling techniques, particularly toxicogenomics, combined with statistical and molecular pathway analysis tools are now widely used in discovery toxicology to predict or monitor adverse effects based on gene signatures or metabolic fingerprints thought to be characteristic for a particular type of toxicity. Additionally, it has been recognized that omics technologies offer great potential for the discovery of novel mechanism-based biomarkers for target organ toxicities. This is achieved by both identification of biological signals related to toxicity and interpretation with respect to regulatory networks and mechanisms. However, the process of identifying and qualifying potential safety markers as valid biomarkers for internal and regulatory decision making is complex and by far exceeds the resources of individual companies. Thus, in recent years, several large-scale collaborative projects involving academic and industrial institutions, as well as regulatory agencies, have emerged with the aim of delivering new tools for improved assessment of the most common problem toxicities. These include hepato- and nephrotoxicity, cardiovascular toxicity, and carcinogenicity (Gallagher et al., 2009).
Through these joint efforts, significant progress has been made, particularly in the field of biomarkers of drug-induced nephrotoxicity. Work by the ILSI Health and Environmental Sciences Institute initiative (http://www.hesiglobal.org), for instance, which set out to identify novel biomarkers for kidney injury based on microarray data, resulted in the discovery of a range of potential preclinical animal model biomarkers (Amin et al., 2004). Several of these markers, among others, were subsequently selected by the Predictive Safety Testing Consortium (http://www.c-path.org/pstc.cfm) as part of a kidney biomarker panel for qualification to determine their effectiveness to better predict drug-induced kidney injury in rats as compared to markers of renal function such as blood urea nitrogen (BUN) and serum creatinine, which are routinely used to measure kidney damage but suffer from lack of sensitivity. While the Food and Drug Administration and the European Medicines Agency recently endorsed the use of a number of these urinary biomarkers for the detection of acute kidney toxicity in the context of nonclinical drug development (http://www.emea.europa.eu/pdfs/human/sciadvice/67971908en.pdf), there are as yet little published data available to the scientific community to demonstrate the effectiveness of novel renal biomarkers for improved detection of drug-induced renal injury.
The Innovative Medicines for Europe (InnoMed) PredTox project (http://www.innomed-predtox.com), a collaborative effort by 12 pharmaceutical companies, two small- and mid-sized enterprises, and three universities, was initiated to assess the value of combining omics technologies with conventional toxicology methods for improved safety assessment of liver and kidney toxicity. Comprehensive data sets, including toxicogenomics, proteomics, and metabonomics, were collected from short-term in vivo experiments in which male Wistar rats were treated with either a model compound or one of the 14 proprietary compounds that previously failed during drug development in part due to toxic effects in the liver or kidney. As an ancillary aim of this project, we were also interested to evaluate the performance of new kidney biomarkers relative to histopathology and routine clinical chemistry. The present report, therefore, focuses on those InnoMed PredTox studies in which nephrotoxic effects occurred and constitutes a thorough assessment of a set of gene-based and urinary kidney biomarkers compared to traditional end points. Putative biomarkers identified from literature and related projects, including kidney injury molecule-1 (Kim-1), clusterin, neutrophil gelatinase-associated lipocalin (NGAL) (lipocalin-2 [Lcn2]/NGAL), osteopontin (OPN), tissue inhibitor of metalloproteinases-1 (Timp-1), vimentin, and heme oxygenase-1 (HO-1), were analyzed in kidney and/or urine samples collected from PredTox studies using a combination of quantitative real-time (qRT) PCR, immunohistochemistry, and ELISA techniques.
MATERIALS AND METHODS
Reagents and antibodies.
All chemicals and reagents were purchased from commercial suppliers. Gentamicin sulfate (test article lot 051K17476) was obtained from Sigma-Aldrich (Taufkirchen, Germany). Primary antibodies used for immunohistochemical analyses were goat anti-mouse clusterin (Santa Cruz, Heidelberg, Germany), goat anti-rat Lcn2 immunoglobulin G (IgG) (R&D Systems, Wiesbaden, Germany), goat anti-rat Kim-1 (R&D Systems), rabbit anti-human Timp-1 (Millipore, Schwalbach, Germany), and mouse anti-human vimentin (Santa Cruz). All secondary antibodies (donkey anti-goat IgG, goat anti-rabbit IgG, and goat anti-mouse IgG) were obtained from Santa Cruz.
Animal studies.
All animal experiments were performed according to the national guidelines for the care and use of laboratory animals. Animal studies were conducted using a harmonized PredTox study protocol designed to mimic in vivo signal generation studies typically conducted within the participating pharmaceutical companies during early phases of the drug discovery process. This comprised the daily administration of male Wistar rats (8–10 weeks of age and 170–200 g at commencement of treatment) with a low and a high dose (five animals per group) of each nephrotoxin (Table 1). Pyrimido[5,4-d]pyrimidin-4-amine, 6-(4-diethylaminomethyl-1-piperidinyl)-N-(3-chloro-4-fluorophenyl)-, hydrochloride (BI-2) (0, 2.5, and 15 mg/kg body weight [bw]), 3-pyrrolidineacetic acid, 5-[[[4′-[imino[(methoxycarbonyl) amino]methyl] [1,1′-biphenyl]-4-yl]oxy]methyl]-2-oxo-, methyl ester,(3S-trans) (BI-3) (0, 100, and 1000 mg/kg bw), gentamicin (0, 25, and 75 mg/kg bw), and O-hydroxyethyl-D-(Ser)8-cyclosporine A (IMM125) (0, 30, and 100 mg/kg bw) were administered for 1, 3, and 14 days by oral gavage (BI-2, BI-3, and IMM125) or sc injection (gentamicin). Urine samples were collected during a period of 16 h starting 8 h after substance administration on days 1, 3, and 12 only from groups of animals treated for a total of 14 days. Blood samples were collected from each animal at the time of necropsy. Rats were sacrificed by humane methods after an overnight fasting period (anesthesia/exsanguination according to standard operating procedures in place in each participating company conducting the animal experiments), followed by necropsy as previously described in detail (Mulrane et al., 2008).
TABLE 1.
Treatment Schedule of Animals
| Animal identification | Treatment | Total no. of doses |
| 01–05 | Vehicle | 1 |
| 06–10 | Low dose | 1 |
| 11–15 | High dose | 1 |
| 16–20 | Vehicle | 3 |
| 21–25 | Low dose | 3 |
| 26–30 | High dose | 3 |
| 26–30 | Vehicle | 14 |
| 31–35 | Low dose | 14 |
| 36–40 | High dose | 14 |
Note. Urine was collected from animals no. 31–45 after 1, 3, and 12 days of dosing.
Clinical chemistry and hematology.
The following hematology and clinical chemistry parameters were measured in urine, blood, and/or serum according to standard operating procedures by the participating companies conducting the animal experiments—Serum: glucose, total cholesterol, triglycerides, BUN, creatinine, aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), γ-glutamyltransferase (GGT), lactate dehydrogenase (LDH), creatinine kinase, total bilirubin, total protein, albumin, globulin, sodium, potassium, chloride, calcium, and inorganic phosphorus.
Urine: color, turbidity, bilirubin, urobilinogen, glucose, protein, urine volume, specific gravity, pH, creatinine, potassium, chloride, calcium, and inorganic phosphorus.
Blood: red and white blood cells, hemoglobin, hematocrit, mean cell hemoglobin (MCH), MCH concentration, mean cell volume, platelet count, neutrophils, lymphocytes, eosinophils, basophils, and monocytes.
Histopathology.
At necropsy, kidneys were removed and cut sagitally. Sections (3–4 mm) of both kidneys were fixed in 10% phosphate-buffered formalin and subsequently embedded in paraffin blocks, sectioned, stained with hematoxylin and eosin, and examined by light microscopy by the respective study pathologist. Histopathological peer review was performed by an independent veterinary pathologist. Based on the peer review, individual terms describing renal alterations involving the proximal tubule, that is, basophilia, degeneration/regeneration, and necrosis, were summarized into a single term “proximal tubule damage” (PTD). Lesions were scored as no lesion (0), minimal (1), mild (2), moderate (3), and severe (4). Animals with a histopathology score of > 1 in both kidneys were defined as showing clear evidence of PTD. A middle section (∼7 mm) of the left lateral lobe of the liver was also fixed in formalin for histopathological examination.
Quantitative real-time PCR.
Total RNA was isolated from frozen kidney tissue using the RNeasy Mini Kit (Quiagen) including DNase digestion according to the manufacturer’s instructions. RNA concentrations were determined by ultraviolet light absorbance at 260 nm. Complementary DNA (cDNA) was generated from 1 μg total RNA using the First Strand Synthesis Kit (Thermo Fisher Scientific, Hamburg, Germany), diluted 1:5 with H2O. qRT PCR was performed using the Roche LightCycler 480 (Roche, Mannheim, Germany) in 20-μl reactions containing 2× mastermix with SYBR Green I (Thermo Fisher Scientific), 2 μl cDNA, and 105nM of each primer. Amplification was carried out using the following temperature profile: 15 min enzyme activation at 95°C followed by 45 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Primer sequences for 18s ribosomal RNA (rRNA) listed in 5′ to 3′ direction were GTAACCCGTTGAACCCCATT (forward) and CCATCCAATCGGTTAGTAGCG (reverse) and for β-actin, clusterin, Lcn2 (Lcn2/NGAL), vimentin, OPN (secreted phosphoproteins 1), heme oxygenase-1 (HMOX1), Timp-1, and Kim-1 (hepatitis A virus cellular receptor 1) were as previously described (Rached et al., 2008; Sieber et al., 2009; Zhou et al., 2008). Gene expression changes relative to controls were determined using the 2−ΔΔ method. Samples were normalized against β-actin (BI-2 and gentamicin) or 18s rRNA where treatment was found to affect β-actin levels (BI-3 and IMM125). Results are presented as mean fold change in messenger RNA (mRNA) expression of five animals per dose group compared to control animals.
