Abstract
The hepatic glutathione S-transferases (Gsts) are critical phase II enzymes in protecting cellular macromolecules against electrophiles and oxidative stress. Little is known about the ontogeny of Gsts and the underlying regulatory mechanisms during liver development. Therefore, in this study, the ontogeny and the regulatory mechanisms of 19 known Gst isoforms were investigated in mouse liver from 2 days before birth to postnatal day 45. With the exception of Gstm5 and MGst2 that showed a progressive decline in postnatal messenger RNA (mRNA) expression, most other Gst isoforms showed a progressive increase in postnatal mRNA expression. Two-way hierarchical clustering revealed three distinct expression patterns of these Gsts isoforms: perinatal, adolescent, and adult enriched. The expression signatures of certain Gst isoforms showed positive association with the ontogeny of critical xenobiotic-sensing transcription factors, including aryl hydrocarbon receptor, pregnane X receptor (PXR), constitutive androstane receptor, peroxisome proliferator–activated receptor α, and NF-E2–related factor-2. Specifically, genome-wide chromatin immunoprecipitation coupled with the next generation sequencing technology (ChIP-Seq) revealed direct PXR-binding sites to the Gsta, Gstm, Gstt, and Gstp polycistron clusters as well as to the Mgst1 gene locus. Chromatin immunoprecipitation-on-chip analysis demonstrated that DNA methylation and histone H3K27-trimethylation (H3K27me3), two-gene expression-suppressing epigenetic marks, were consistently low around the Gstz1 gene locus. In contrast, enrichment of histone H3K4-dimethylation (H3K4me2), a hallmark for gene activation, increased 60% around the Gstz1 gene locus from prenatal to the young adult period. Regression analysis revealed a strong correlation between the enrichment of H3K4me2 and Gstz1 mRNA expression (r = 0.76). In conclusion, this study characterized three distinct ontogenic expression signatures of the 19 Gst isoforms and examined some genetic and epigenetic mechanisms inducing their transcription during liver development.
Keywords: GST, ontogeny, PXR, ChIP-Seq, epigenetics, histone methylation
The glutathione S-transferases (Gsts) are thought to play important roles in protecting macromolecules against electrophiles and products of oxidative stress, thus providing an efficient detoxification mechanism. The ability of Gst(s) to metabolize cancer chemotherapeutic drugs, insecticides, herbicides, and carcinogens suggests that their expression can influence the efficacy and detoxification capacity of drugs as well as an individual’s susceptibility to cancer (Board et al., 1997; Hayes and Pulford, 1995). Previous studies in this laboratory have examined the tissue distribution and chemical induction of various Gst isoforms in adult mice (Knight et al., 2007, 2008). For example, multiple Gsts are enriched in adult mouse liver (Gsta3, k1, m1, m4, m6, p1/2, t1, z1, and Mgst1). In addition, many hepatic Gst messenger RNAs (mRNAs) are inducible by ligands of critical xenobiotic-sensing transcription factors. For example, Gstm1 by the aryl hydrocarbon receptor (AhR); Gsta1/2, m1, m2, m3, m4, and t1 by the constitutive androstane receptor (CAR); Gsta1/2, m1, m2, m3, m4, m5, m6, and MGst1 by the pregnane X receptor (PXR); Gstk1, m5, t1, t2, z1, MGst1, and MGst3 by the peroxisome proliferator–activated receptor α (PPARα); and Gsta1/2, a4, m1, m2, m3, m4, m6, o1, t1, MGst1, and MGst3 by the NF-E2–related factor-2 (Nrf2) (Knight et al., 2008). In addition, it has been shown that the mouse hepatic Gstp1 and p2 gene expression was induced in an Nrf2-dependent manner (Satoh et al., 2002; Yeager et al., 2009).
Maturation of the drug-metabolizing capacity of the liver is essential during liver development to protect the child from environmental toxicants that they may be exposed to. It is known that developing embryos, fetuses, and newborns all face challenges from the environment that are different from that of adults. For example, in utero growth and development of eutherian mammals require flow of nutrients from the mother through the placenta. But after birth, the newborn is gradually exposed to various xenobiotics through food and drink, and the expression of drug-metabolizing enzymes and transporters is not the same between newborns and adults.
It is becoming increasingly evident that gene expression during development is also tightly regulated by epigenetic mechanisms, such as DNA methylation and histone modifications (Jaenisch and Bird, 2003; Kiefer, 2007). In general, changes in DNA methylation profiles and histone code determine whether there is a permissive chromatin state for the transcription machinery to access gene promoter regions and initiate transcription. DNA methylation is a covalent modification resulting in stable gene silencing (Bird, 2002; Reik, 2007). Histone modifications such as histone H3 lysine-4 dimethylation (H3K4me2) is present in promoters and transcribed regions of many active genes, and is positively associated with gene transcription (Bernstein et al., 2005; Kim et al., 2005; Roh et al., 2006), whereas H3 lysine-27 trimethylation (H3K27me3) is usually associated with suppression of gene transcription because H3K27me3 is a target for the chromodomain protein Polycomb, which silences genes by yet unknown mechanisms (Boyer et al., 2006; Kiefer, 2007; Lee et al., 2006). The epigenetic regulation of the Gst ontogenic expression by DNA and histone modifications has not been investigated; thus, such studies are needed to fill the critical knowledge gap in understanding the epigenetic mechanisms underlying the maturation of drug-metabolizing capacity of the developing liver.
