Abstract
Acetaminophen (APAP), a widely used analgesic and antipyretic that is considered to be relatively safe at recommended doses, is the leading cause of drug-induced liver failure in the United States. 3′-Hydroxyacetanilide (AMAP), a regioisomer of APAP, is useful as a comparative tool for studying APAP-induced toxicity because it is nontoxic relative to APAP. Transforming growth factor-alpha transgenic mouse hepatocytes were treated with both isomers to investigate mitogen-activated protein kinase (MAPK) cascades in order to differentiate their toxicological outcomes. Posttranslational modifications of MAPK signaling were assessed using immunoblotting and Bioplex technology, whereas gene expression changes were measured using Affymetrix Mouse Gene 1.0 ST arrays. APAP treatment led to higher levels of glutathione depletion at 6 and 24 h compared with AMAP in mitochondria. Glutathione depletion was preceded by increased levels of c-Jun N-terminal kinase (JNK) phosphorylation at 2 and 6 h after APAP treatment compared with AMAP, whereas AMAP treatment led to increased extracellular signal–regulated protein kinase (ERK) phosphorylation at 2 and 6 h compared with APAP. Furthermore, APAP treatment significantly upregulated jun oncogene (c-Jun) gene expression, which was confirmed by Western blotting for both the phosphorylated and the nonphosphorylated forms of c-Jun protein. Transfection with JNK siRNA attenuated APAP toxicity after 24 h, suggesting that higher levels of APAP-induced activation of JNK were related to higher rates of cell death. In summary, genomic regulation of MAPK-related transcription factors coupled with posttranslational activation of their upstream kinases is critical in differentiating the toxicities of APAP and AMAP.
Keywords: kinase regulation, acetaminophen, 3′-hydroxyacetanilide, c-Jun N-terminal kinase, jun oncogene
Acetaminophen (N-acetyl-p-aminophenol, APAP) is a widely used analgesic and antipyretic that is considered to be relatively safe at recommended doses. However, overdose cases are fairly common because of the widespread availability of APAP (Lee, 2004). The majority of an APAP dose is metabolized to relatively nontoxic products via conjugation reactions, such as sulfation and glucuronidation (Watari et al., 1983). The remaining dose is predominantly metabolized in the liver by cytochrome P4502E1 to the presumed reactive intermediate, N-acetyl-p-benzoquinoneimine (NAPQI) (James et al., 2003; Nelson and Bruschi, 2007). At therapeutic doses, low levels of P450-generated NAPQI are cleared by glutathione and glutathione-S-transferases (Coles et al., 1988). However, in APAP overdose situations, higher concentrations of NAPQI deplete cellular glutathione pools leading to arylation of intracellular nucleophiles by NAPQI. Modification of these nucleophilic residues within the cell can lead to oxidative/electrophilic stress resulting in hepatocellular injury (Jollow et al., 1973). A comparative tool useful for studying APAP-induced toxicity is its regioisomer, 3′-hydroxyacetanilide (N-acetyl-m-aminophenol, AMAP) (Holme et al., 1991; Myers et al., 1995; Pierce et al., 2002; Rashed et al., 1990; Roberts et al., 1990; Tirmenstein and Nelson, 1989). AMAP is nontoxic relative to APAP, despite the fact that its quinone and quinoneimine intermediates are inherently more reactive and form more covalent protein adducts than those formed during APAP metabolism (Holme et al., 1991).
The fact that overall covalent adduct formation does not provide a definitive explanation for the differences in APAP and AMAP toxicity has led to the exploration of other pathways and biomarkers to differentiate the toxicity of these isomers. One pathway of interest is the mitogen-activated protein kinase (MAPK) cascade. Characterizing MAPK modulation with respect to treatment with APAP and AMAP may give insight into differentiating their toxicities. MAPKs are proteins responsible for the relay of cellular signals to the nucleus in order to elicit genomic responses. This phosphorelay cascade involves activation of a MAPK kinase kinase kinase (MAP4K), which phosphorylates a MAPK kinase kinase (MAP3K), which phosphorylates a MAPK kinase (MAP2K), followed by phosphorylation of a MAPK, which in turn phosphorylates specific nuclear targets resulting in gene expression modulation (Johnson et al., 2005). MAPKs can be divided into six major groups based on sequence homology: (1) the extracellular signal–related kinases (ERK1/2), (2) c-Jun N-terminal kinases (JNKs), (3) p38s, (4) ERK5, (5) ERK3s, and (6) ERK7s (Turjanski et al., 2007). MAPKs have been implicated in a wide array of physiological processes, including cell proliferation, differentiation, and survival (Su and Karin, 1996).
Treatment with APAP has already been shown to induce JNK activity, leading to increased translocation of proapoptotic Bax into the mitochondria (Gunawan et al., 2006). Furthermore, treatment with JNK inhibitors seems to protect hepatocytes against APAP-induced hepatocellular injury by inhibiting JNK-mediated jun oncogene protein (c-Jun) phosphorylation (Bennett et al., 2001) and preventing mitochondrial dysfunction via inhibition of the mitochondrial permeability transition (Latchoumycandane et al., 2007). Although the role of JNK during APAP administration has typically been associated with pathologic consequences, it is worth noting that recent evidence suggests JNK2 may play a cytoprotective role in APAP-induced liver injury (Bourdi et al., 2008). TGF-alpha transgenic mouse hepatocytes (TAMH) were used as a cell culture model to differentiate the toxicities of APAP and AMAP because they not only retain CYP2E1 and CYP3A protein expression but also show characteristic markers of APAP-mediated cell death processes (Pierce et al., 2002). We hypothesize that JNK activation plays a major role in differentiating the toxicities of the regioisomers in TAMH cells.
MATERIALS AND METHODS
Reagents.
Acetaminophen, 3′-hydroxyacetanilide, glycine, dexamethasone, nicotinamide, soybean trypsin inhibitor, and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were obtained from Sigma-Aldrich (St Louis, MO). Gentamicin, trypsin, Dulbecco’s modified Eagle’s medium/Ham’s F12 (1:1), Dulbecco’s PBS (DPBS), Hank’s balanced salt solution, Trizol reagent, NP40 cell lysis buffer, and 4-plex antibody beads were purchased from Invitrogen (Carlsbad, CA). ITS premix was obtained from BD Bioscience (Bedford, MA). Dimethyl sulfoxide and chloroform were purchased from MP Biomedicals (Solon, OH). NaCl was purchased from Fisher (Pittsburgh, PA). All reagents used in the determination of total glutathione content were supplied in the BioVision ApoGSH Glutathione Colorimetric Detection Kit (Mountain View, CA). All reagents used in the processing of total RNA for microarray analysis were supplied in the One-Cycle Target Labeling and Control Reagents Kit from Affymetrix (Santa Clara, CA).