Immunohistochemistry.
Immunohistochemical analyses were performed on tissue microarrays (TMAs) containing tissue cores from each of the 45 individual animals used in each study. TMAs of rat kidney specimens were constructed using an TMA-I arrayer (Beecher Instruments), and a similar array format was used for all preclinical studies. Specifically, cores (1 mm diameter) were punched from donor blocks of rat kidney tissue and placed into an empty paraffin block, leaving a space of 2 mm between horizontal cores and 2.5 mm between vertical cores. A reference core was placed at position (1, 1) to provide orientation when scanning. Kidney TMAs were constructed with duplicate cores taken from outer cortex, inner cortex, corticomedullary junction, medullary, and papillary regions, set across 10 separate TMAs. Kidney sections (5 μm) were prepared from TMA blocks and mounted onto glass slides. Tissues were deparaffinized, rehydrated, and washed in PBS. Heat-induced antigen retrieval was achieved by 4 min autoclaving in 10mM citrate buffer, pH 6.0. For detection of Timp-1 and Lcn2/NGAL, tissues were then treated with 0.1% trypsin for 2 min at 37°C. After washing the tissue pieces in PBS, they were blocked with 5% donkey serum for 1 h. Endogenous peroxidase was subsequently blocked by incubation with 3% H2O2 in PBS for 10–15 min, followed by two additional blocking steps with 0.001% avidin in PBS for 15 min and 0.001% biotin for 15 min. Between all steps, tissues were washed several times with PBS. They were then incubated with the primary antibody diluted in 5% serum (species of secondary antibody) in PBS overnight at 4°C. Antibodies were applied in the following concentrations—anti-clusterin: 1 μg/ml, anti-NGAL: 1.3 μg/ml, anti-OPN: 0.25–5 μg/ml, anti-Kim-1: 0.5–1 μg/ml, anti-Timp-1: 2.5 μg/ml, and anti-vimentin: 0.25 μg/ml. After three wash steps, tissues were incubated with the biotinylated secondary antibody (goat anti-mouse IgG, goat anti-rabbit IgG, or donkey anti-goat IgG) diluted 1:200 in PBS for 45 min at room temperature and subsequently washed with PBS. Following 30 min incubation with the complex of avidin and biotinylated horseradish peroxidase (Vectastain Elite ABC Reagent, Vector Laboratories, Peterborough, United Kingdom), enzyme activity was visualized using 3,3′-diaminobenzidine (DAB) (DAB Substrate Kit, Vector Laboratories). Depending on the antigen, staining was performed for 1–2 min. Tissues were counterstained with hematoxylin and eosin, dehydrated, and mounted in Eukitt mounting medium (Sigma-Aldrich).
Enzyme-linked immunosorbent assay.
A commercially available Rat ELISA Kit for detection of clusterin in serum and urine was used (Biovendor Laboratory Medicine Inc.). Kim-1 was measured as previously described (Vaidya et al., 2006). Timp-1 was determined using the rat Timp-1 DouSet ELISA Development Systems (R&D Systems) according to the manufacturer’s instructions. For quantification of NGAL in urine and serum, a sandwich ELISA was developed using two monoclonal antibodies (BioPorto Diagnostics). Microtitre plates (Nunc-Immuno Maxisorp, Denmark) were coated overnight at 4°C with purified mouse anti-rNGAL (125 nanograms per well) in carbonate buffer (0.1M Na2CO3/NaHCO3, pH 9.6). After repeated washing with phosphate buffered saline-Tween 20 (0.05% Tween-20, pH 7.4), nonspecific protein-binding sites were blocked by incubating with 1% bovine serum albumin/PBS (200 microliters per well) for 2 h at room temperature. After washing, 100 μl of urine (diluted 1:1600 or 1:6400), serum (diluted 1:400 or 1:800), or standards (recombinant rNGAL in a concentration range 156.3–5000 pg/ml) were added to the wells and incubated for 2 h at room temperature. Following several washing steps, bound antigens were detected by addition of biotinylated anti-rNGAL antibody diluted 1:4000 for 90 min and streptavidin-horseradish peroxidase (Vector Laboratories) for 30 min at room temperature. Finally, peroxidase activity was determined using the chromogenic substrate (3,3′,5,5′-tetramethylbenzidine, 30 min). The reaction was stopped by adding 100 μl 1M sulfuric acid (Sigma-Aldrich), and absorbance was measured by a microplate reader (SpectraMax 190, Molecular Devices, Sunnyvale, CA) at 450 nm. In-house validation of this assay was evaluated by measuring the sensitivity, specificity, reference range, reproducibility, recovery, and dilutional linearity. The lower limit of detection (LLD) is the lowest NGAL concentration that can be reliably detected from the assay blank. The LLD was 78 pg/ml, and the coefficient of variation (CV) was < 15% (n = 6; p < 0.001 by Student’s t-test). This assay is specific for rat NGAL protein and does not detect mouse or human NGAL. The range of the assay was determined by establishing a standard curve in the range from 5000, 2500, 1250, 625, 312.5, to 156.3 pg/ml. Intra- and interassay CV was < 6.19% (n = 8) and < 9.38% (n = 4). The recovery rate of NGAL ranged from 101 to 132% in urine of rats spiked with 0, 200, or 400 ng/ml recombinant NGAL. Dilution linearity: NGAL concentration was measured in two rat urine and plasma samples by diluting the samples 1:200, 1:400, 1:800, and 1:1600 (n = 2). Variation of the results was estimated by multiplying the reading with the dilution factor and ranged from 94 to 110%.
Concentrations of urinary biomarkers were normalized to urinary creatinine, which was not significantly changed in response to drugs associated with PTD (Table 3).
TABLE 3.
Clinical Chemistry Parameters of Male Wistar Rats after Repeated Administration of BI-2, BI-3, Gentamicin, or IMM125 for 1, 3, and 12 (urine) or 14 (serum) Days
| Days of treatment | BI-2 |
BI-3 |
Gentamicin |
IMM125 |
|||||||||
| 0 mg/kg | 2.5 mg/kg | 15 mg/kg | 0 mg/kg | 100 mg/kg | 1000 mg/kg | 0 mg/kg | 25 mg/kg | 75 mg/kg | 0 mg/kg | 30 mg/kg | 100 mg/kg | ||
| Urine | |||||||||||||
| Creatinine (mg/16 h) | 1 | 4.3 | 3.9 | 4.4 ± 0.8 | 7.9 ± 1.3 | 8.5 ± 0.8 | 6.8 ± 1.6 | 2.4 ± 0.2 | 2.6 ± 0.3 | 2.3 ± 0.1 | 4.3 ± 1.1 | 4.7 ± 0.7 | 5.3 ± 0.7 |
| 3 | 5.1 ± 0.5 | 4.0 ± 0.0* | 3.7 ± 0.1* | 8.1 ± 1.5 | 8.9 ± 1.3 | 6.3 ± 1.1 | 2.7 ± 0.3 | 2.7 ± 0.3 | 2.6 ± 0.3 | 4.8 ± 1.4 | 5.5 ± 0.8 | 6.0 ± 1.1 | |
| 12 | 5.6 ± 0.6 | 4.8 ± 0.7 | 4.5 ± 0.3 | 7.7 ± 1.2 | 7.7 ± 2.0 | 5.5 ± 1.8 | 3.9 ± 0.7 | 3.8 ± 0.6 | 3.8 ± 0.3 | 6.1 ± 1.0 | 6.9 ± 1.2 | 7.0 ± 0.6 | |