Differential expression of cytochrome p-450s and xenobiotic transporters during developmental stages has been reported (Buist et al., 2002; Gonzalez et al., 1986; Hakkola et al., 1998; Li et al., 2002; Omiecinski et al., 1990; Pineau et al., 1991; Slitt et al., 2002). However, little is known about the ontogenic expression of various Gst isoforms and its genetic and epigenetic regulatory mechanisms. Therefore, the purposes of this study were to characterize the ontogenic expression of 19 known Gst isoforms in mouse liver and to determine the genetic and epigenetic mechanisms for the ontogeny of Gsts. For the genetic regulation of Gst ontogeny, direct PXR-binding signatures to all the Gst gene loci were characterized by chromatin immunoprecipitation coupled with the next generation sequencing technology (ChIP-Seq), a recently developed high-throughput technique to identify genome-wide transcription factor–binding sites. The choice of PXR is dictated by the fact that it is instrumental in the regulation of many important genes associated with drug metabolism and transport. For the epigenetic regulation of Gst gene expression, Gstz1, the Gst isoform most highly expressed in liver (Knight et al., 2007) was used as a model for investigating the association between its developmental expression and specific epigenetic signatures of gene expression, namely DNA methylation and histone modifications (H3K4me2 and H3K27me3).
MATERIALS AND METHODS
Reagents.
Anti-PXR antibody (sc-25381) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibody against 5-methyl-cytosine (ab51552) was purchased from Abcam (Cambridge, MA), and polyclonal antibody against methylated H3K4 and H3K27 was purchased from Millipore Upstate (Billerica, MA).
Animals.
Male and female C57BL/6 mice (Charles River Laboratories, Inc., Wilmington, MA) were housed according to the American Animal Association Laboratory Animal Care guidelines and were bred under standard conditions at the University of Kansas Medical Center in an environmentally controlled room with a 12-h light/dark cycle and allowed free access to feed and water. All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee at University of Kansas Medical Center. To obtain newborn mice, breeding cages were set up between 5 P.M. and 6 P.M. Each breeding cage contained two female mice and one male mouse. The following morning, vaginal plugs were checked and mice with vaginal plugs were placed in separate cages. Liver samples were collected from mice at birth and after birth at different ages, day 0 (birth), 5, 10, 15, 22, 30, and 45 (n = 12 per age), and also from fetuses that were obtained 2 days before birth, i.e., on gestation day 17 (designated throughout the text as −2, i.e., prenatal day −2). The livers of pups from each age were randomly sampled from different litters to obtain six males and six females per age. Livers from male and female were not conducted separately at day −2 of age, whereas after birth, genders were distinguished by visual inspection of the genital area and livers from male and female pups were collected separately. All tissues were snap-frozen in liquid nitrogen and stored at −80°C until use.
RNA extraction.
Total RNA was extracted using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX) as per the manufacturer’s instructions. The entire liver of the fetal mice was used to achieve the desired amount for RNA isolation. At older ages (after day 10 of age), about 50 mg of liver was used for RNA isolation. RNA concentrations were determined spectrophotometrically at A260, and the integrity of RNA was determined by gel electrophoresis.
Branched DNA signal amplification assay.
The mRNA expression of all the Gst isoforms was determined by the single-plex branched DNA (bDNA) technology. Mouse Gst gene sequences were obtained from GenBank. Oligonucleotide probe sets were designed using Probe Designer software, version 1.0 (Bayer Diagnostics, East Walpole, MA). Because of >90% similarity, one probe set was designed to recognize both Gsta1 and Gsta2 isoforms; for the same reason, one probe set was designed to recognize both Gstp1 and Gstp2 isoforms. The sequences of various capture extender and label extender probes were presented previously (Knight et al., 2007). Each probe was designed with a Tm of ∼63°C to ensure optimal hybridization conditions. Probe sets were submitted to the National Center for Biotechnology Information for nucleotide comparison by the basic logarithmic alignment search tool (BLASTn) to ensure minimal cross-reactivity with mouse genomic sequences and expressed sequence tags.
The lyophilized oligonucleotide probe sets for Gsts were reconstituted in Tris-EDTA buffer, pH 8.0, as per the manufacturer’s instructions (Quantigene bDNA Signal Amplification Kit; Panomics/Affymetrix, Fremont, CA). Total RNA (1 μg/μl; 10 μl = 10 μg) was added to each well of a 96-well plate containing 50 μl capture hybridization buffer and 50 μl of each diluted probe set. Total RNA was allowed to hybridize overnight at 53°C in a hybridization oven. Hybridization and subsequent wash steps were carried out according to the manufacturer’s protocol. Luminescence was quantified using Quantiplex 320 bDNA luminometer, interfaced with Quantiplex Data Management Software version 5.02.
The ontogenic expression of mRNAs of the five xenobiotic-sensing transcription factors (AhR, CAR, PXR, PPARα, and Nrf2) was determined by the multiplex suspension bDNA technology (Panomics/Affymetrix). Individual bead-based oligonucleotide probe sets specific for each gene examined were developed by Panomics/Affymetrix Inc. (panel ID: 2058, www.panomics.com). Samples were analyzed using a Bio-Plex 200 System Array reader with Luminex 100 X-MAP technology; the data were acquired using a Bio-Plex Data Manager Software version 5.0 (Bio-Rad, Hercules, CA). Assays were performed according to the manufacturer’s protocol. Data are expressed as the ratio of relative light units specific to the mRNA expression and normalized to 10 μg of total RNA.
ChIP-Seq analysis.