Cell culture.
TAMH cells were grown in serum-free Dulbecco’s modified Eagle’s medium/Ham’s F12 (1:1) media supplemented with 100nM dexamethasone, 10nM nicotinamide, 0.1% (vol/vol) gentamicin, and an ITS premix containing insulin (5 mg/ml), transferrin (5 mg/ml), and selenium (5 ng/ml). Cell passages between 25 and 35 were grown in a humidified incubator with 5% CO2 and 95% air at 37°C. During passages, cells were incubated for 1 min with trypsin. Cell detachment was monitored using a microscope. Once detachment was complete, 5 ml of 0.5 mg/ml soybean trypsin inhibitor in Hank’s balanced salt solution was added before plating.
Cell viability and glutathione depletion assays.
APAP and AMAP were dissolved directly into the culture media on the same day as the dosing regimen. Two hundred microliter of the treatment solutions was added to the confluent monolayers plated on 96-well plates. When incubation of the hepatocytes with APAP or AMAP was completed, 50 μl of a 2 mg/ml MTT solution in DPBS was added per well and incubated in the dark at 37°C for 2 h. The MTT dye solution was then removed by aspiration, and 200 μl dimethyl sulfoxide and 25 μl of Sorensen’s glycine buffer (0.1M glycine, 0.1M NaCl, pH 10.5) was added per well. Plates were then read at 560 nm using a Thermo microplate reader and i-Control software. The viability of drug-treated cells was expressed as the percent of the viability of control cells. For the adenosine triphosphate (ATP) depletion assays, confluent monolayers plated on 96-well plates were treated in the same manner as described above for the MTT assay. ATP levels were measured using the Promega CellTiter-Glo Luminescent Cell Viability Assay (Madison, WI) by following the manufacturer’s protocol. Luminescence was then measured and analyzed with Phenix PlateLumino (Hayward, CA). Total glutathione levels were measured from whole-cell and mitochondrial preparations of TAMH cells grown to confluence on 6-well plates (whole cell) or 150-mm2 tissue culture dishes (mitochondria) and dosed. Mitochondrial fractions were isolated in the absence of dithiothreitol using the protocol for Biovision’s Mitochondria/Cytosol Fractionation Kit (Mountain View, CA). Glutathione content was then analyzed according to the manufacturer’s protocol.
MAPK phosphoprotein analysis.
TAMH cells were treated with 2mM APAP, 2mM AMAP, or control culture media for 2 and 6 h and processed in duplicate for simultaneous measurement of phosphoprotein levels using an Akt, JNK1/2, ERK1/2, and p38 MAPK Phospho 4-plex panel (Invitrogen) according to the manufacturer’s instructions. Briefly, treated cells were rinsed with DPBS and harvested by scraping. They were then lysed with proprietary NP40 cell lysis buffer (Biosource) supplemented with 1mM phenylmethylsulfonyl fluoride and protease cocktail inhibitor cocktail (Roche, Palo Alto, CA) for 30 min on ice. The lysates were placed in a microcentrifuge and spun at 13,000 × g for 10 min at 4°C. Supernatants were collected and pretreated with twofold dilution of the manufacturer’s sample treatment buffer and eightfold dilution of assay diluent. Then, 100 μl of each standard from the series of twofold dilution and treated samples were incubated with target capturing beads on a 96-well plate for 2 h, followed by 1 h of incubation with specific biotinylated detection antibodies. Next, goat anti-rabbit IgG-R-Phycoerythrin was added to each well and incubated for 30 min. Filtration and washing were incorporated after each incubation step with wash buffer using vacuum manifold. Samples were incubated at room temperature with shaking, resuspended in wash buffer, and read using the Luminex 100 (Austin, TX) suspension array reader. Total protein levels of Akt, JNK1/2, ERK1/2, and p38 MAPK were measured simultaneously with the same procedure steps above using an Akt, JNK1/2, ERK1/2, and p38 MAPK total 4-plex panel (Invitrogen).
JNK and c-Jun Western analysis.
TAMH cells were grown to confluence on 6-well tissue culture plates and dosed with 2mM APAP, 2mM AMAP, or control culture media for 2 and 6 h. After treatment was complete, cells were lysed by adding 1× SDS sample buffer and harvested by scraping. Lysates from 1 × 105 cells were loaded per well and resolved by denaturing electrophoresis using 8% SDS-polyacrylamide gel electrophoresis in the Bio-Rad Mini-PROTEAN II system (Hercules, CA). Transfer to nitrocellulose membranes was performed using the Bio-Rad TransBlot SD Semi-Dry Transfer Cell (Hercules, CA) for 90 min at 15 V. Membranes were blocked in a 1:1 ratio of Tris-buffered saline (25mM Tris base, pH 8) and Li-Cor Blocking Buffer (Lincoln, NE) for 1 h at room temperature. Membranes were incubated overnight at 4°C with specific primary antibodies diluted (1:1000) in Li-Cor Blocking Buffer with 0.2% (vol/vol) Tween-20. All primary antibodies were purchased from Cell Signaling Technology (Danvers, MA). Membranes were then incubated in secondary IR Dye-800 Conjugated Affinity Purified anti-Rabbit IgG from Rockland (Gilbertsville, PA) diluted in Li-Cor Blocking Buffer with 0.2% (vol/vol) Tween-20 for 1 h at room temperature (1:20000 for phospho-JNK and 1:10000 for c-Jun and phospho-c-Jun). Detection and quantification of the immunofluorescence bands was accomplished using the Odyssey Infrared Imaging System and Software (Li-Cor).
Inhibition of JNK1 with siRNA.
TAMH cells were passaged regularly to maintain exponential growth. Prior to plating in 96-well plates, a reverse transfection method was utilized to improve transfection efficiency. Transient JNK1 siRNA transfection was performed using MAPK8 Silencer Select siRNA at a final concentration of 30nM and following the protocol for Ambion’s Silencer siRNA Starter Kit (Austin, TX). Cell viability was measured by MTT assay as described previously in the manuscript.