| Protein (mg/mg creatinine) | 1 | Not determined | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.7 ± 0.6 | 1.0 ± 0.1 | 0.9 ± 0.2 | 1.1 ± 0.3 | 0.1 ± 0.1 | 0.5 ± 0.4 | 0.6 ± 0.5 | ||
| 3 | Not determined | 0.5 ± 0.1 | 0.7 ± 0.2 | 1.1 ± 0.9 | 1.0 ± 0.2 | 1.0 ± 0.3 | 1.3 ± 0.3 | 0.3 ± 0.2 | 0.3 ± 0.3 | 0.4 ± 0.7 | |||
| 12 | Not determined | 0.7 ± 0.6 | 1.1 ± 0.9 | 0.6 ± 0.4 | 1.4 ± 0.3 | 2.3 ± 0.7 | 2.7 ± 0.6* | 0.4 ± 0.2 | 0.3 ± 0.1 | 0.5 ± 0.2 | |||
| Serum | |||||||||||||
| Creatinine (mg/dl) | 1 | 0.43 ± 0.13 | 0.42 ± 0.05 | 0.37 ± 0.06 | 0.51 ± 0.03 | 0.46 ± 0.03 | 0.47 ± 0.03 | 0.17 ± 0.02 | 0.16 ± 0.02 | 0.16 ± 0.02 | 0.28 ± 0.05 | 0.29 ± 0.07 | 0.22 ± 0.02 |
| 3 | 0.41 ± 0.04 | 0.44 ± 0.05 | 0.43 ± 0.07 | 0.52 ± 0.04 | 0.45 ± 0.04* | 0.48 ± 0.04 | 0.19 ± 0.01 | 0.19 ± 0.04 | 0.21 ± 0.02 | 0.25 ± 0.04 | 0.27 ± 0.05 | 0.30 ± 0.08 | |
| 14 | 0.46 ± 0.07 | 0.41 ± 0.05 | 0.45 ± 0.04 | 0.52 ± 0.07 | 0.53 ± 0.05 | 0.65 ± 0.32 | 0.21 ± 0.03 | 0.22 ± 0.03 | 0.26 ± 0.02* | 0.32 ± 0.02 | 0.32 ± 0.04 | 0.35 ± 0.04 | |
| BUN (mg/dl) | 1 | 25 ± 3 | 27 ± 2 | 25 ± 2 | 49 ± 6 | 53 ± 17 | 46 ± 9 | 41 ± 4 | 39 ± 5 | 43 ± 6 | 13 ± 3 | 13 ± 4 | 13 ± 3 |
| 3 | 25 ± 4 | 28 ± 3 | 31 ± 7 | 50 ± 7 | 48 ± 7 | 67 ± 19 | 47 ± 7 | 46 ± 7 | 48 ± 7 | 14 ± 2 | 14 ± 3 | 15 ± 4 | |
| 14 | 26 ± 2 | 26 ± 1 | 29 ± 4 | 45 ± 6 | 60 ± 6 | 110 ± 72 | 38 ± 6 | 41 ± 5 | 48 ± 4* | 15 ± 1 | 17 ± 3 | 17 ± 3 | |
| GGT (U/l) | 1 | 0.40 ± 0.14 | 1.95 ± 2.19 | 0.18 ± 0.40 | 0.00 ± 0.00 | 0.01 ± 0.03 | 1.75 ± 1.38** | < LOD | < LOD | < LOD | 1.86 ± 1.90 | 1.02 ± 1.42 | 1.54 ± 2.21 |
| 3 | 1.00 ± 0.85 | 0.88 ± 0.29 | 0.35 ± 0.21 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.60 ± 2.44 | < LOD | < LOD | < LOD | 1.60 ± 3.58 | 2.54 ± 3.69 | 2.20 ± 3.73 | |
| 14 | 1.15 ± 0.21 | 0.04 ± 0.09 | 0.62 ± 0.30 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.65 ± 1.31** | < LOD | < LOD | < LOD | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | |
| Total protein (g/l) | 1 | 55 ± 2 | 54 ± 4 | 54 ± 2 | 59 ± 1 | 58 ± 2 | 61 ± 2 | 54 ± 2 | 54 ± 2 | 54 ± 1 | 61 ± 1 | 58 ± 2* | 56 ± 2** |
| 3 | 55 ± 1 | 55 ± 1 | 53 ± 2 | 59 ± 1 | 58 ± 3 | 56 ± 3 | 56 ± 2 | 56 ± 1 | 57 ± 0 | 64 ± 2 | 59 ± 3* | 52 ± 3*** | |
| 14 | 56 ± 3 | 55 ± 2 | 52 ± 2* | 58 ± 2 | 55 ± 2 | 45 ± 6*** | 54 ± 1 | 55 ± 2 | 55 ± 3 | 64 ± 2 | 58 ± 2*** | 53 ± 1*** | |
| AST (U/l) | 1 | 53 ± 12 | 61 ± 32 | 60 ± 12 | 193 ± 37 | 167 ± 38 | 298 ± 195 | 94 ± 18 | 93 ± 7 | 102 ± 12 | 132 ± 10 | 107 ± 13* | 80 ± 17*** |
| 3 | 66 ± 14 | 75 ± 40 | 62 ± 8 | 190 ± 25 | 172 ± 22 | 215 ± 43 | 94 ± 10 | 91 ± 11 | 87 ± 15 | 108 ± 15 | 90 ± 10 | 69 ± 26* | |
| 14 | 62 ± 20 | 65 ± 9 | 62 ± 6 | 157 ± 19 | 151 ± 27 | 144 ± 40 | 89 ± 16 | 88 ± 19 | 97 ± 17 | 138 ± 22 | 95 ± 7** | 72 ± 9*** | |
| ALP (U/l) | 1 | 99 ± 21 | 98 ± 10 | 104 ± 25 | 182 ± 25 | 149 ± 32 | 513 ± 289* | 577 ± 129 | 536 ± 105 | 592 ± 156 | 213 ± 49.3 | 228 ± 61.8 | 191 ± 26 |
| 3 | 120 ± 30 | 87 ± 8* | 89 ± 10* | 197 ± 47 | 177 ± 36 | 425 ± 77*** | 712 ± 179 | 666 ± 241 | 561 ± 169 | 245 ± 69.1 | 155 ± 27.1* | 123 ± 12** | |
| 14 | 86 ± 17 | 87 ± 25 | 57 ± 5 | 114 ± 11 | 116 ± 13 | 262 ± 90** | 510 ± 120 | 455 ± 94 | 398 ± 131 | 176 ± 20.5 | 116 ± 24.9** | 95 ± 24*** | |
| ALT (U/l) | 1 | 26 ± 4 | 28 ± 8 | 27 ± 6 | 31 ± 5 | 28 ± 14 | 169 ± 185 | 25 ± 4 | 24 ± 5 | 23 ± 1 | 57 ± 9 | 49 ± 6 | 33 ± 12** |
| 3 | 29 ± 6 | 45 ± 41 | 26 ± 7 | 24 ± 4 | 22 ± 2 | 43 ± 14** | 24 ± 4 | 21 ± 2 | 22 ± 4 | 54 ± 11 | 22 ± 9*** | 22 ± 6*** | |
| 14 | 34 ± 15 | 30 ± 6 | 27 ± 2 | 24 ± 4 | 28 ± 10 | 24 ± 4 | 26 ± 5 | 24 ± 2 | 31 ± 10 | 52 ± 7 | 46 ± 8 | 34 ± 8** | |
| Bilirubin (mg/dl) | 1 | 0.10 ± 0.02 | 0.09 ± 0.05 | 0.09 ± 0.02 | Not determined | 0.08 ± 0.02 | 0.08 ± 0.02 | 0.08 ± 0.01 | 0.07 ± 0.02 | 0.09 ± 0.02 | 0.17 ± 0.05** | ||
| 3 | 0.11 ± 0.04 | 0.11 ± 0.03 | 0.11 ± 0.01 | Not determined | 0.08 ± 0.01 | 0.07 ± 0.01 | 0.09 ± 0.01 | 0.09 ± 0.03 | 0.14 ± 0.03 | 0.45 ± 0.10*** | |||
| 14 | 0.07 ± 0.03 | 0.04 ± 0.02 | 0.06 ± 0.02 | Not determined | 0.06 ± 0.02 | 0.08 ± 0.02 | 0.09 ± 0.01* | 0.10 ± 0.03 | 0.17 ± 0.02* | 0.28 ± 0.07*** | |||
| Glucose (mg/dl) | 1 | 175 ± 14 | 188 ± 9 | 180 ± 29 | 147 ± 11 | 132 ± 23 | 152 ± 20 | 123 ± 9 | 119 ± 14 | 122 ± 13 | 95 ± 16 | 114 ± 12 | 120 ± 23 |
| 3 | 154 ± 9 | 176 ± 17* | 169 ± 13 | 144 ± 20 | 143 ± 8 | 122 ± 6* | 138 ± 22 | 126 ± 16 | 132 ± 12 | 74 ± 4 | 84 ± 9 | 101 ± 14** | |
| 14 | 171 ± 35 | 191 ± 21 | 192 ± 14 | 148 ± 14 | 163 ± 4 | 128 ± 47 | 123 ± 25 | 117 ± 12 | 130 ± 11 | 80 ± 14 | 92 ± 15 | 111 ± 8** | |
Note. Data are presented as mean ± SD (n = 5). Statistical analysis was performed by ANOVA and Dunnett’s post hoc test; LOD, limit of detection.
*p < 0.05, **p < 0.01, and ***p < 0.001.
Statistical analyses.
Data are presented as individual animals or mean ± SD (n = 5 animals per group). Statistical analysis was performed by ANOVA and Dunnett’s test. Values significantly different from control are indicated as *p < 0.05, **p < 0.01, and ***p < 0.001. Inclusion receiver operator characteristics (ROC) analysis was performed using biomarker data of all animals with a corresponding histopathology readout (kidney marker genes: days 1, 3, and 14; urinary markers: day 12/14 only). ROC curves were plotted by entering data obtained from animals with a PTD histopathology score > 1 versus vehicle controls and treated animals without clear evidence of renal injury (histopathology score ≤ 1) using the Graph Pad Prism 5 software package (GraphPad Software Inc., La Jolla, CA).
RESULTS
Histopathology and Clinical Chemistry
Following administration of BI-2, which was previously found to produce papillary necrosis within 4 weeks of treatment, very minor histopathological effects in the form of hyaline droplets in renal tubule epithelium were evident in kidneys of high-dose animals after 14 days (Table 2), while no histopathological changes were seen in kidneys of rats treated with the low dose of BI-2. Clinical chemistry did not reveal clear signs of nephrotoxicity, although a slight decrease in the excretion of urinary creatinine was evident in both dose groups on day 3 (Table 3). However, a significant increase in the number of neutrophils (0.59 ± 0.17 × 109 in controls vs. 1.98 ± 0.29 × 109 in high-dose animals) was observed after 14 days of treatment, indicative of a systemic inflammatory response to BI-2 at 15 mg/kg bw. No changes were evident in livers.
TABLE 2.