Livers of 8-week-old C57/BL6 male mice were used for ChIP-Seq experiments. Fragments of DNA were tagged by 35-nucleotide identifiers and subjected to sequencing by the Illumina Genome Analyzer Sequencer based on the Solexa Technology (Illumina, San Diego, CA). Preprocessing of the ChIP-Seq data was performed by Genpathway (San Diego, CA). Briefly, the tags identified were mapped to the genome using Eland Software, which resulted in a list of their chromosome coordinates. Only tags that mapped uniquely and that have no more than one mismatch were retained. Because the 5′-end of the sequence tags represented the end of ChIP fragments, the tags were extended in silico using Genpathway software at their 3′-ends to a length of 110 bp, which was the average fragment length in the size-selected library. To identify the density of fragments (extended tags) along the mouse genome, the genome was divided into 32-nucleotide bins, and the number of fragments in each bin was determined and stored together in a Binary Analysis Results (BAR) file. The BAR files were then viewed in the Affymetrix Integrated Genome Browser (IGB) for PXR binding in the mouse genome. The locations of fragment density peaks, defined by chromosome number and a start and end coordinate, were termed as “intervals.” For each BAR file, intervals were calculated using the Affymetrix Tiling Analysis Software and compiled into Browser Extensible Data file. Three parameters of intervals were identified: threshold, MaxGap, and MinRun. The threshold was set at 20-fold over background signal, which is adjusted depending on the number of tags sequenced, information on positive and negative test sites, and estimation of false discovery rate as per the company’s recommendation (Genpathway). The PXR binding to Gst genes was analyzed and visualized in the IGB. In addition, because Cyp3a11 is a prototypical direct target of the PXR protein, PXR binding to the entire Cyp3a gene cluster was also determined as a positive control. MaxGap and MinRun were set at 100 bp. The exact locations of intervals along with their proximities to gene annotations and other genomic features were then determined. In addition, average and peak fragment densities within intervals were compiled.
ChIP-on-chip assay of DNA methylation and histone methylation.
Genpathway’s chromatin immunoprecipitation (ChIP)-on-chip assays using Affymetrix GeneChip Mouse Tiling 2.0R E array were used to determine the following epigenetic profiles: DNAme, H3K4me2, and H3K27me3 as described previously (Cui et al., 2009). The detection threshold value was set at 3.0-fold above the background input for DNAme, and 4.0-fold for H3K4me2 and H3K27me3, based on the calculation of false discovery rate estimated by the “negative peaks” approach as previously described (Johnson et al., 2006). The raw and processed data are stored in the Gene Expression Omnibus database with the accession number GSE14620.
Motif analysis for PXR binding.
All chromosome coordinates of the positive PXR-binding sites within ±10 kb of the Gst gene loci were retrieved from the ChIP-Seq database and submitted to the University of California Santa Cruz Genome Browser, which returned a series of DNA sequences. These ChIP-DNA sequences were then analyzed by NHR-scan software (Sandelin and Wasserman, 2004) for putative nuclear receptor–binding sites (DR-3, DR-4, ER-6, ER-8, and IR-0) as described previously (Kliewer et al., 2002; Sonoda et al., 2002). The combined probability of entering match states was set at 0.05.
Statistical analysis.
Statistical differences in Gst mRNA expression between male and female were determined using Student’s t-test, with significance set at p ≤ 0.05. The mRNA ontogeny of all the Gst isoforms, as well as AhR, CAR, PXR, PPARα, and Nrf2, was analyzed by a two-way hierarchical clustering method (JMP v. 7.0) using Ward’s minimum variance and visualized by a dendrogram. Distances between genes reflect significance of associations. Red color represents higher and blue color represents lower expression levels, respectively.
RESULTS
Three Expression Signatures of Gst mRNAs in Developing Mouse Liver
The Gsta family.
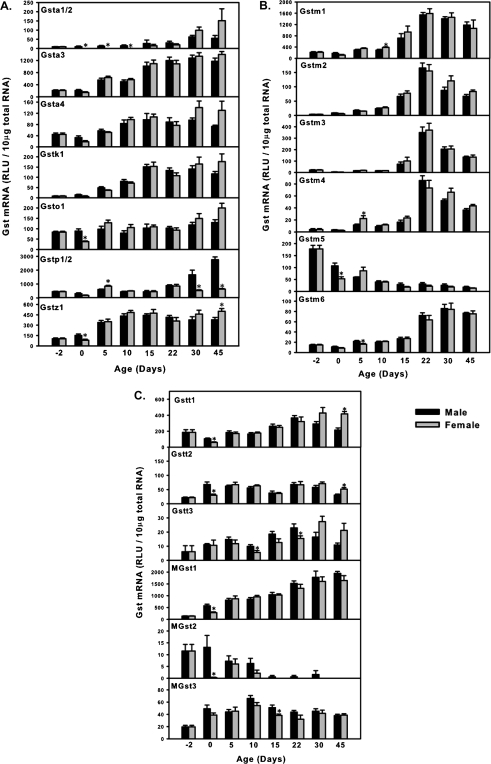
As shown in Figure 1A, the mRNAs of all four Gsta family members (Gsta1/2, a3, and a4) showed a similar expression pattern, which was low before birth but gradually increased and peaked from day 15 to 45 of age in both male and female livers. Interestingly, male-predominant expression patterns were observed for Gsta1/2 mRNA from day 0 to 10 of age, whereas the female Gsta1/2 mRNA expression was very low for the first 15 days of age.
FIG. 1.
The mRNA ontogenic expression of the 19 Gst isoforms. (A) Ontogeny of Gsta polycistron cluster (Gsta1/2, a3, and a4) as well as Gstk1, o1, Gstp1/2 cluster, and Gstz1 in male and female livers from day −2 to 45 of age. (B) The mRNA ontogenic expression of the Gstm polycistron cluster (Gstm1, m2, m3, m4, m5, and m6) in male and female livers from day −2 to 45 of age. (C) The mRNA ontogenic expression of the Gstt polycistron cluster (Gst1, t2, and t3) as well as microsomal Gsts (MGst1, MGst2, and MGst3) in male and female livers from day −2 to 45 of age. Total RNA was isolated from liver at each age and analyzed by the single-plex bDNA assay as described in “Materials and Methods” section. Data are presented as mean relative light unit ± SEM (n = 6 animals per gender, i.e., n = 12 per age). Asterisks represent significant differences (p < 0.05) between male and female mRNAs at each age.
Gstk1, o1, p1/2, and Gstz1.