RNA isolation.
RNA was isolated from TAMH cells dosed with 2mM APAP, 2mM AMAP, or control culture media for 2-, 6-, and 24-h timepoints. For each treatment, cells were grown to confluence in two 150-mm2 tissue culture dishes and dosed. At the end of each treatment, cells were harvested using a rubber scraper. Cell pellets were collected by centrifugation and washed using ice-cold DPBS. Trizol reagent was added to the DPBS-washed pellet, vortexed, and passed through a 22-G needle multiple times to ensure complete cell lysis. Chloroform was then added, and the mixture was spun in a microcentrifuge at 8200 × g. The aqueous phase was isolated and dissolved in 70% ethanol. The resulting mixture was loaded onto a Qiagen RNeasy column (Valencia, CA). Purified total RNAs were eluted in sterile water according to the manufacturer’s protocol.
Gene expression analysis.
RNA integrity was assessed using the Agilent 2100 Bioanalyzer and only samples passing quality control were further processed. The manufacturer’s protocol was then followed for the determination of gene expression data using either nine Affymetrix Mouse Genome 430 2.0 arrays (n = 1 per group) or 28 Affymetrix Mouse Gene 1.0 ST arrays (n = 3 per group). Following hybridization and washing, Affymetrix arrays were scanned with an Affymetrix GeneChip 3000 scanner. Image generation and feature extraction were performed using GCOS Software. Only data from arrays that passed the manufacturer’s quality specifications were used for further analysis. All microarray data have been deposited in the Gene Expression Omnibus Database under accession number GSE18614 (http://www.ncbi.nlm.nih.gov/geo/). Gene Pathway analysis was performed using PathwayStudio software version 6.2 (Ariadne Genomics, Rockville, MD). Gene expression data containing log2 fold changes and p values comparing APAP with AMAP treatments at all three timepoints were uploaded. The gene list was then filtered to include only genes with a p value < 0.01 and a |fold change| > 1.5. A Fisher’s exact test was then performed to identify gene subnetworks enriched by these significant expression changes.
Statistical analysis.
Data are presented as mean ± SEM. Comparisons between multiple groups were performed with Stata 10 using one-way ANOVA and subsequent Bonferroni t-tests; p < 0.05 was considered significant. For gene expression data, statistical analysis and data normalization were carried out with Bioconductor software as described previously (Coe et al., 2007).
RESULTS
Cytotoxicity of Acetaminophen and Its Regioisomer
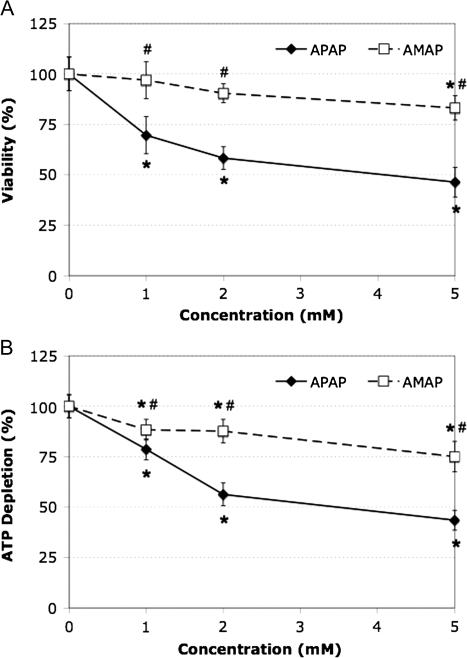
TAMH cells were exposed to various concentrations (1–5mM) of APAP and AMAP for 24 h (Fig. 1). Significant differences in cytotoxicity and ATP depletion were observed between APAP and AMAP over all tested concentrations. The viability of cells treated with 2mM APAP decreased by 42 and 32% compared with control and AMAP treatments, respectively (Fig. 1A). This coincided with a 44 and 32% decrease in ATP levels as compared with control and AMAP treatments, respectively (Fig. 1B). Time-course studies were then performed to determine the rate at which cell death occurs. By MTT assay, significant differences in cytotoxicity were observed between the regioisomers at 6 and 24 h (Fig. 2A). These results again were confirmed using the ATP depletion assay with significant differences observed at all three timepoints, 2, 6, and 24 h (Fig. 2B). These data are consistent with previous reports from our lab confirming that APAP administration leads to changes in TAMH cell morphology indicative of toxicity (6). Taken together, these results suggest that APAP causes more toxicity than AMAP in TAMH cells at 1, 2, and 5mM treatments. In addition, these differences are manifested by 6 h during treatment with 2mM APAP and AMAP.
FIG. 1.
Dose-related toxicity comparison between APAP and AMAP treatments in TAMH. Cells were treated with either no drug or varying concentrations of APAP or AMAP (1–5mM) over a 24-h period. Cell viability was measured by (A) MTT (n = 4) and (B) ATP depletion (n = 6) assays. *p < 0.05 (compared with controls); #p < 0.05 (compared with APAP).
FIG. 2.
Time-related toxicity comparison between APAP and AMAP treatments in TAMH cells. TAMH cells were treated with no drug, 2mM APAP, or 2mM AMAP for 2, 6, and 24 h. Cell viability was measured by (A) MTT (n = 12) and (B) ATP depletion (n = 12) assays. *p < 0.05 (compared with controls); #p < 0.05 (compared with APAP).
Glutathione Depletion Following Treatment with APAP and AMAP
To confirm whether glutathione depletion was related to a loss in cell viability, total glutathione levels were measured in both whole cells and isolated mitochondria and are displayed in Table 1. Six- and 24-h treatment with either regioisomer led to significantly lower levels of glutathione in whole-cell preparations, whereas mitochondrial glutathione pools were significantly depleted at all three timepoints compared with controls. Although no significant differences in whole-cell glutathione depletion existed between the regioisomers, APAP treatment caused significantly more glutathione depletion than AMAP at 6 and 24 h in isolated mitochondria. These data suggest that glutathione depletion in the mitochondria, and not the whole cell, is related to the higher levels of ATP depletion and cell death (Figs. 2B and 2A, respectively) observed after APAP treatment.
TABLE 1.