Summary of Histopathological Observations in Kidneys of Male Wistar Rats Treated with BI-2, BI-3, Gentamicin, or IMM125 for 1, 3, and 14 Days
| Days of treatment | Histopathological change | BI-2 |
BI-3 |
Gentamicin |
IMM125 |
||||||||
| 0 mg/kg | 2.5 mg/kg | 15 mg/kg | 0 mg/kg | 100 mg/kg | 1000 mg/kg | 0 mg/kg | 25 mg/kg | 75 mg/kg | 0 mg/kg | 30 mg/kg | 100 mg/kg | ||
| 1 | PTD | − | − | +(1/5) | − | +(1/5) | − | − | − | − | − | +(1/5) | +(1/5) |
| Infiltrate: mononuclear cell | − | − | − | − | − | − | − | +(1/5) | +(2/5) | − | − | − | |
| Hyaline droplet: renal tubule | − | − | − | − | − | − | − | − | − | +(2/5) ++(1/5) | +(2/5) | +(1/5) ++(2/5) | |
| 3 | PTD | − | − | − | +(2/5) | +(2/5) +(1/5) | +(1/5) | − | − | − | − | +(1/5) | − |
| Infiltrate: mononuclear cell | − | − | − | − | +(1/5) | − | − | − | − | − | +(4/5) | +(1/5) | |
| Hyaline droplet: renal tubule | − | − | − | − | − | − | − | − | − | +(3/5) ++(/5) | +(2/5) | +(1/5) | |
| 14 | PTD | − | − | − | − | − | +++(2/5) ++++(1/5) | − | +(2/5) | ++(3/5) +++(2/5) | − | +(1/5) | +(1/5) ++(2/5) +++(2/5) |
| Infiltrate: mononuclear cell | − | − | − | − | − | − | − | +(3/5) | +(4/5) ++(1/5) | − | +(4/5) ++(1/5) | +(3/5) ++(1/5) +++(1/5) | |
| Hyaline droplet: renal tubule | − | − | +++(5/5) | − | − | − | − | − | − | +(4/5) | +(2/5) ++(1/5) +++(1/5) | +(5/5) | |
Note. −, lesion not observed; +, minimal; ++, mild; +++, moderate; ++++, high severity of lesion. The summary term PTD includes (a) basophilic tubule: proximal (BI-3 and IMM125), (b) degeneration/regeneration: renal tubule (gentamicin), and (c) necrosis: proximal tubule (BI-3 and gentamicin).
In contrast, treatment with BI-3 resulted in histopathological changes in both kidney and liver. Liver effects in the form of cholangitis and bile duct hyperplasia were observed in both dose groups with increasing severity related to time and dose. Cholestasis was evident after 14 days of treatment. In the kidney, marked basophilia of proximal tubules accompanied by proximal tubule cell necrosis and associated acute inflammation was observed in three high-dose animals (41, 43, and 45) after 14 days of treatment (Table 2). At the same time, a slight increase in serum creatinine and BUN indicative of impaired kidney function was observed in the high-dose group, although this effect was not statistically significant due to the large interanimal variability. Consistent with the hepatobiliary changes, clinical chemistry revealed a significant increase in GGT and ALP activities and a decrease in total protein in serum of high-dose animals treated for 14 days (Table 3). No significant changes in clinical chemistry parameters were seen at the low dose of BI-3. Similar to BI-2, a marked increase in the number of neutrophils was observed in high-dose animals after 14 days treatment (Supplementary table 1).
Treatment with the aminoglycoside antibiotic gentamicin for 14 days led to dose-dependent histopathological alterations in kidney cortex, consisting of minimal (low dose) to mild (high dose) tubular degeneration and regeneration and an interstitial mononuclear cell infiltration (Table 2). No evidence of PTD was observed at earlier time points. Traditional clinical pathology revealed increased serum creatinine, BUN, and proteinuria in rats treated with the high dose for 14 days (Table 3). There were no compound-related microscopic findings in the liver throughout the study.
Treatment with IMM125 resulted in histopathological alterations in both kidney and liver. Within the kidney proximal tubule, basophilia was noted in four of five high-dose animals treated for 14 days along with an increased incidence of interstitial mononuclear cell infiltrates (Table 2), but no clinical chemistry signs of nephrotoxicity were recorded. Histopathological changes in the liver included increased incidence and severity of hepatocellular vacuolation after 3 and 14 days high-dose treatment associated with a dose-dependent decrease in total protein and in the activity of liver enzymes AST, ALP, ALT (Table 3), and LDH (Supplementary table 1). In contrast, bilirubin (Table 3) and peripheral blood monoyctes (Supplementary table 1) were elevated in high-dose animals throughout the treatment period.
Altered Expression of Candidate Kidney Injury Marker Genes
Consistent with the absence of renal injury, qRT-PCR analyses provided no indication of gene expression changes associated with PTD following treatment with BI-2 (Table 4). No treatment-related effects on the expression of Kim-1, clusterin, Lcn2, Timp-1, OPN, HO-1, or vimentin were observed throughout the study.
TABLE 4.
Changes in mRNA Expression of a Panel of Novel Renal Biomarkers in Kidneys of Male Wistar Rats Repeatedly Dosed with BI-2, BI-3, Gentamicin, or IMM125 for 1, 3, or 14 Days
| Days of treatment | BI-2 fold change (relative to control) |
BI-3 fold change (relative to control) |
Gentamicin fold change (relative to control) |
IMM125 fold change (relative to control) |
|||||||||
| Gene | 0 mg/kg | 2.5 mg/kg | 15 mg/kg | 0 mg/kg | 100 mg/kg | 1000 mg/kg | 0 mg/kg | 25 mg/kg | 75 mg/kg | 0 mg/kg | 30 mg/kg | 100 mg/kg | |
| Kim-1 (hepatitis A virus cellular receptor 1) | 1 | 1.0 ± 0.2 | 0.8 ± 0.4 | 0.9 ± 0.4 | 1.0 ± 0.5 | 1.1 ± 0.8 | 0.9 ± 0.6 | 1.0 ± 0.5 | 0.9 ± 0.2 | 0.9 ± 0.3 | 1.0 ± 0.2 | 2.5 ± 1.3 | 3.4 ± 2.1* |
| 3 | 1.0 ± 0.4 | 1.0 ± 0.3 | 0.9 ± 0.3 | 1.0 ± 0.4 | 0.9 ± 0.2 | 11.2 ± 23.5 | 1.0 ± 0.3 | 1.1 ± 0.3 | 2.1 ± 1.2 | 1.0 ± 0.3 | 2.9 ± 3.5 | 9.9 ± 6.0** | |
| 14 | 1.0 ± 0.3 | 0.8 ± 0.3 | 0.7 ± 0.1 | 1.0 ± 0.5 | 0.5 ± 0.2 | 392.5 ± 450.5 | 1.0 ± 0.2 | 91.2 ± 92.0 | 191.7 ± 100.8** | 1.0 ± 0.3 | 3.1 ± 2.1 | 51.6 ± 29.9** | |
| Clusterin | 1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.4 | 1.1 ± 0.5 | 1.0 ± 0.4 | 1.0 ± 0.2 | 1.1 ± 0.2 | 1.0 ± 0.1 | 1.0 ± 0.3 | 0.7 ± 0.3 | 0.6 ± 0.3 |
| 3 | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.2 | 1.0 ± 0.3 | 2.8 ± 4.4 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.3 ± 0.2 | 1.0 ± 0.1 | 2.0 ± 1.3 | 1.9 ± 1.2 | |
| 14 | 1.0 ± 0.1 | 1.1 ± 0.2 | 0.9 ± 0.0 | 1.0 ± 0.4 | 1.3 ± 0.3 | 39.4 ± 43.1 | 1.0 ± 0.2 | 2.1 ± 1.2 | 1.6 ± 0.2 | 1.0 ± 0.4 | 1.7 ± 0.5 | 5.7 ± 3.0** | |
| Lcn2 (NGAL) | 1 | 1.0 ± 0.8 | 0.5 ± 0.1 | 0.6 ± 0.1 | 1.0 ± 0.6 | 0.8 ± 0.7 | 0.7 ± 0.4 | 1.0 ± 0.4 | 1.4 ± 0.2 | 1.1 ± 0.3 | 1.0 ± 0.9 | 2.6 ± 1.4 | 3.8 ± 2.4 |
| 3 | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.2 | 1.0 ± 0.4 | 1.0 ± 0.4 | 1.5 ± 1.5 | 1.0 ± 0.1 | 0.9 ± 0.4 | 1.0 ± 0.3 | 1.0 ± 0.2 | 3.6 ± 3.0 | 4.1 ± 2.6 | |
| 14 | 1.0 ± 0.4 | 0.7 ± 0.1 | 0.7 ± 0.1 | 1.0 ± 0.3 | 1.7 ± 0.3 | 81.6 ± 151.4 | 1.0 ± 0.3 | 2.4 ± 1.8 | 4.9 ± 2.5* | 1.0 ± 0.4 | 2.3 ± 1.2 | 6.2 ± 2.6** | |
| Timp-1 | 1 | 1.0 ± 0.3 | 1.2 ± 0.5 | 1.3 ± 0.3 | 1.0 ± 0.4 | 0.9 ± 0.4 | 0.7 ± 0.3 | 1.0 ± 0.5 | 1.2 ± 0.6 | 1.2 ± 0.5 | 1.0 ± 0.4 | 2.7 ± 1.1 | 3.7 ± 2.1 |
| 3 | 1.0 ± 0.2 | 1.2 ± 0.2 | 1.1 ± 0.6 | 1.0 ± 0.2 | 0.8 ± 0.5 | 1.4 ± 1.5 | 1.0 ± 0.4 | 1.0 ± 0.4 | 1.2 ± 0.5 | 1.0 ± 0.5 | 2.9 ± 1.7 | 2.3 ± 1.4 | |
| 14 | 1.0 ± 0.4 | 1.1 ± 0.3 | 1.1 ± 0.3 | 1.0 ± 0.3 | 1.0 ± 0.3 | 12.7 ± 12.1* | 1.0 ± 0.5 | 1.9 ± 1.2 | 1.5 ± 0.6 | 1.0 ± 0.9 | 2.9 ± 1.6 | 5.5 ± 2.9** | |
| OPN (secreted phosphoprotein 1) | 1 | 1.0 ± 0.6 | 0.7 ± 0.2 | 0.8 ± 0.3 | 1.0 ± 0.5 | 1.1 ± 0.7 | 1.0 ± 0.5 | 1.0 ± 0.5 | 1.1 ± 0.7 | 1.0 ± 0.5 | 1.0 ± 1.0 | 1.5 ± 0.8 | 2.6 ± 2.1 |
| 3 | 1.0 ± 0.1 | 1.2 ± 0.3 | 0.9 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.8 | 1.0 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.8 | 1.0 ± 0.2 | 2.6 ± 2.3 | 3.6 ± 2.4 | |
| 14 | 1.0 ± 0.2 | 0.8 ± 0.2 | 0.8 ± 0.2 | 1.0 ± 0.3 | 1.3 ± 0.5 | 21.3 ± 21.6* | 1.0 ± 0.0 | 2.4 ± 1.2 | 5.1 ± 1.6*** | 1.0 ± 0.7 | 1.6 ± 0.6 | 4.8 ± 2.6** | |
| HO-1 (HMOX1) | 1 | 1.0 ± 0.1 | 1.0 ± 0.2 | 0.9 ± 0.1 | 1.0 ± 0.6 | 1.4 ± 1.2 | 0.9 ± 0.6 | 1.0 ± 0.1 | 1.1 ± 0.3 | 0.8 ± 0.2 | 1.0 ± 0.4 | 3.0 ± 0.8 | 7.1 ± 5.8 |
| 3 | 1.0 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.0 ± 0.2 | 1.2 ± 0.3 | 1.2 ± 0.3 | 1.0 ± 0.4 | 0.7 ± 0.2 | 0.9 ± 0.1 | 1.0 ± 0.3 | 11.8 ± 9.9 | 13.7 ± 9.4* | |
| 14 | 1.0 ± 0.2 | 1.0 ± 0.2 | 1.3 ± 0.2 | 1.0 ± 0.3 | 1.5 ± 0.3 | 7.1 ± 8.2 | 1.0 ± 0.1 | 1.8 ± 0.3* | 3.4 ± 0.7*** | 1.0 ± 0.5 | 13.4 ± 7.6** | 14.7 ± 2.0** | |
| Vimentin | 1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.4 | 1.0 ± 0.4 | 0.9 ± 0.3 | 1.0 ± 0.2 | 0.9 ± 0.1 | 0.9 ± 0.2 | 1.0 ± 0.3 | 1.3 ± 0.5 | 1.3 ± 0.4 |
| 3 | 1.0 ± 0.1 | 1.0 ± 0.2 | 0.9 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.3 | 1.1 ± 0.3 | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.3 | 3.9 ± 2.5 | 3.3 ± 2.4 | |
| 14 | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.0 | 1.0 ± 0.2 | 1.3 ± 0.4 | 3.4 ± 3.0 | 1.0 ± 0.2 | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.0 ± 0.4 | 3.8 ± 1.8** | 3.4 ± 0.6* | |
Note. Data are presented as mean ± SD of five individual animals per dose group. Statistical analysis was performed by ANOVA and Dunnett’s test.