The Gstk1, o1, and z1 are all isolated Gsts, which do not form clusters with other Gst isoforms, and all these three Gsts gradually increased during liver development (Fig. 1A). Gsto1 mRNA was relatively higher in males than in females at birth. Because of high homology, the bDNA probes were unable to differentiate Gstp1 and p2 in the same cluster; therefore, the expression of these two genes was combined together as “Gstp1/2.” Gstp1/2 showed a distinct male-predominant expression pattern from day 30 onwards in adult mice. The expression in females was low, and it showed little variation at all time points studied.
The Gstm family.
Gstm1, m2, m3, m4, and m6, which are all transcribed from the minus strand, displayed similar expression patterns, a gradual increase in their mRNA expression during liver development (Fig. 1B). In contrast, Gstm5, which is the only Gstm transcribed from the plus strand, i.e., in the opposite direction, showed a progressive decrease in its mRNA expression with age. Gender differences were observed for Gstm4 mRNA at day 5 (female predominant), Gstm5 at day 0 (male predominant), and Gstm6 at day 5 of age (male predominant).
The Gstt family.
Gstt1, t2, and t3 are members of the Gstt polycistron cluster on mouse chromosome 10. Interestingly, all these three genes showed a postnatal enrichment pattern (Fig. 3C). For Gstt1 and t2, male-predominant expression was observed at day 0 of age, whereas for Gstt3, male-predominant pattern was observed at day 10 and 22 of age.
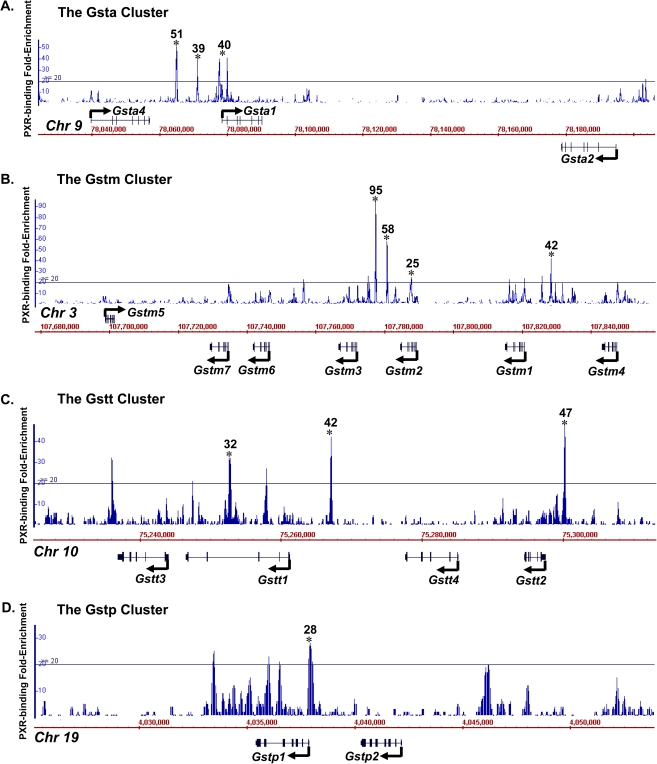
FIG. 3.
Location and fold enrichment of PXR-binding sites to various Gst polycistron clusters in adult male mouse liver by ChIP-Seq as described in “Materials and Methods” section: (A) the Gsta cluster on chromosome (chr) 9, (B) the Gstm cluster on chr 3, (C) the Gstt cluster on chr 10, and (D) the Gstp cluster on chr 19. Image was generated by the Affymetrix IGB. Line, 20-fold of background signal was used as the threshold value based on calculations of false discovery rate. Asterisks represent positive enrichment of PXR bindings at a certain genomic location.
The microsomal Gsts.
The three microsomal Gsts (MGsts) in mice are located on different chromosomes and do not form clusters with each other or other Gst isoforms (data not shown). These three MGsts showed three distinct expression patterns (Fig. 3C). Whereas MGst1 mRNA progressively increased with age in both male and female livers, MGst2 displayed a perinatal-enriched pattern in both male and female mouse livers that decreased with age, and MGst3 was highly expressed in every postnatal time point studied with no distinct expression patterns. For MGst2, a large difference in expression between male and female livers was observed in day 0, the expression in females being negligible. The regulatory mechanism for such a temporal drop in expression in females is yet to be identified.
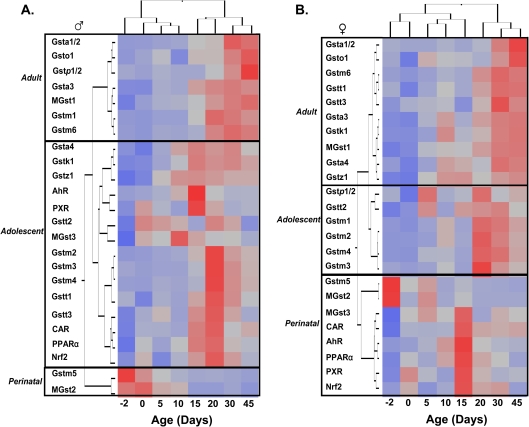
In order to perform an unbiased classification of the expression patterns of the mouse Gst isoforms, as well as to identify critical transcription factors for the developmental regulation of these Gsts in mouse liver, the mRNA ontogenic expression of the 19 Gst isoforms, as well as the mRNAs of five important xenobiotic-sensing transcription factors, namely AhR, CAR, PXR, PPARα, and Nrf2, were analyzed in developing mouse liver by a two-way hierarchical clustering method (JMP v. 7.0) and visualized as heatmaps. As shown in Figure 2A, male Gst isoforms had three distinct ontogenic patterns: (1) two perinatal-enriched Gst isoforms, Gstm5 and MGst2; (2) 10 adolescent-enriched Gst isoforms, Gsta4, k1, z1, t2, t3, m2, m3, m4, t1, and t3; and (3) seven adult-enriched Gst isoforms, Gsta1/2, o1, p1/2, a3, MGst1, m1, and m6. Interestingly, all the five transcription factors showed an adolescent-enriched pattern in developing liver of males, suggesting their potential functions in regulating the ontogeny of the Gsts in postnatal period.