APAP- and AMAP-Induced Depletion of Glutathione in TAMH Cells
| Time (h) | Cellular glutathione (μmol/g)a | Mitochondrial glutathione (μmol/g) | ||||
| Control | APAP | AMAP | Control | APAP | AMAP | |
| 2 | 3.21 ± 0.06 | 3.22 ± 0.03 | 3.03 ± 0.08 | 3.51 ± 0.17 | 2.68 ± 0.14* | 2.86 ± 0.21* |
| 6 | 3.29 ± 0.06 | 1.98 ± 0.09* | 2.40 ± 0.12* | 3.40 ± 0.02 | 2.04 ± 0.14* | 2.42 ± 0.26*# |
| 24 | 3.23 ± 0.06 | 1.70 ± 0.14* | 2.17 ± 0.14* | 3.58 ± 0.10 | 1.79 ± 0.18* | 2.28 ± 0.11*# |
Units represent micromole glutathione per gram protein.
*p < 0.05 (compared with controls); #p < 0.05 (compared with APAP).
APAP- and AMAP-Induced Changes in MAPK Phosphorylation
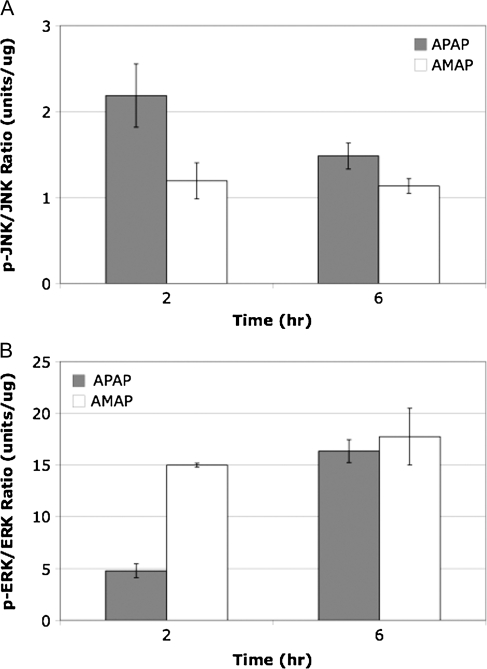
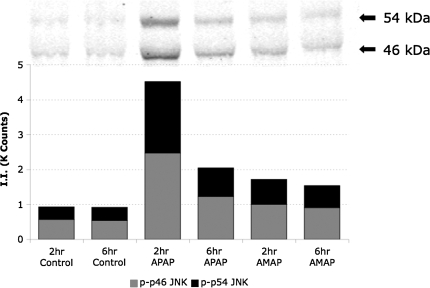
In order to determine whether the differences in cytotoxicity and glutathione depletion were related to differences in MAPK activation, levels of phosphorylated protein were analyzed using the Bioplex system. Cell lysates from TAMH cells treated with 2mM APAP and AMAP for 2 and 6 h were collected, and protein samples were analyzed using a 4-plex phospho-assay that measured phosphorylation ratios of both JNK1/2 and ERK1/2 (Fig. 3). JNK phosphorylation was greater at both timepoints after APAP treatment compared with AMAP (Fig. 3A). Western blotting was then used to confirm these results (Fig. 4). In contrast, ERK phosphorylation was greater at both timepoints after AMAP treatment compared with APAP (Fig. 3B). This data suggest that the greater degree of APAP-induced JNK phosphorylation may lead to greater activation of JNK-mediated cell death pathways, whereas AMAP-induced ERK activation may play a role in attenuating toxicity.
FIG. 3.
Comparison of MAPK phosphorylation in APAP- or AMAP-treated TAMH cells relative to controls. TAMH cells were treated with 2mM APAP or AMAP for 2 and 6 h. Samples were then analyzed in duplicate using a multiplex assay with the Bioplex system. Average ratios of phosphorylated to total MAPK protein were then calculated for (A) JNK and (B) ERK relative to control.
FIG. 4.
JNK1/2 phosphorylation following APAP or AMAP treatment. TAMH cells were treated with 2mM APAP or AMAP for 2 and 6 h. Total cell lysates were blotted with a polyclonal anti-phospho-SAPK/JNK (thr-183/tyr-185) antibody and quantified. Bars represent quantified fluorescence using Odyssey software as described in the “Materials and Methods” section.
APAP- and AMAP-Induced Changes in c-Jun Phosphorylation
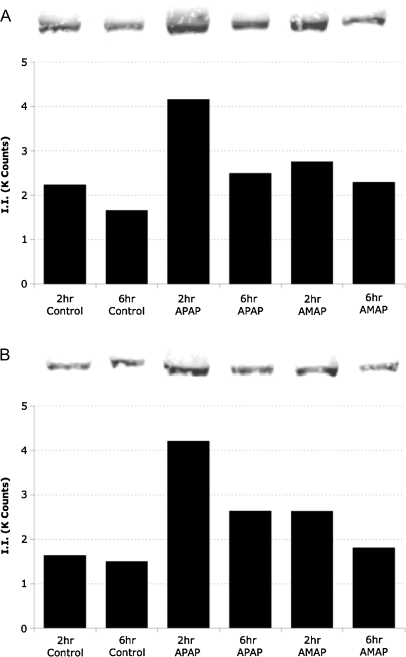
In order to further confirm APAP-induced JNK activation, phosphorylation of c-Jun was measured. TAMH cells were treated with 2mM APAP and AMAP and probed for phosphorylated-c-Jun (Fig. 5A). Compared with control, phosphorylation was greatest after 2-h APAP treatment (1.9-fold increase), and by 6 h, a 1.5-fold increase in c-Jun phosphorylation was observed. Increased c-Jun phosphorylation after AMAP treatment was also observed but at levels more similar to the controls, 1.6- and 1.2-fold increases at 2 and 6 h, respectively. Levels of nonphosphorylated c-Jun were also investigated in the same cell lysates (Fig. 5B). Compared with control, 2.6- and 1.8-fold increases in c-Jun protein were observed after 2- and 6-h APAP treatment, respectively. AMAP treatment also leads to an upregulation of c-Jun but to a lesser extent, as a 1.6-fold increase in c-Jun protein was observed after 2-h AMAP treatment, followed by a 1.2-fold increase after 6 h.
FIG. 5.