*p < 0.05, **p < 0.01, and ***p < 0.001.
In contrast, a dramatic increase in the expression of kidney injury marker genes was observed following treatment with BI-3, although some of these changes were not statistically significant due to high interanimal variability. However, it is important to point out that altered expression of marker genes was restricted to those animals showing clear signs of PTD and thus correlated with the renal histopathological changes evident in three high-dose animals (41, 43, and 45) after 14 days (Table 4). Interestingly, after 3 days of treatment, one of the high-dose animals (29) also showed an increase in the expression of these marker genes in the absence of renal histopathological changes.
qRT-PCR revealed a marked increase in the expression of HO-1, Lcn2, OPN, and Kim-1 in high-dose animals after 14 days of treatment with gentamicin, whereas no or only minimal effects on clusterin, Timp-1, and vimentin were evident throughout the study. Compared to BI-3, the gene expression changes in response to gentamicin were less pronounced, thus reflecting the degree of renal injury (mild to moderate vs. moderate to severe with gentamicin and BI-3, respectively). Several marker genes were also found to be upregulated in individual animals after 14 days treatment with the low dose of gentamicin. For example, animal 36 (low dose, 14 days) showed increased mRNA expression of OPN, Lcn2, Kim-1, clusterin, and Timp-1 (Table 4), correlating with the minimal histopathological changes in this animal (Table 2).
Significant dose-dependent changes in the expression of putative kidney biomarkers were evident as early as 1 day after treatment with the high dose of IMM125 (e.g., Kim-1, up 3.4-fold; HO-1, up 7.1-fold; Timp-1, up 3.7-fold; and Lcn2, up 4.7-fold). In addition, these genes were found to be upregulated even in low-dose animals after single administration despite the absence of statistical significance due to the large variability between animals. In contrast, alterations in clusterin, OPN, and vimentin expression were only observed after treatment for 14 days. Overall, the most pronounced changes occurred in kidneys of rats exhibiting mild to moderate PTD, that is, four of five high-dose animals treated for 14 days.
Protein Expression and Immunolocalization of Renal Markers in Kidneys
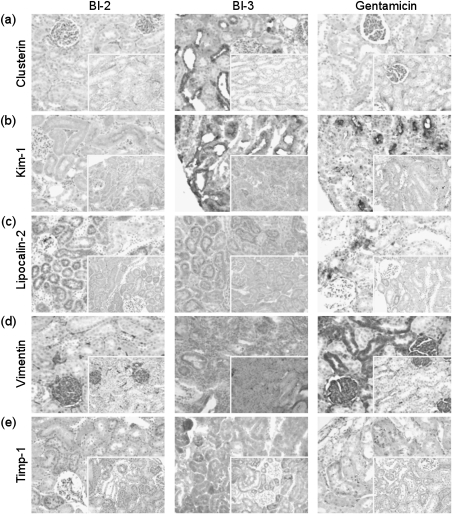
In line with the absence of gene expression changes, immunohistochemical analysis of Kim-1, clusterin, vimentin, and Timp-1 (Figs. 1a, b, d, and e) in kidneys of rats treated with BI-2 revealed no alterations in the protein level or subcellular localization of these markers compared to controls (insets). Surprisingly, a marked increase in Lcn2 was evident in proximal tubule cells of the outer cortex of two high-dose animals (41 and 43) in response to 14 days of treatment (Fig. 1c). At this time point, increased Lcn2 staining was also evident in kidneys of the remaining high-dose animals, albeit to a lesser degree.
FIG. 1.
Immunohistochemical analysis showing expression and immunolocalization of (a) clusterin, (b) Kim-1, (c) Lcn2, (d) vimentin, and (e) Timp-1 in kidneys of male Wistar rats treated with BI-2, BI-3, or gentamicin. Representative images from cortical regions of controls (insets) and high-dose animals after 14 days of treatment (3 days for BI-3 and Lcn2) are shown. Images were acquired at ×200 magnifications using an Olympus CH-2 microscope fitted with an Olympus E330 digital camera.
Immunohistochemical changes in response to BI-3 were restricted to those animals showing PTD. For example, Kim-1 was markedly increased at the apical side of proximal tubule epithelial cells in the outer cortex of animals 41, 43, and 45 (high dose, 14 days treatment) (Fig. 1b), but no changes in Kim-1 expression were observed in high-dose animals 42 and 44, consistent with the lack of effects on the mRNA level. Weak Kim-1–positive tubule cells were also observed in kidney tissue of animal 29 (high dose, 3 days treatment) (data not shown), which is in good agreement with qRT-PCR (Table 4) and histopathology data (Table 2). Enhanced expression of clusterin was also observed in proximal tubule cells of affected high-dose animals after 14 days (Fig. 1a). Similarly, Lcn2 was found to be upregulated in cortical regions of two high-dose animals (29 and 30) after 3 days and in two high-dose animals after 14 days treatment (41 and 43) (Fig. 1c), whereas animal number 45 showed no immunohistochemical staining of Lcn2, correlating well with the absence of Lcn2 gene expression change in this animal. Increased expression of vimentin was also detected in high-dose animals showing histopathology alterations after 14 days of treatment in comparison to control animals (Fig. 1d). Few Timp-1–positive tubules were seen in high-dose animals after treatment with BI-3 for 14 days compared to the uniform punctuate vesicular staining of Timp-1 in tubule epithelial cells in controls (Fig. 1e).
Immunohistochemical staining of candidate biomarkers in kidneys of gentamicin-treated rats confirmed the marked induction of Kim-1 in proximal tubule epithelial cells in the outer cortex, the target site of gentamicin toxicity (Fig. 1b). Furthermore, a slight increase in Lcn2 expression was observed in tissue of high-dose animals after 14 days of treatment (Fig. 1c). In contrast, no effects in the localization or staining intensity of clusterin were evident (Fig. 1a), consistent with the lack of significant changes on the mRNA level (Table 4). Localization of Timp-1 changed from equal distribution in small vesicles in controls to more intensely stained larger vesicles associated with affected tubules in gentamicin-treated rats, but no changes in absolute expression level were apparent (Fig. 1e). Consistent with studies demonstrating that vimentin protein expression is regulated at the translational level (Thomas, 1986), increased immunoreactivity of vimentin was observed (Fig. 1d) in the absence of changes at the mRNA level (Table 4).
Unfortunately, immunohistochemical analysis of kidney sections of rats treated with IMM125 and their corresponding controls showed no immunoreactivity against a range of kidney biomarkers. Vimentin and Lcn2 are normally present even in untreated controls, and slides of other studies run in parallel yielded good results, suggesting that tissues from this study (processed by the participating company) may have been overfixed and thus unsuitable for immunohistochemistry (data not shown).