FIG. 2.
Heatmaps of the mRNA ontogeny of all the Gst isoforms as well as five xenobiotic-sensing transcription factors (AhR, CAR, PXR, PPARα, and Nrf2) in male (A) and female (B) mouse liver. The ontogenic expression of these mRNAs from day −2 to 45 of age was analyzed by a two-way hierarchical clustering method (JMP v. 7.0) using Ward’s minimum variance and visualized by a dendrograph, which revealed three distinct patterns: perinatal, adolescent, and adult enriched. Distances between genes reflect significance of associations. Red color represents relative high expression, and blue color represents relative low expression.
The female-predominant Gst isoforms also showed three distinct ontogenic patterns (Fig. 2B). However, the perinatal-enriched Gsts in female livers include not only Gstm5 and MGst2 but also MGst3. Fewer Gst isoforms were adolescent-enriched in females compared with males and these include Gstp1/2, t2, m1, m2, m3, and m4. Most Gsts in females were adult enriched in adulthood in female livers, including Gsta1/2, o1, m6, t1, t3, a3, k1, MGst1, Gsta4, and Gstz1. Interestingly, all the five transcription factors showed a perinatal-enriched pattern in female livers, with only three Gst isoforms in the same category.
In summary, there were three distinct expression patterns of the mouse Gst mRNAs in developing mouse liver. Gst genes in the same polycistron clusters tended to have similar expression patterns. Most Gst isoforms were enriched postnatally in both male and female livers. However, the ontogeny of the five transcription factors only positively associated with most Gst mRNAs in male but not in female livers.
PXR-Mediated Regulation of Gst mRNA Expression in Developing Mouse Liver
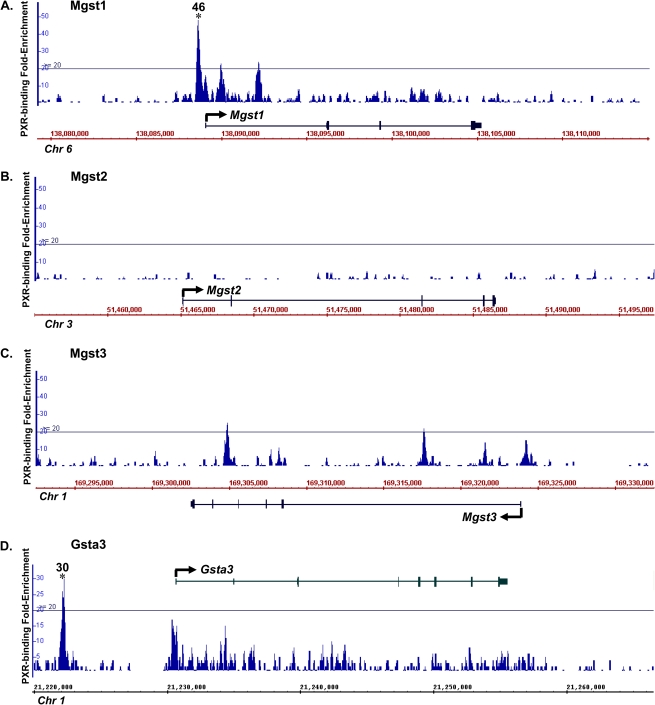
To further elucidate the mechanisms of the transcription factor–mediated ontogenic expression of Gsts, the major xenobiotic sensor PXR was selected to examine whether it directly transactivates Gst expression. Young adult mice were selected (8 weeks old) to determine the bindings of PXR to Gst genes in liver because both the PXR and the Gst genes are highly expressed at this age. Using genome-wide ChIP-Seq analysis revealed three positive PXR-binding sites within the Gsta1/a4 polycistron cluster, which were 51-, 39-, and 40-fold above background values, respectively (threshold: 20-fold) (Fig. 3A). Gsta2 is transcribed from the opposite strand and is located more than 100 kb from the closest PXR-binding site. For the Gstm cluster (Fig. 3B), one positive PXR-binding site was observed between Gstm1 and m4 (41-fold) and three sites between Gstm2 and m3 (95-, 58-, and 25-fold). To note, Gstm5 gene locus is on the boundary of the Gstm cluster, which is ∼80 kb away from the closest PXR-binding site. Multiple PXR-binding sites were also observed within the Gstt polycistron cluster (Fig. 3C), one upstream of Gstt2 (47-fold), one between Gstt4 and t1 (42-fold), and one in the intronic region of Gstt1 (32-fold). For the Gstp cluster, one positive PXR-binding site was observed at the promoter region of Gstp1, which is downstream of Gstp2 (Fig. 3D). For the Gst genes that are not part of a cluster, a PXR-binding site 418 bp upstream of the MGst1 gene locus was observed (Fig. 4A) but not in any regions within ±10 kb of the MGst2 or MGst3 gene loci (Figs. 4B–C). Interestingly, only MGst1 is highly expressed in liver, which corresponds to enriched PXR binding. In addition, we have also identified one positive PXR-binding site 8.5 kb upstream of the Gsta3 gene locus (30-fold) (Fig. 4D). There were no observed PXR bindings to Gstk1, o1, and z1 (data not shown). NHR-scan revealed 18 consensus PXR-DNA binding motifs (DR-3, DR-4, ER-6, ER-8, or IR-0) present in 9 of the 13 ChIP-DNA sequences (3 DR-3, 7 DR-4, 4 ER-6, 3 ER-8, and 1 IR-0) within ±10 kb of Gst gene loci with positive PXR bindings (Supplementary table 1).