APAP- and AMAP-induced changes in both total and phosphorylated c-Jun protein levels. TAMH cells were treated with 2mM APAP or AMAP for 2 and 6 h. Total cell lysates were blotted with either (A) monoclonal anti-phospho-c-Jun (ser-73) or (B) monoclonal anti-c-Jun antibodies and quantified. Bars represent quantified fluorescence using Odyssey software as described in the “Materials and Methods” section.
Cytotoxicity of Acetaminophen and Its Regioisomer in a Transient JNK1 Knockdown Model
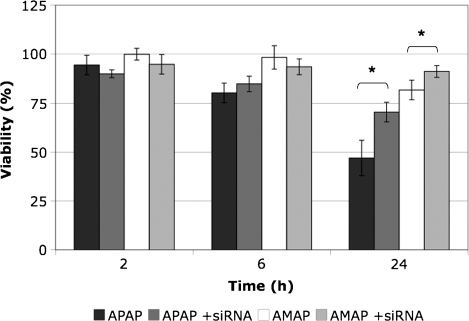
In an attempt to further elucidate the role that JNK activity plays in APAP-induced toxicity, TAMH cells were reverse transfected with JNK1 siRNA. After 24 h of treatment, significant differences were observed between TAMH cells and JNK1 knockdowns for the same set of conditions (Fig. 6). Inhibition of JNK1 led to a 23% increase in survival following treatment with APAP, whereas inhibition of JNK1 led to only a 9% increase in survival following treatment with AMAP. These data suggest that JNK1 activation plays a greater role in propagating the toxicity of APAP compared with AMAP.
FIG. 6.
Effects of JNK1 inhibition on APAP- and AMAP-induced toxicity using siRNA. TAMH cells were reverse transfected with JNK1 siRNA and treated with 2mM APAP or AMAP for 2, 6, and 24 h. Transfected cells (n = 4) were compared with controls not exposed to siRNA (n = 12), and cell viability was measured by MTT assay. *p < 0.05 (compared with controls); #p < 0.05 (compared with APAP).
Microarray Comparison of APAP- and AMAP-Induced Changes in Gene Regulation
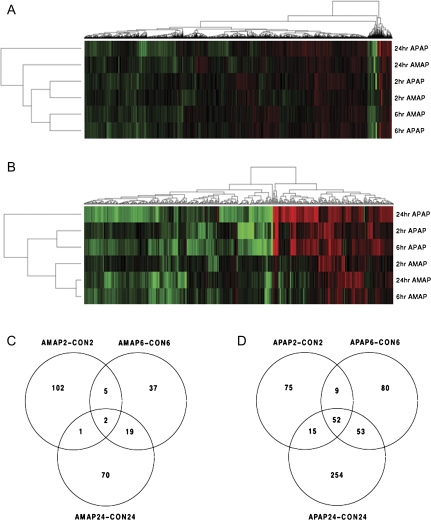
In order to identify transcriptional changes that may be responsible for observed differences in toxicity, messenger RNA was isolated from TAMH cells exposed to therapeutically relevant concentrations of 2mM APAP and AMAP. Microarray analysis was then performed using Affymetrix Mouse Gene 1.0 ST arrays (n = 3 per group). Figure 7A is a heatmap of the 9121 genes whose expression was significantly altered (p < 0.05) in at least one of the timepoints/treatments relative to the vehicle controls. Red indicates increased gene expression relative to the control, whereas green represents downregulation. Overall, APAP and AMAP cause many similar gene expression changes. This is evidenced by the fact that transcriptional changes cluster more closely with respect to dosing time rather than drug treatment. However, when a heatmap was generated for the 524 genes found to be both significant (p < 0.05) and relatively large (|fold change| > 1.5) in at least one of the timepoints/treatments relative to the vehicle controls, gene expression patterns clustered more closely with respect to drug treatment rather than dosing time (Fig. 7B). This suggests that overall gene expression after APAP or AMAP treatment is quite similar but that the regioisomers have very different effects on genes whose expression is altered the most. Significant differences (|fold change| > 1.5, p < 0.05) between AMAP and control existed for 236 genes on the array (Fig. 7C), 2 of which were common among all three timepoints. In comparison, the number of significant gene differences nearly doubled (538) between APAP and control treatments (Fig. 7D), 52 of which were common among all three timepoints. Taken together, this suggests that APAP alters the expression of more genes to a greater extent than AMAP.
FIG. 7.
Array analysis of TAMH cells treated with APAP or AMAP compared with control. TAMH cells were treated with 2mM APAP and AMAP for 2, 6, and 24 h and analyzed for genome expression using Affymetrix Mouse Gene 1.0 ST arrays (n = 3). Heatmaps were generated using two-dimensional hierarchical clustering for genes that either (A) underwent significant changes in expression in at least one of the timepoints/treatments relative to the vehicle controls (p < 0.05) or (B) underwent changes that were both significant (p < 0.05) and considered large (|fold change| > 1.5). Red genes represent upregulation, whereas green genes represent downregulation. Venn diagrams represent the commonality and exclusivity in significant (p < 0.05) and large (|fold change| > 1.5) gene expression changes for (C) AMAP and (D) APAP over all three timepoints compared with control.
MAPK-Related Gene Regulation Following Treatment with APAP or AMAP
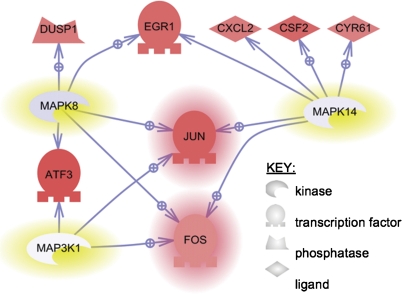
In order to determine whether early changes in MAPK protein levels corresponded with early changes in genomic expression, the 2-h AMAP and APAP microarray data sets were imported into PathwayStudio 6.2. This software tool is designed to facilitate the discovery of biological themes in microarray data. One feature of this software is to identify so called subnetworks in the data. Each subnetwork consists of a single “regulator” gene and its targets. The result of the analysis is the identification of regulators that can explain the gene expression changes observed in a given experiment. A Fisher’s exact test evaluates the statistical significance of the resulting subnetworks. The regulators that were identified for the three most significant subnetworks were MAP3K1 (MEKK1), MAPK8 (JNK1), and MAPK14 (p38-alpha) (Supplementary table 1), suggesting that MAPK-related gene expression plays a major role in differentiating the effects of AMAP/APAP toxicity. The software also allowed for investigating relationships between MEKK1, JNK1, and p38-alpha and this is shown in Figure 8. Table 2 gives quantitative fold changes and p values that correspond to the genes in Figure 8. Two-hour APAP treatment led to significant increases in expression (p < 0.01) for all genes listed in Table 2 (excluding the three kinases) compared with the 20-h AMAP treatment. Gene expression changes caused by APAP and AMAP relative to control are listed in Supplementary table 2. Of interest is the fact that c-Jun and its AP-1 dimeric partner c-Fos were the only two genes found to be related to all three regulators. Furthermore, increased c-Jun protein levels (both phosphorylated and nonphosphorylated) were previously confirmed by immunoblotting (Fig. 5).