ELISA Analysis of Candidate Biomarkers in Urine
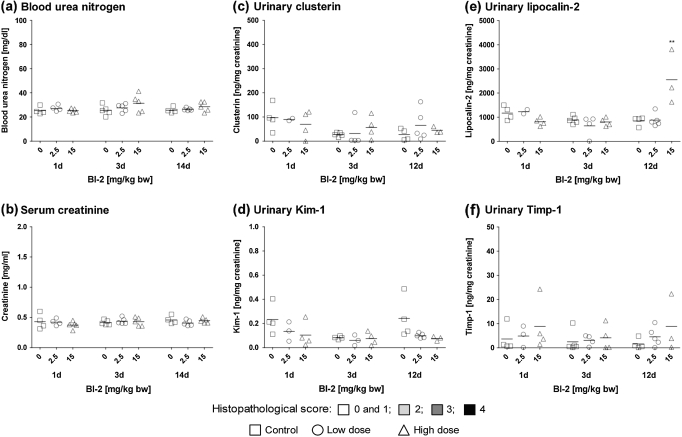
Consistent with immunohistochemical analyses showing enhanced immunoreactivity against Lcn2 in kidney tubule cells of rats treated with BI-2 (Fig. 1c), urinary concentrations of Lcn2 were significantly increased in high-dose animals (Fig. 2). In contrast to Lcn2, no changes in urinary concentration of Kim-1, clusterin, or Timp-1 were observed (Fig. 2), confirming the gene and protein expression data.
FIG. 2.
Analysis of (a) BUN, (b) serum creatinine, (c) urinary clusterin, (d) urinary Kim-1, (e) urinary Lcn2, and (f) urinary Timp-1 in male Wistar rats treated with BI-2 for 1, 3, or 12/14 days. Data are presented as individual animals and color coded according to histopathology scores for PTD. Mean values of five individual animals per dose group are indicated by a black line. Statistical analysis was performed by ANOVA and Dunnett’s test. Statistical changes are indicated by *p < 0.05, **p < 0.01, and ***p < 0.001. Note: for urinary markers after 1 and 3 days of treatment, histopathology scores shown are from groups of rats treated in parallel, that is, animals no. 1–15 and 16–30, respectively.
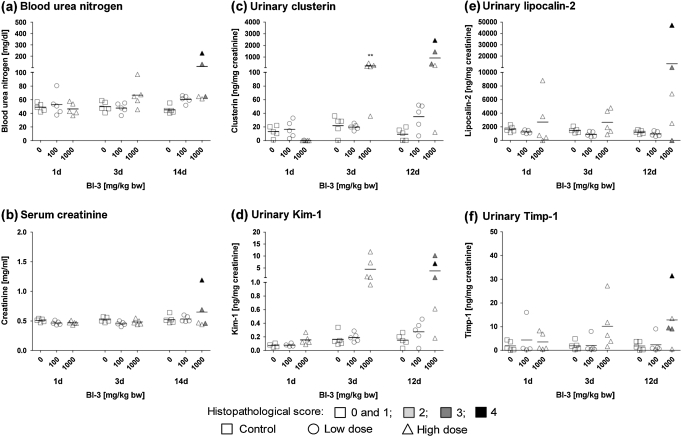
Following treatment with BI-3, a marked increase in urinary Kim-1 and clusterin was present in several high-dose animals after 3 days of treatment (Fig. 3). Thus, changes in these biomarkers appeared to be more sensitive than gene and protein expression and histopathology performed in another group of animals treated in parallel. Enhanced urinary excretion of Lcn2 and Timp-1 was also observed in individual high-dose animals, but the results were not statistically significant due to the high variability in-between animals (Fig. 3).
FIG. 3.
Determination of (a) BUN, (b) serum creatinine, (c) urinary clusterin, (d) urinary Kim-1, (e) urinary Lcn2, and (f) urinary Timp-1 in male Wistar rats treated with BI-3 for 1, 3, or 12/14 days. Data are presented as individual animals and color coded according to histopathology scores for PTD. Mean values of five individual animals per dose group are indicated by a black line. Statistical analysis was performed by ANOVA and Dunnett’s test. Statistical changes are indicated by *p < 0.05, **p < 0.01, and ***p < 0.001. Note: for urinary markers on days 1 and 3, histopathology scores shown are from groups of rats treated in parallel, that is, animals no. 1–15 and 16–30, respectively.
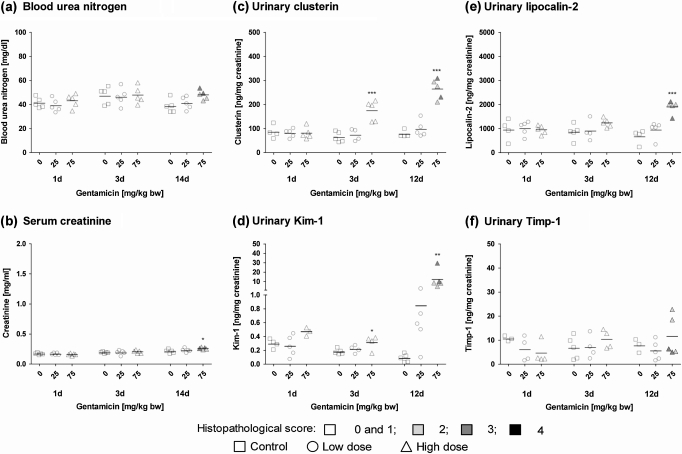
Kim-1 was significantly increased in urine of animals treated with a high dose of gentamicin (75 mg/kg bw) for 3 or 12 days and in individual low-dose animals after 12 days of treatment (Fig. 4). Similarly, but in contrast to the gene and protein expression data, urinary clusterin was significantly increased in high-dose animals, even after 3 days of treatment (Fig. 4). No histopathological alterations were observed at this time point in animals treated with the same regimen in parallel but terminated after 3 days of dosing. Enhanced urinary excretion of Lcn2 was observed in high-dose animals treated for 12 days, but no changes in the concentration of Timp-1 were evident (Fig. 4), consistent with the gene expression and immunohistochemical analyses.
FIG. 4.
Analysis of (a) BUN, (b) serum creatinine, (c) urinary clusterin, (d) urinary Kim-1, (e) urinary Lcn2, and (f) urinary Timp-1 in male Wistar rats treated with gentamicin for 1, 3, or 12/14 days. Data are presented as individual animals and color coded according to histopathology scores for PTD. Mean values of five individual animals per dose group are indicated by a black line. Statistical analysis was performed by ANOVA and Dunnett’s test. Statistical changes are indicated by *p < 0.05, **p < 0.01, and ***p < 0.001. Note: for urinary markers after 1 and 3 days of treatment, histopathology scores shown are from groups of rats treated in parallel, that is, animals no. 1–15 and 16–30, respectively.
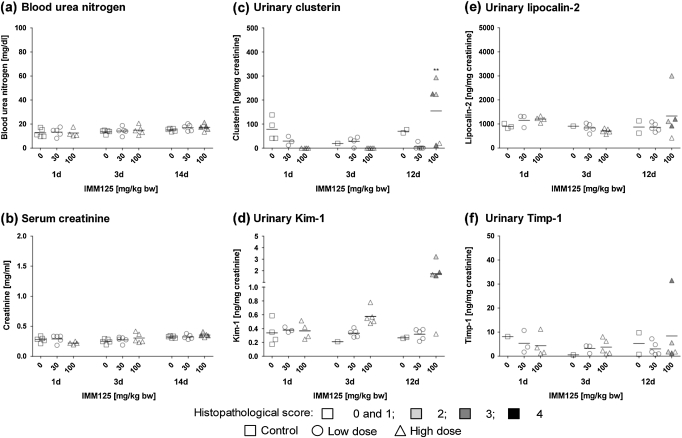
Following treatment with IMM125, urinary excretion of Kim-1, clusterin, and Lcn2 was increased in individual high-dose animals after 12 days of treatment (Fig. 5). While these effects correlated with the histopathological alterations and gene expression changes at this time point, enhanced mRNA expression of kidney biomarkers at earlier time points was not reflected on the level of the respective proteins in urine. Despite a significant increase in Timp-1 mRNA expression following 14 days of treatment with a high dose of IMM125, no effects on urinary Timp-1 were observed.
FIG. 5.
Determination of (a) BUN, (b) serum creatinine, (c) urinary clusterin, (d) urinary Kim-1, (e) urinary Lcn2, and (f) urinary Timp-1 in male Wistar rats treated with IMM125 for 1, 3, or 12/14 days. Data are presented as individual animals and color coded according to histopathology scores for PTD. Mean values of five individual animals per dose group are indicated by a black line. Statistical analysis was performed by ANOVA and Dunnett’s test. Statistical changes are indicated by *p < 0.05, **p < 0.01, and ***p < 0.001. Note: for urinary markers after 1 and 3 days of treatment, histopathology scores shown are from groups of rats treated in parallel, that is, animals no. 1–15 and 16–30, respectively.
ROC Analysis
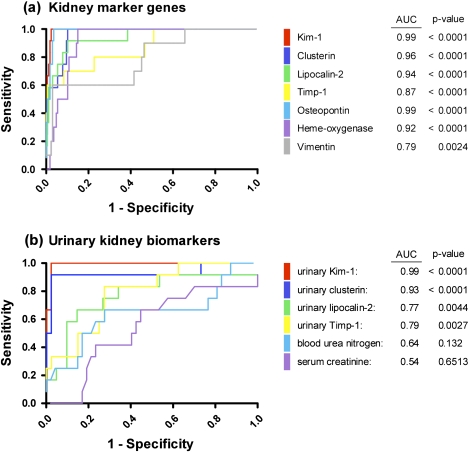
ROC curves were plotted to compare the performance of the novel marker genes and urinary biomarkers versus traditional clinical chemistry parameters, whereby the area under the ROC curve serves as a measure for the overall ability of a biomarker to discriminate control and treated animals without signs of PTD from those with a histopathology score > 1. These analyses revealed high specificity and sensitivity of Kim-1, clusterin, Lcn2, OPN, and HO-1 mRNA expression as kidney marker genes (area under the curve [AUC] > 0.9) and confirmed urinary Kim-1 (AUC = 0.99) and clusterin (AUC = 0.93) as the most sensitive and specific noninvasive indicators of PTD. Although urinary Lcn2 and Timp-1 were less sensitive, they still outperformed indicators of renal function, that is, BUN and serum creatinine, which were found to be the least responsive markers of renal injury in this study (Fig. 6).