FIG. 4.
Location and fold enrichment of PXR-binding sites to MGst1 (A), MGst2 (B), MGst3 (C), and Gsta3 (D) in adult male mouse liver by ChIP-Seq as described in “Materials and Methods” section. Image was generated by the Affymetrix IGB. Line, 20-fold of background signal was used as the threshold value based on calculations of false discovery rate. Asterisks represent positive enrichment of PXR bindings at a certain genomic location.
Because Cyp3a11 is a prototypical target gene of PXR, we analyzed the PXR bindings to the Cyp3a gene cluster as a positive control and identified 19 active regions with enriched PXR bindings in the Cyp3a gene loci (Supplementary figure 1), and specifically, two positive PXR-binding sites were identified around the Cyp3a11 gene locus, which showed a 36-fold (site 7) and 66-fold (site 8) enrichment of PXR binding over background (threshold = 20).
In summary, in addition to identifying strong associations between the postnatal-enriched PXR mRNA and multiple Gst mRNAs, this study also demonstrated that the Gsta, Gstm, Gstt, and Gstp clusters, as well as MGst1, are direct target genes of the PXR protein.
Epigenetic Aspects of Regulation of Gst mRNA Expression in Developing Mouse Liver
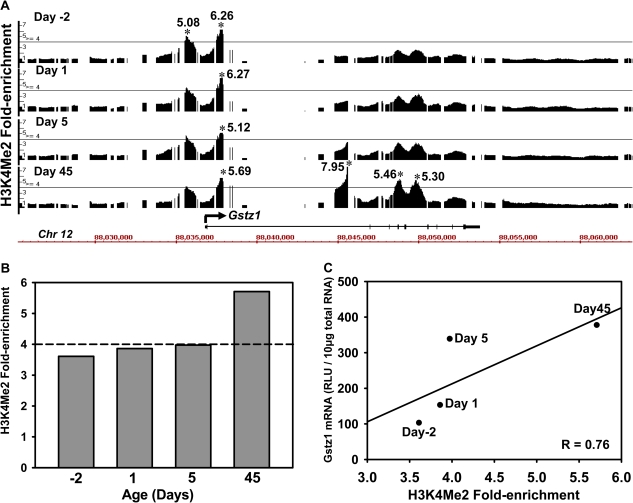
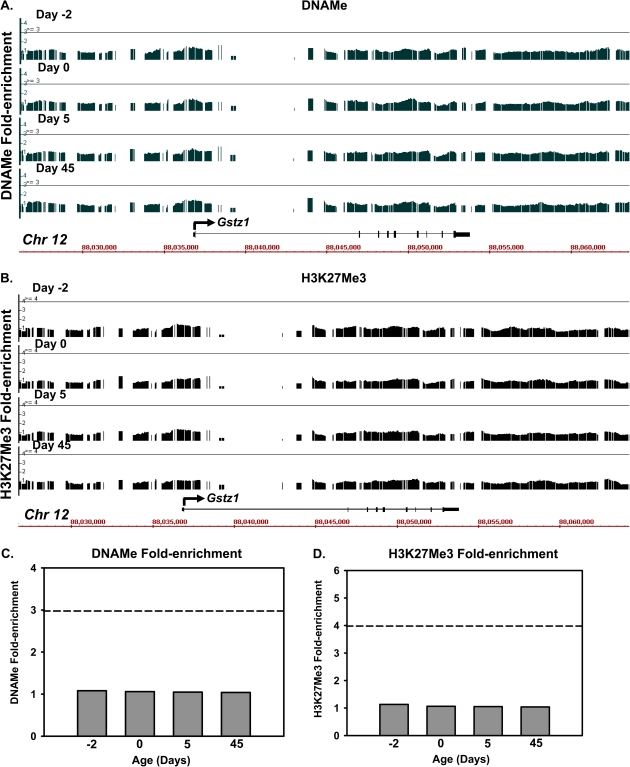
To identify whether the ontogeny is also regulated by epigenetic mechanisms, three distinct epigenetic signatures, namely DNA methylation, histone H3K4 dimethylation (H3K4me2), and histone H3K27 trimethylation (H3K27me3), were characterized by ChIP-on-chip around the Gstz1 locus, which is highly expressed in liver. Positive enrichment of the gene activation signal H3K4me2 was observed at all four selected ages (day −2, 1, 7, and 45, threshold: 4.0-fold), with more H3K4me enrichment sites at day 45 in the adult liver (Fig. 5A). The overall average H3K4me2 enrichment gradually increased during liver maturation (Fig. 5B), which was strongly associated with the postnatal-enriched pattern of Gstz1 mRNA (r = 0.76) (Fig. 5C). In contrast, there was no enrichment in the two suppression signals, DNA methylation or histone H3K27me3, within ±10 kb of the Gstz1 gene locus (Fig. 6). Together, these data indicate that epigenetic modifications, such as the absence of transcription-repressing epigenetic marks, combined with the presence of H3K4me2 (transcription-activating epigenetic mark), might play a role in facilitating a permissive chromatin state that activates Gstz1 gene transcription during mouse liver development.
FIG. 5.
Dimethylation of histone H3 at lysine-4 (H3K4me2) at the Gstz1 gene locus during mouse liver development. (A) Histone H3K4me2 fold changes at the Gstz1 gene locus at day −2, 1, 5, and 45 of age (equal amount of pooled samples from n = 5 at each age). Solid lines through the signal enrichment peaks indicate the threshold value (4.0) for enriched intervals. Bars under the peaks of each age indicate the existence and length of active regions for H3K4me2. Asterisks indicate the peak center. (B) Average peak values of H3K4me2 at day −2, 1, 5, and 45 of age. The dashed line indicates the threshold value (4.0) for enriched intervals. (C) Regression analysis of the correlation (r) between Gstz1 mRNA and the fold changes of the three epigenetic marks (DNA and histone dimethylation and trimethylation) at day −2, 1, 5, and 45 of age during liver development in mice.