FIG. 8.
Union of MAPK8 (JNK1), MAPK14 (p38-alpha), and MAP3K1 (MEKK1) subnetworks using PathwayStudio. Subnetworks were enriched with gene expression data considered significant (p < 0.01) and large (|fold change| > 1.5) between 2-h APAP and AMAP treatments. The three most significant subnetworks were combined in order to identify shared genes. Regulators of the three most significant subnetworks are highlighted in yellow and genes common to all three regulators are highlighted in red. Genes shaded in red represent an upregulation in expression, whereas blue represents downregulation. Relationships between regulators and target genes are indicated by arrows and are supported by more than two references in the literature; a standard arrow indicates unknown regulation, whereas a standard arrow overlaid with a plus sign indicates positive regulation.
TABLE 2.
Comparison of APAP and AMAP Gene Expression Data from the Most Significant Subnetworks after 2-h Treatment
| Name (common alias) | APAP2_AMAP2 (fold change) | APAP2_AMAP2 (p value) | Affy ID |
| MAP3K1 (MEKK1) | 0.99 | 9.35E-01 | 10412100 |
| MAPK8 (JNK1) | 0.93 | 2.43E-01 | 10418879 |
| MAPK14 (p38-alpha) | 0.98 | 7.03E-01 | 10443391 |
| JUN (c-Jun)a | 2.27 | 4.24E-05 | 10514466 |
| FOS (c-Fos)a | 1.61 | 1.77E-05 | 10397346 |
| EGR1b | 2.08 | 2.51E-06 | 10454782 |
| ATF3b | 2.54 | 4.01E-05 | 10361091 |
| DUSP1 (MKP-1)c | 1.80 | 3.29E-04 | 10449284 |
| CXCL2c | 1.78 | 1.66E-03 | 10523151 |
| CSF2c | 2.13 | 5.62E-03 | 10385912 |
| CYR61 (CCN1)c | 1.73 | 6.99E-03 | 10502655 |
Target shared by all three of top three regulators.
Target shared by two of top three regulators.
Target linked to one of top three regulators.
It is worth noting that while c-Jun upregulation occurred following treatment with either regioisomer compared with control, APAP-induced c-Jun upregulation was significantly greater in magnitude (p < 0.0001) at all three timepoints compared with AMAP (1.18, 1.13, and 1.43 at 2, 6, and 24 h, respectively). The fact that APAP-induced gene expression of MEKK1, JNK1, and p38-alpha is relatively unchanged compared with AMAP, whereas gene expression of their subnetworks is significantly upregulated (Fig. 8), suggests that genomic regulation of MAPK targets, and not genomic regulation of the MAPKs themselves, plays an important role along with MAPK-related posttranslational modifications in differentiating the effects of APAP and AMAP treatment.
DISCUSSION
The study of model compounds, like APAP, helps to lay groundwork for understanding how xenobiotics affect biological systems mechanistically. Despite an extensive body of existing literature, much of the mechanistic toxicology of APAP is still poorly understood (Jaeschke and Bajt, 2006; James et al., 2003; Nelson and Bruschi, 2007). By comparing APAP with its nontoxic regioisomer, AMAP, it may be possible to reveal certain routes of toxicity that differ between the two drugs.
The first step in doing this was to confirm a cell-based system that behaved in a similar way to in vivo systems after APAP and AMAP exposure. Previously, we have shown that the TAMH line is both susceptible to APAP-induced toxicity and relatively resistant to AMAP-induced toxicity (Pierce et al., 2002). These results were confirmed by treating TAMH cells with APAP and AMAP over a number of drug concentrations ranging from 1 to 5mM (Fig. 1). Toxicity at this concentration of APAP is of interest because similar plasma concentrations have been reported in APAP overdose patients (Smith et al., 2008).
Once significant differences in toxicity were observed between APAP and AMAP at therapeutically relevant concentrations, time-course assays were performed to determine when toxicity was occurring. Significant (p < 0.05), albeit small, differences in toxicity were observed as early as 2 h posttreatment (Fig. 2). By 6 h, APAP-induced cell death was ∼20% greater with similar increases in ATP depletion compared with AMAP (Fig. 2). These differences in toxicity and ATP levels were also related to significant differences in the depletion of mitochondrial glutathione by the regioisomers (Table 1). Taken together, these data confirm previous reports in the literature (Tirmenstein and Nelson, 1989) that APAP treatment depletes glutathione pools in the mitochondria to a greater extent than AMAP, resulting in greater levels of cell death.
Because of the fact that APAP-induced glutathione depletion and mitochondrial dysfunction have been associated with JNK pathway activation (Hanawa et al., 2008), differences in MAPK phosphorylation profiles for the regioisomers were investigated. In order to accomplish this, the Bioplex assay was utilized to determine the extent of JNK and ERK phosphorylation. Both APAP and AMAP led to increased levels of JNK phosphorylation compared with controls with APAP-induced JNK phosphorylation occurring to a greater extent at both timepoints compared with AMAP (Figs. 3A and 4). Perhaps AMAP-induced JNK phosphorylation is not extensive enough to clear thresholds for full activation of JNK-mediated cell death processes. To test this, c-Jun phosphorylation levels were measured as a downstream marker of JNK activity (Turjanski et al., 2007). c-Jun phosphorylation was most apparent after 2-h APAP treatment (Fig. 5A), directly correlating with peak JNK phosphorylation (Figs. 3A and 4). This suggests that the greater extent of APAP-induced JNK phosphorylation may overcome thresholds leading to increased levels of c-Jun phosphorylation, whereas smaller AMAP-induced increases in JNK phosphorylation were not significant enough to extensively phosphorylate c-Jun. In addition, knockdown studies of JNK1 significantly attenuated APAP-induced toxicity to a greater extent than APAP 24 h after treatment (Fig. 6). This is consistent with reports in the literature in which inhibition of JNK has been shown to protect against APAP-induced toxicity in both in vitro (Matsumaru et al., 2003) and in vivo systems (Gunawan et al., 2006). With respect to ERK, AMAP-induced phosphorylation was greater at both 2 and 6 h compared with APAP (Fig. 3B). The dramatic increase in the AMAP-induced ERK phosphorylation ratio at 2 h may be critical for an early ERK-mediated survival response. This suggests not only that AMAP treatment increases levels of ERK phosphorylation but also that this prosurvival response occurs at an early enough timepoint to attenuate cell death.