FIG. 6.
ROC curves for (a) gene expression of candidate biomarkers and (b) urinary biomarkers compared to traditional clinical chemistry parameters. The area under the ROC curve, which serves as measure for the overall ability of a biomarker to discriminate healthy versus injured animals, is shown in the legend.
DISCUSSION
For decades, assessment of nephrotoxicity has relied primarily on the detection of impaired kidney function by a rise in serum creatinine or BUN. However, it has long been recognized that—due to the functional reserve of the kidney—these commonly used clinical markers are late and, therefore, unreliable indicators of acute kidney injury (AKI). Thus, the need for earlier and more accurate detection of renal toxicity both in the clinical and in preclinical safety assessment has stimulated intense research into the development of novel biomarkers with the ability to measure subtle effects on renal tubule integrity rather than reduced kidney function. Advances in biomedical research, including high-throughput omics technologies, have revolutionized the field of biomarker discovery and have succeeded in identifying a number of promising marker genes and gene products, which are rapidly induced and/or leak into urine in response to tissue injury. In this study, we applied a subset of emerging biomarkers to real-life preclinical toxicity studies with drug candidates that were dropped from further development to assess the value of these novel markers for improved detection of drug-induced kidney toxicity compared to the current gold standard. While an ideal biomarker for use in both clinical and preclinical settings should be reliable, easily measured, and preferably noninvasive (i.e., urinary), we were also interested in linking biomarker concentrations in urine to their expression/localization in the kidney to provide anchoring to the target organ and site of injury.
Treatment-related renal histological changes in the form of PTD, (degeneration, necrosis, and regeneration), increased interstitial mononuclear cell infiltrations, or hyaline droplet accumulation in renal tubules were observed in the course of 4 of 16 short-term toxicity studies in rats conducted within the frame of the InnoMed PredTox project. Although minimal alterations were sometimes evident at earlier time points (days 1–3) or at lower doses, more severe lesions involving the renal tubules were generally restricted to animals treated with a high dose of BI-3, gentamicin, or IMM125 for 14 days, with significant interanimal variability and accompanying hepatotoxicity in some studies. Treatment with BI-2, which was expected to produce papillary necrosis based on a previous study, resulted in hyaline droplet accumulation in kidney cortex, but no signs of tubule cell degeneration or regeneration were evident. Importantly, with the exception of 14 days treatment with gentamicin, no significant alterations in clinical chemistry parameters indicative of nephrotoxicity were observed. ROC analysis confirmed the poor sensitivity of the functional markers BUN and serum creatinine to diagnose PTD, with areas under the ROC curve of 0.64 and 0.54 for BUN and serum creatinine, respectively.
In contrast, putative kidney marker genes were clearly upregulated in response to drugs associated with proximal tubule injury, and—despite the lack of statistical significance due to large interanimal variability—a time- and dose-dependent trend of increasing marker gene expression was sometimes seen before histopathological alterations were recorded. This suggests that even very minor lesions, which may not be detected by standard histopathology assessment, may cause a pronounced transcriptional response that can be picked up by sensitive techniques, such as qRT-PCR. Based on ROC analysis, Kim-1 and OPN appeared to be the most sensitive and specific marker genes of PTD, followed by clusterin, Lcn2, and HO-1. Although Timp-1 and vimentin, which are known to be regulated in part at the translational level (Thomas, 1986), exhibited slightly lower AUC values than the other markers, ROC analysis still suggests that alterations in mRNA expression of these two genes may serve as good markers of renal injury.
As expected, immunohistochemical staining of kidney biomarkers was a less rigorous measure as compared to gene expression analysis but increased our mechanistic understanding and confidence of the candidate biomarkers by allowing us to map the expression change to the site of injury. These analyses demonstrated a good correlation between Kim-1 and clusterin gene and protein expression in affected renal tubules. However, it is important to emphasize that a marked increase in Lcn2 immunoreactivity was observed in renal tubules in response to treatment with BI-2 in the absence of histopathological findings and mRNA expression changes. In contrast to kidneys of rats administered with gentamicin or BI-3, in which individual injured tubules stained positive for Lcn2, uniform distribution of Lcn2 was observed throughout the kidney cortex in BI-2–treated rats. Similar to controls, Lcn2 was localized to vesicular structures in renal tubules of these animals, but staining was far more intense than in control rats from the same study. In light of these somewhat unexpected findings, it is important to note that—as a member of the lipocalin superfamily implicated in the transport of iron—Lcn2, which may be expressed not only by epithelial cells but also by neutrophils (Katano et al., 2009), may undergo glomerular filtration and reabsorption into proximal tubule cells via receptor-mediated endocytosis (Yang et al., 2003). Thus, it appears plausible to assume that the increase in Lcn2 protein in rat kidney and in urine of BI-2–treated rats may be linked to the increase in peripheral blood neutrophils seen in these animals and subsequent renal handling of Lcn2, rather than tubule damage. Collectively, these and other data demonstrating increased urinary concentrations of Lcn2 in response to treatment with hepatotoxic compounds (Smyth et al., 2008) (Adler and Mally, unpublished data) suggest that urinary Lcn2 may not be as specific for kidney injury as generally assumed but may also occur in response to systemic inflammation or tissue damage at other sites or organs. This may have important implications for the use of Lcn2 in predicting AKI in preclinical safety studies as well as in the clinical setting, where Lcn2 has already been proposed as the “renal troponin” (Bennett et al., 2008), referring to cardiac troponin as a highly sensitive and specific marker of acute myocardial injury. While Lcn2 may be rapidly induced and may thus represent an early and sensitive marker of pathological conditions including kidney toxicity, a rise in urinary Lcn2 per se does not allow one to draw conclusions with regard to the cause and site of injury.
In a previous study in which rats were administered gentamicin at 60 and 120 mg/kg bw per day for 7 days, we found that urinary Lcn2 performed better than Kim-1 in detecting aminoglycoside-induced nephrotoxicity, as shown by a slightly larger area under the ROC curve of 0.95 for Lcn2 versus 0.92 for Kim-1 (Sieber et al., 2009). Although our present data on gentamicin also demonstrate a significant increase in urinary Lcn2 and thus support our previous findings, the lower area under the ROC curve for urinary Lcn2 (0.77), which was calculated using combined data from BI-2, BI-3, gentamicin, and IMM125, reflects both the lack of specificity for renal injury (i.e., increased urinary Lcn2 in the absence of tubule damage following treatment with BI-2) and the failure to respond to IMM125-induced nephrotoxicity.
In contrast, urinary Kim-1 and clusterin performed extremely well in this study, with marked increases in affected animals and areas under the ROC curve of 0.99 and 0.93, respectively. Our data, therefore, support previous studies in rats and in patients with AKI, which demonstrated significantly higher sensitivity and specificity of urinary Kim-1 and clusterin as markers of renal injury than traditional clinical chemistry parameters (Eti et al., 1993; Han et al., 2008, 2009; Horstrup et al., 2002; Ichimura et al., 2004; Liangos et al., 2009; Prozialeck et al., 2007; Vaidya et al., 2008; Zhou et al., 2008). In our study, a rise in urinary Kim-1 and/or clusterin was occasionally observed at lower doses or earlier time points than nephrotoxic changes were evident by histopathological evaluation. However, given that the markers under investigation present biomarkers that are mechanistically linked to degeneration and regeneration, we do not propose that biomarker responses precede tubule damage. Rather, it seems possible that the presence of few damaged cells in an entire organ, which may go unnoticed when evaluating random tissue sections, may give rise to a prominent increase in gene expression and subsequent increase in urinary biomarker concentration and may thus be a more objective and quantifiable measure. On the other hand, it needs to be pointed out that urine was collected from rats treated for a total of 14 days, while histopathological evaluation of kidney lesions after 1 and 3 days of treatment was performed using separate groups of animals, which were treated in parallel. Thus, even though drug-induced nephrotoxicity was thoroughly assessed at these early time points, there may be minor differences with regard to the actual degree of nephrotoxic change in those individuals from which urinary biomarker data were collected.
Enhanced urinary excretion of Timp-1 was seen in individual high-dose animals, but the overall performance of Timp-1 as an early indicator of PTD was poor compared to Kim-1 and clusterin, although previous studies reported elevated urinary excretion of Timp-1 in patients with renal disease (Horstrup et al., 2002; Liu et al., 2006; Sanders et al., 2007; Wasilewska and Zoch-Zwierz, 2008). At present, we can only speculate that these discordant findings may reflect species differences in biomarker sensitivity or differences in chemically induced kidney injury versus disease processes. However, this may have important implications for the value of Timp-1 as a bridging biomarker to directly link responses between species in the preclinical and clinical setting.
In our study, OPN gene expression correlated well with the renal histopathological lesions. While studies demonstrating increased urinary levels of OPN in children with steroid-resistant nephrotic syndrome, as well as in mice suffering from focal segmental glomerulosclerosis, supported the use of urinary OPN as a promising kidney marker (Lorenzen et al., 2008; Shui et al., 2007), attempts to measure OPN in urine samples from our study using a commercially available assay (TiterZyme EIA rat OPN, Biomol, Hamburg, Germany) remained unsuccessful. This may be due to the presumed instability of OPN in urine and the need to include stabilizers at the time of urine collection.