FIG. 6.
(A) DNA methylation status of hepatic Gstz1 gene locus at day −2, 1, 5, and 45 of age. Solid lines through the signal enrichment peaks indicate the threshold value (3.0) for positive DNA methylation. (B) Trimethylation of histone H3 at lysine-27 (H3K27me3) of the hepatic Gstz1 gene locus at day −2, 1, 5, and 45 of age. Solid lines through the signal enrichment peaks indicate the threshold value (4.0) for enriched intervals. (C) Average peak values of DNA methylation at day −2, 1, 5, and 45 of age. The dashed line indicates the threshold value (3.0) for enriched intervals. (D) Average peak values of H3K27me3 at the Ahr gene locus at day −2, 1, 5, and day 45 of age. The dashed line indicates the threshold value (4.0) for enriched intervals.
DISCUSSION
This study is among the first to identify the age-specific mRNA expression signatures of the 19 Gst isoforms in developing mouse liver. Using recent technological advancements, such as ChIP-Seq and ChIP-on-chip, this study has provided new insights in characterizing the Gst genes that are direct targets of the critical xenobiotic sensor PXR and revealed potential epigenetic regulators for the ontogeny of Gstz1 in liver maturation. Our data suggest that the developmental regulation of Gst genes is a sequential event regulated by transcription factors and alterations of the chromatin structure. Both the genetic and the epigenetic factors may play important roles in forming the age-specific expression signatures of Gst mRNA in mouse liver.
It has been proposed that changes in expression of liver-specific proteins generally occur at three specific developmental stages: (1) late gestation, (2) at or directly after birth, or (3) just before weaning (Greengard, 1970). During these periods, the liver undergoes significant anatomical and physiological changes associated with maturation. Thus, investigation of the age-specific ontogenic expression of Gsts will help understand differences in chemical detoxification abilities between adult and neonatal livers. In the current study, hepatic Gst mRNA expression was lowest or absent at prenatal day −2 and at birth except for Gstm5 and MGst2, which showed highest expression at prenatal day −2 followed by a gradual decrease over age. Embryonic development and fetal growth depends on nutrients obtained from the mother and such exchange occurs through the placenta. Although placenta is capable of metabolism and detoxification of xenobiotics and endogenous chemicals, it does not entirely exclude or metabolize all chemicals in the mother’s blood. Consequently, many drugs and other xenobiotic or endobiotic metabolites reach fetal organs from the mother’s blood. The unique expression profiles of Gstm5 and MGst2 need further study because their expression profiles suggest that they may have special functional significance during gestation.
Our findings of lower Gst expression at birth and early postnatal period are consistent with an earlier report in rats where a low total hepatic Gst enzyme activity was observed in neonatal period but was higher in adults (Tee et al., 1992). In that study, total Gst activity was determined using CDNB (1-chloro-2,4-dinitrobenzene) as the universal Gst substrate. Perhaps the most intriguing results are the marked differences in the expression profiles of the Gsts between day 15 and 20. One can envision that these gene expression changes observed around weaning may be in response to dietary changes because mice transition from milk to chow during this period. Dietary factors have been shown to be critical regulators for liver gene expression. For example, both the mRNA and the activities of lipogenic enzymes increase in rat liver after weaning in response to a high-carbohydrate diet (Girard et al., 1994). If the hypothesis that dietary factors regulate Gst gene expression is true, it can be argued that they may serve as critical signaling molecules to initiate a cascade whereby the liver becomes more capable of detoxifying electrophilic chemicals and reactive oxygen species, highlighting the importance of diet on drug-metabolizing enzyme gene expression patterning and detoxification capacity.
Interestingly, Tee et al. (1992) reported that in rats, Gstps are abundant in 18-day fetal livers but are almost absent postnatally. In contrast, in this study in mice, postnatal hepatic Gstp1/2 expression increased between 15 and 45 days of age in male mice, whereas female Gstp1/2 expression was similar to that in male mice until 22 days of age but did not increase thereafter as in male mice. Such differences in Gstp gene ontogeny could be because of species differences and/or detection methods. For example, for species differences, it has been demonstrated that rat GST-P gene has a strong enhancer element GST-P enhancer-1 (GPE1) that specifically regulates the GST-P gene by interacting with certain transcription factors, such as CCAAT/enhancer binding protein alpha and Nrf2/v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K. In contrast, the mouse GST-P1 gene does not contain a GPE1 or related element (Sakai and Muramatsu, 2007). For differences in detection method, the study by Tee et al. in rats used immunocytochemistry and high performance liquid chromatography identification of Gst subunits, whereas the study by Raijmakers et al. (2001) in humans used Western blot to study specific isoforms. Tee et al. also used northern blot to determine the mRNA expression of Gst alpha, mu, and pi class, using longer complementary DNA probes. Obviously, the ability to distinguish all 19 isoforms using such long probes was limited. In contrast, this study reveals additional information because of the ability to investigate the expression of individual isoforms by using isoform-specific short oligonucleotide probes.
Among all the 19 Gst isoforms, the most predominant gender difference was observed in the ontogeny of the Gstp1/2 gene, which had higher levels of mRNA in livers of male than female mice at 30 and 45 days of age. Our observation for the male-predominant expression of Gstp1/2 is consistent with previous findings (Bammler et al., 1994; Knight et al., 2007; Townsend et al., 2009). Gender differences were also observed at early postnatal ages, with Gsta1/2, o1, z1, m5, and p1/2. Such neonatal gender differences suggest that distinct molecular mechanisms might also exist in regulating the gender-divergent gene expression at early ages. The gender-specific gene expression of Gsts suggests that males and females may have different capacities in metabolizing and detoxifying chemicals that are Gst substrates.