Differential modulation of MAPK phosphorylation appeared to play a role in differentiating the toxicity between APAP and AMAP. Of additional interest was to determine whether these changes at the protein level correlated well with genomic regulation. A number of genomic experiments using both in vitro and in vivo systems have been published, identifying genes that may play significant roles influencing APAP-induced toxicity (Harrill et al., 2009; Jeong et al., 2006; Reilly et al., 2001). Furthermore, in a multicenter microarray study, researchers compared APAP- and AMAP-induced gene expression in mice, identifying possible targets that included JNK-related genes identified in this in vitro study, such as c-Jun and c-Fos (Beyer et al., 2007). Overall, significant gene changes were relatively conserved between APAP and AMAP (Fig. 7A), which was expected since APAP and AMAP are structurally alike and have been shown to form similar metabolites and have similar therapeutic effects (Nelson, 1980; Rashed et al., 1990; Roberts et al., 1990). APAP treatment led to more changes in gene expression than treatment with AMAP compared with controls (Figs. 7C and 7D). This is likely because of the fact that while both regioisomers caused changes in expression because of nontoxic aminophenol-related genomic response, only APAP treatment caused additional genomic regulation related to toxicity. Significant differences in gene expression were observed between APAP and AMAP when the microarray data set was limited to large (|fold change| > 1.5) and significant (p < 0.05) gene changes (Fig. 7B). Expectations are that within this subset of genes, changes in expression are critical in differentiating the toxicity of the regioisomers.
Because of the fact that many of the observed differences in protein regulation occurred at early timepoints, the 2-h gene expression data were analyzed using PathwayStudio to compare whether these changes in protein expression and posttranslational modifications were present at the genomic level. The data set was enriched for significant (p < 0.01) and large (|fold change| > 1.5) changes in gene expression and the most significant subnetworks were identified (Supplementary data). Two of the three most significant regulators of subnetworks differentially modulated by APAP and AMAP are the stress-activated JNK1 and p38-alpha. Furthermore, the third most significantly modulated regulator was MEKK1, a known upstream MAP3K for both JNK and p38-alpha (Davis, 2000). The only two genes associated with all three MAPKs are the heterodimeric partners, c-Jun and c-Fos, that form the AP-1 transcription factor (Curran and Franza, 1988). Both these genes were found to be significantly upregulated by APAP compared with AMAP (Fig. 8, Table 2). In the case of c-Jun, the genomic data correlate well with earlier experiments at the protein level in which greater expression of c-Jun protein was observed after APAP treatment compared with AMAP (Fig. 5B). The genomic upregulation and increased protein expression of c-Jun coupled with higher levels of p-JNK-mediated c-Jun phosphorylation (Fig. 5A) would lead to increased c-Jun transcriptional activity after APAP treatment. The fact that APAP-induced modulation of JNK-related gene expression correlates well with protein data (Figs. 3–5) suggests that this pathway is heavily implicated in differentiating the toxicity of the regioisomers.
It is apparent from cell viability data, based both on dose (Fig. 1) and on time (Fig. 2), that APAP is more toxic than AMAP in TAMH cells at concentrations therapeutically relevant to humans in APAP overdose cases. These differences in cell survival are likely because of modulation of MAPK signaling, involving extensive JNK cascade activation by APAP compared with AMAP, and modulation of ERK phosphorylation, which acts as a prosurvival signal attenuating AMAP-induced toxicity. Future work in our laboratory will focus on a more detailed investigation into the effects that AMAP (and APAP) treatment have on ERK pathway regulation. These studies should help determine whether ERK modulation is related to or independent from the mitochondrial effects and JNK activation outlined in this manuscript. In summary, these results suggest that APAP-induced modulation of the JNK cascade at two levels, posttranslational (phosphorylation of JNK and c-Jun) and genomic (upregulation of c-Jun as well as other proapoptotic MAPK-related transcription factors), leads to high rates of cell death accompanied by depletion of glutathione, whereas the same dose of AMAP is not sufficient to activate these pathways.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (GM32165); University of Washington National Institute of Environmental Health Sciences sponsored Center for Ecogenetics and Environmental Health (P30ES07033); Amgen Corp.
Supplementary Material
Acknowledgments
We thank Dr Nelson Fausto from the University of Washington Department of Pathology for generously providing us with TAMH cells for experimentation and Erin Stamper from the University of Washington Department of Environmental and Occupational Health Sciences for her assistance in data analysis.