In summary, repeated administration of compounds causing PTD (BI-3, gentamicin, and IMM125) resulted in alterations in the expression of candidate biomarkers. Effects on marker gene and protein expression generally correlated well with the renal histopathology alterations and were frequently detected at earlier time points or lower doses than the traditional clinical parameters BUN and serum creatinine, which indicate impaired kidney function rather than tissue injury. Urinary Kim-1 and clusterin reflected changes in mRNA and protein expression and histopathological alterations in the target organ in the absence of functional changes, confirming clusterin and Kim-1 as early and sensitive, noninvasive markers of renal injury and supporting their value for improved detection of drug-induced kidney injury in preclinical safety assessment. Although Lcn2 may not be specific for kidney toxicity, its rapid response to inflammation and tissue damage in general may reinforce its use in routine toxicity testing.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Partially funded by the 6th Research Framework Program of the European Union (InnoMed PredTox, LSHB-CT-2005-518170); National Institutes of Health (ES016723 to V.S.V., DK39773, DK72831, DK74099 to J.V.B.).
Supplementary Material
Acknowledgments
The contribution of all members of the PredTox Consortium (www.innomed-predtox.com) is gratefully acknowledged. The authors would also like to thank Michaela Bekteshi and Ursula Tatsch, University of Würzburg, for excellent technical assistance. PredTox Consortium: Nycomed, Bayer Schering Pharma, Boehringer-Ingelheim, Johnson & Johnson, Lilly, Merck-Serono, Novartis, Novo-Nordisk, Schering-Plough, Roche, Sanofi-Aventis, Servier, University of Würzburg, University College Dublin, University Hacettepe, Bio-Rad, and Genedata.
References
- Amin RP, Vickers AE, Sistare F, Thompson KL, Roman RJ, Lawton M, Kramer J, Hamadeh HK, Collins J, Grissom S, et al. Identification of putative gene based markers of renal toxicity. Environ. Health Perspect. 2004;112:465–479. doi: 10.1289/ehp.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Dent CL, Ma Q, Dastrala S, Grenier F, Workman R, Syed H, Ali S, Barasch J, Devarajan P. Urine NGAL predicts severity of acute kidney injury after cardiac surgery: a prospective study. Clin. J. Am. Soc. Nephrol. 2008;3:665–673. doi: 10.2215/CJN.04010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eti S, Cheng CY, Marshall A, Reidenberg MM. Urinary clusterin in chronic nephrotoxicity in the rat. Proc. Soc. Exp. Biol. Med. 1993;202:487–490. doi: 10.3181/00379727-202-43564. [DOI] [PubMed] [Google Scholar]
- Gallagher WM, Tweats D, Koenig J. Omic profiling for drug safety assessment: current trends and public-private partnerships. Drug Discov. Today. 2009;14:337–342. doi: 10.1016/j.drudis.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Han WK, Wagener G, Zhu Y, Wang S, Lee HT. Urinary biomarkers in the early detection of acute kidney injury after cardiac surgery. Clin. J. Am. Soc. Nephrol. 2009;4:873–882. doi: 10.2215/CJN.04810908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han WK, Waikar SS, Johnson A, Betensky RA, Dent CL, Devarajan P, Bonventre JV. Urinary biomarkers in the early diagnosis of acute kidney injury. Kidney Int. 2008;73:863–869. doi: 10.1038/sj.ki.5002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstrup JH, Gehrmann M, Schneider B, Ploger A, Froese P, Schirop T, Kampf D, Frei U, Neumann R, Eckardt KU. Elevation of serum and urine levels of TIMP-1 and tenascin in patients with renal disease. Nephrol. Dial. Transplant. 2002;17:1005–1013. doi: 10.1093/ndt/17.6.1005. [DOI] [PubMed] [Google Scholar]
- Ichimura T, Hung CC, Yang SA, Stevens JL, Bonventre JV. Kidney injury molecule-1: a tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am. J. Physiol. Renal Physiol. 2004;286:F552–F563. doi: 10.1152/ajprenal.00285.2002. [DOI] [PubMed] [Google Scholar]
- Katano M, Okamoto K, Arito M, Kawakami Y, Kurokawa MS, Suematsu N, Shimada S, Nakamura H, Xiang Y, Masuko K, et al. Implication of GM-CSF induced neutrophil gelatinase-associated lipocalin in pathogenesis of rheumatoid arthritis revealed by proteome analysis. Arthritis Res. Ther. 2009;11:R3. doi: 10.1186/ar2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, Sagartz JE, Morris DL. The application of discovery toxicology and pathology towards the design of safer pharmaceutical lead candidates. Nat. Rev. Drug Discov. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- Liangos O, Tighiouart H, Perianayagam MC, Kolyada A, Han WK, Wald R, Bonventre JV, Jaber BL. Comparative analysis of urinary biomarkers for early detection of acute kidney injury following cardiopulmonary bypass. Biomarkers. 2009;14:423–431. doi: 10.1080/13547500903067744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BC, Zhang L, Lv LL, Wang YL, Liu DG, Zhang XL. Application of antibody array technology in the analysis of urinary cytokine profiles in patients with chronic kidney disease. Am. J. Nephrol. 2006;26:483–490. doi: 10.1159/000096871. [DOI] [PubMed] [Google Scholar]
- Lorenzen J, Shah R, Biser A, Staicu SA, Niranjan T, Garcia AM, Gruenwald A, Thomas DB, Shatat IF, Supe K, et al. The role of osteopontin in the development of albuminuria. J. Am. Soc. Nephrol. 2008;19:884–890. doi: 10.1681/ASN.2007040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrane L, Rexhepaj E, Smart V, Callanan JJ, Orhan D, Eldem T, Mally A, Schroeder S, Meyer K, Wendt M, et al. Creation of a digital slide and tissue microarray resource from a multi-institutional predictive toxicology study in the rat: an initial report from the PredTox group. Exp. Toxicol. Pathol. 2008;60:235–245. doi: 10.1016/j.etp.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Prozialeck WC, Vaidya VS, Liu J, Waalkes MP, Edwards JR, Lamar PC, Bernard AM, Dumont X, Bonventre JV. Kidney injury molecule-1 is an early biomarker of cadmium nephrotoxicity. Kidney Int. 2007;72:985–993. doi: 10.1038/sj.ki.5002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rached E, Hoffmann D, Blumbach K, Weber K, Dekant W, Mally A. Evaluation of putative biomarkers of nephrotoxicity after exposure to ochratoxin a in vivo and in vitro. Toxicol. Sci. 2008;103:371–381. doi: 10.1093/toxsci/kfn040. [DOI] [PubMed] [Google Scholar]
- Sanders JS, Huitema MG, Hanemaaijer R, van Goor H, Kallenberg CG, Stegeman CA. Urinary matrix metalloproteinases reflect renal damage in anti-neutrophil cytoplasm autoantibody-associated vasculitis. Am. J. Physiol. Renal Physiol. 2007;293:F1927–F1934. doi: 10.1152/ajprenal.00310.2007. [DOI] [PubMed] [Google Scholar]
- Shui HA, Ka SM, Yang SM, Lin YF, Lo YF, Chen A. Osteopontin as an injury marker expressing in epithelial hyperplasia lesions helpful in prognosis of focal segmental glomerulosclerosis. Transl. Res. 2007;150:216–222. doi: 10.1016/j.trsl.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Sieber M, Hoffmann D, Adler M, Vaidya VS, Clement M, Bonventre JV, Zidek N, Rached E, Amberg A, Callanan JJ, et al. Comparative analysis of novel noninvasive renal biomarkers and metabonomic changes in a rat model of gentamicin nephrotoxicity. Toxicol. Sci. 2009;109:336–349. doi: 10.1093/toxsci/kfp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth R, Lane CS, Ashiq R, Turton JA, Clarke CJ, Dare TO, York MJ, Griffiths W, Munday MR. Proteomic investigation of urinary markers of carbon-tetrachloride-induced hepatic fibrosis in the Hanover Wistar rat. Cell Biol. Toxicol. 2008;25:499–512. doi: 10.1007/s10565-008-9104-8. [DOI] [PubMed] [Google Scholar]
- Thomas G. Translational control of mRNA expression during the early mitogenic response in Swiss mouse 3T3 cells: identification of specific proteins. J. Cell Biol. 1986;103:2137–2144. doi: 10.1083/jcb.103.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya VS, Ramirez V, Ichimura T, Bobadilla NA, Bonventre JV. Urinary kidney injury molecule-1: a sensitive quantitative biomarker for early detection of kidney tubular injury. Am. J. Physiol. Renal Physiol. 2006;290:F517–F529. doi: 10.1152/ajprenal.00291.2005. [DOI] [PubMed] [Google Scholar]
- Vaidya VS, Waikar SS, Ferguson MA, Collings FB, Sunderland K, Gioules C, Bradwin G, Matsouaka R, Betensky RA, Curhan GC, et al. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin. Transl. Sci. 2008;1:200–208. doi: 10.1111/j.1752-8062.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska AM, Zoch-Zwierz WM. Urinary levels of matrix metalloproteinases and their tissue inhibitors in nephrotic children. Pediatr. Nephrol. 2008;23:1795–1802. doi: 10.1007/s00467-008-0881-3. [DOI] [PubMed] [Google Scholar]
- Yang J, Mori K, Li JY, Barasch J. Iron, lipocalin, and kidney epithelia. Am. J. Physiol. Renal Physiol. 2003;285:F9–F18. doi: 10.1152/ajprenal.00008.2003. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Vaidya VS, Brown RP, Zhang J, 2 BA, Thompson KL, Miller TJ, Bonventre JV, Goering PL. Comparison of kidney injury molecule-1 and other nephrotoxicity biomarkers in urine and kidney following acute exposure to gentamicin, mercury, and chromium. Toxicol. Sci. 2008;101:159–170. doi: 10.1093/toxsci/kfm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.