References
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer RP, Fry RC, Lasarev MR, McConnachie LA, Meira LB, Palmer VS, Powell CL, Ross PK, Bammler TK, Bradford BU, et al. Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol. Sci. 2007;99:326–337. doi: 10.1093/toxsci/kfm150. [DOI] [PubMed] [Google Scholar]
- Bourdi M, Korrapati MC, Chakraborty M, Yee SB, Pohl LR. Protective role of c-Jun N-terminal kinase 2 in acetaminophen-induced liver injury. Biochem. Biophys. Res. Commun. 2008;374:6–10. doi: 10.1016/j.bbrc.2008.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe KJ, Jia Y, Ho HK, Rademacher P, Bammler TK, Beyer RP, Farin FM, Woodke L, Plymate SR, Fausto N, et al. Comparison of the cytotoxicity of the nitroaromatic drug flutamide to its cyano analogue in the hepatocyte cell line TAMH: evidence for complex I inhibition and mitochondrial dysfunction using toxicogenomic screening. Chem. Res. Toxicol. 2007;20:1277–1290. doi: 10.1021/tx7001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles B, Wilson I, Wardman P, Hinson JA, Nelson SD, Ketterer B. The spontaneous and enzymatic reaction of N-acetyl-p-benzoquinonimine with glutathione: a stopped-flow kinetic study. Arch. Biochem. Biophys. 1988;264:253–260. doi: 10.1016/0003-9861(88)90592-9. [DOI] [PubMed] [Google Scholar]
- Curran T, Franza BR. Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Gunawan BK, Liu ZX, Han D, Hanawa N, Gaarde WA, Kaplowitz N. c-Jun N-terminal kinase plays a major role in murine acetaminophen hepatotoxicity. Gastroenterology. 2006;131:165–178. doi: 10.1053/j.gastro.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Hanawa N, Shinohara M, Behnam S, Gaarde WA, Han D, Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J. Biol. Chem. 2008;283:13565–13577. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AE, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme JA, Hongslo JK, Bjorge C, Nelson SD. Comparative cytotoxic effects of acetaminophen (N-acetyl-p-aminophenol), a non-hepatotoxic regioisomer acetyl-m-aminophenol and their postulated reactive hydroquinone and quinone metabolites in monolayer cultures of mouse hepatocytes. Biochem. Pharmacol. 1991;42:1137–1142. doi: 10.1016/0006-2952(91)90299-k. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, Bajt ML. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol. Sci. 2006;89:31–41. doi: 10.1093/toxsci/kfi336. [DOI] [PubMed] [Google Scholar]
- James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug. Metab. Dispos. 2003;31:1499–1506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- Jeong SY, Lim JS, Park HJ, Jo JW, Rana SV, Yoon S. Effects of acetaminophen on hepatic gene expression in mice. Physiol. Chem. Phys. Med. NMR. 2006;38:77–83. [PubMed] [Google Scholar]
- Johnson GL, Dohlman HG, Graves LM. MAPK kinase kinases (MKKKs) as a target class for small-molecule inhibition to modulate signaling networks and gene expression. Curr. Opin. Chem. Biol. 2005;9:325–331. doi: 10.1016/j.cbpa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Jollow DJ, Mitchell JR, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. II. Role of covalent binding in vivo. J. Pharmacol. Exp. Ther. 1973;187:195–202. [PubMed] [Google Scholar]
- Latchoumycandane C, Goh CW, Ong MM, Boelsterli UA. Mitochondrial protection by the JNK inhibitor leflunomide rescues mice from acetaminophen-induced liver injury. Hepatology. 2007;45:412–421. doi: 10.1002/hep.21475. [DOI] [PubMed] [Google Scholar]
- Lee WM. Acetaminophen and the U.S. acute liver failure study group: lowering the risks of hepatic failure. Hepatology. 2004;40:6–9. doi: 10.1002/hep.20293. [DOI] [PubMed] [Google Scholar]
- Matsumaru K, Ji C, Kaplowitz N. Mechanisms for sensitization to TNF-induced apoptosis by acute glutathione depletion in murine hepatocytes. Hepatology. 2003;37:1425–1434. doi: 10.1053/jhep.2003.50230. [DOI] [PubMed] [Google Scholar]
- Myers TG, Dietz EC, Anderson NL, Khairallah EA, Cohen SD, Nelson SD. A comparative study of mouse liver proteins arylated by reactive metabolites of acetaminophen and its nonhepatotoxic regioisomer, 3-hydroxyacetanilide. Chem. Res. Toxicol. 1995;8:403–413. doi: 10.1021/tx00045a012. [DOI] [PubMed] [Google Scholar]
- Nelson EB. The pharmacology and toxicology of meta-substituted acetanilide I: acute toxicity of 3-hydroxyacetanilide in mice. Res. Commun. Chem. Pathol. Pharmacol. 1980;28:447–456. [PubMed] [Google Scholar]
- Nelson SD, Bruschi SA. Mechanisms of acetaminophen-induced liver disease. In: Kaplowitz N, DeLeve LD, editors. Drug-Induced Liver Disease. New York: Informa Healthcare; 2007. pp. 353–388. [Google Scholar]
- Pierce RH, Franklin CC, Campbell JS, Tonge RP, Chen W, Fausto N, Nelson SD, Bruschi SA. Cell culture model for acetaminophen-induced hepatocyte death in vivo. Biochem. Pharmacol. 2002;64:413–424. doi: 10.1016/s0006-2952(02)01180-2. [DOI] [PubMed] [Google Scholar]
- Rashed MS, Myers TG, Nelson SD. Acetaminophen and a non-hepatotoxic regioisomer, 3'-hydroxyacetanilide, in the mouse. Drug Metab. Dispos. 1990;18:765–770. [PubMed] [Google Scholar]
- Reilly TP, Bourdi M, Brady JN, Pise-Masison CA, Radonovich MF, George JW, Pohl LR. Expression profiling of acetaminophen liver toxicity in mice using microarray technology. Biochem. Biophys. Res. Commun. 2001;282:321–328. doi: 10.1006/bbrc.2001.4576. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Price VF, Jollow DJ. Acetaminophen structure-toxicity studies: in vivo covalent binding of a nonhepatotoxic analog, 3-hydroxyacetanilide. Toxicol. Appl. Pharmacol. 1990;105:195–208. doi: 10.1016/0041-008x(90)90181-s. [DOI] [PubMed] [Google Scholar]
- Smith SW, Howland MA, Hoffman RS, Nelson LS. Acetaminophen overdose with altered acetaminophen pharmacokinetics and hepatotoxicity associated with premature cessation of intravenous N-acetylcysteine therapy. Ann. Pharmacother. 2008;42:1333–1339. doi: 10.1345/aph.1K680. [DOI] [PubMed] [Google Scholar]
- Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- Tirmenstein MA, Nelson SD. Subcellular binding and effects on calcium homeostasis produced by acetaminophen and a nonhepatotoxic regioisomer, 3-hydroxyacetanilide, in mouse liver. J. Biol Chem. 1989;264:9814–9819. [PubMed] [Google Scholar]
- Turjanski AG, Vaque JP, Gutkind JS. MAP kinases and the control of nuclear events. Oncogene. 2007;26:3240–3253. doi: 10.1038/sj.onc.1210415. [DOI] [PubMed] [Google Scholar]
- Watari N, Iwai M, Kaneniwa N. Pharmacokinetic study of the fate of acetaminophen and its conjugates in rats. J. Pharmacokinet. Biopharm. 1983;11:245–272. doi: 10.1007/BF01061867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.