Yijia et al. (2008) studied the expression of membrane-associated proteins in eicosanoid and glutathione metabolism in an in vitro system of mouse embryonic stem cell–derived hepatic tissue. The protein expression of MGst1 was not detected until postnatal day 14 and gradually increased with the maturation of hepatic tissue. This finding is consistent with MGst1 mRNA expression observed in this study. The membrane-associated enzyme activities involved in eicosanoid and glutathione metabolism are important in both inflammation and cell protection and therefore a part of the tissue’s defense mechanism.
Certain Gst/GSTs exist as polycistron clusters in both mice and humans. For example, Gsta1 and a2 cluster together on chromosome 9 in mice and transcribe from the opposite strands, whereas Gsta1 transcribes from the plus strand and Gsta2 transcribes from the minus strand. In humans, five members of the GSTA family (GSTA1–A5) also form a polycistron cluster on chromosome 6; the human GSTA5 is homologous to both Gsta1 and a2 in mice (Supplementary figure 2). Other examples of Gst/GST family members forming polycistron clusters include Gstm/GSTM clusters on mouse chromosome 3 and human chromosome 1, with multiple homologous genes between the two species (Supplementary figure 3), as well as Gstt/GSTT and Gstp/GSTP clusters in mice and humans (Supplementary figure 4). From an evolutionary standpoint, the formation of Gst/GST polycistron clusters suggests a possible fine-tuning of gene expression through coordinate regulation, which should produce cluster-specific gene expression signatures. Therefore, the ontogenic expression signatures of the clustered Gst isoforms were investigated in this study.
Previous studies in this laboratory have shown that several Gsts were induced following pregnenolone 16α-carbonitrile administration (Knight et al., 2008), but only a few genes have been shown to be direct PXR targets. This study is among the first to identify that multiple Gst genes are direct PXR targets, including a few genes in the Gsta, Gstm, Gstt, Gstp clusters, and the microsomal MGst1. Similar ontogenic expression of genes in the same Gst polycistron clusters indicate that they share common regulatory mechanisms, likely mediated by PXR protein binding. Gstm5 is the only Gstm isoform that showed a different ontogenic pattern. This might be because of the fact that Gstm5 transcribes from the opposite strand and thus may be under separate regulatory control.
Recently, a large body of data has been generated to characterize histone modifications in the genomes of various organisms (Hawkins and Ren, 2006). A current area of research is to understand how histone marks correlate and/or regulate gene transcription. High-resolution profiling of histone methylations in the entire human genome has demonstrated that active genes are characterized by high levels of H3K4me2; in contrast, inactive genes are characterized by low or negligible levels of H3K4me2 of the promoter regions and high levels of H3K27me3 (Barski et al., 2007). It has been shown that the H3K4me2 signals are usually localized close to transcription start sites, providing a permissive chromatin environment to trigger gene transcription (Barski et al., 2007). In this study, the strong postnatal enrichment of H3K4me2 in the close vicinity of the Gstz1 gene promoter indicates that H3K4Me2 is likely a mechanism to trigger the increase in Gstz1 gene activation during mouse liver development.
In conclusion, this study identified three distinct ontogenic expression patterns among the 19 hepatic Gst isoforms in both male and female mice, characterized the occurrence of gender differences in the expression of certain Gsts during liver maturation, identified that certain Gsts are direct target genes by the xenosensor PXR, and revealed positive associations between the Gstz1 ontogeny and enrichment of the epigenetic mark H3K4Me2. By combining genome-scale investigations and Gst gene expression profiling, our study has provided novel insights in understanding the genetic and epigenetic mechanisms in regulating the maturation of drug metabolism in developing liver.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Health (ES-07078, ES-09649, ES-09716, ES-013714, and ES-021940) to C.D.K.
Supplementary Material
Acknowledgments
The authors would like to thank all the graduate students and postdoctoral fellows in Dr Curtis Klaassen’s laboratory for proofreading the manuscript. The opinions expressed in this article are the authors’ personal opinions and do not necessarily reflect those of FDA, DHHS, or the Federal Government.
References
- Bammler TK, Smith CA, Wolf R. Isolation and characterization of two mouse Pi-class glutathione S-transferase genes. Biochem. J. 1994;298:385–390. doi: 10.1042/bj2980385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Board PG, Baker RT, Chelvanayagam G, Jermiin LS. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem. J. 1997;328(Pt 3):929–935. doi: 10.1042/bj3280929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Buist SC, Cherrington NJ, Choudhuri S, Hartley DP, Klaassen CD. Gender-specific and developmental influences on the expression of rat organic anion transporters. J. Pharmacol. Exp. Ther. 2002;301:145–151. doi: 10.1124/jpet.301.1.145. [DOI] [PubMed] [Google Scholar]
- Cui YJ, Yeager RL, Zhong XB, Klaassen CD. Ontogenic expression of hepatic Ahr mRNA is associated with histone H3K4 di-methylation during mouse liver development. Toxicol. Lett. 2009;189:184–190. doi: 10.1016/j.toxlet.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J, Perdereau D, Foufelle F, Prip-Buus C, Ferre P. Regulation of lipogenic enzyme gene expression by nutrients and hormones. FASEB J. 1994;8:36–42. doi: 10.1096/fasebj.8.1.7905448. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Song BJ, Hardwick JP. Pregnenolone 16 alpha-carbonitrile-inducible P-450 gene family: gene conversion and differential regulation. Mol. Cell. Biol. 1986;6:2969–2976. doi: 10.1128/mcb.6.8.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard O. In: Biochemical Action of Hormones. Litwack G, editor. Vol. 1. New York: Academic Press; 1970. pp. 53–87. [Google Scholar]
- Hakkola J, Tanaka E, Pelkonen O. Developmental expression of cytochrome P450 enzymes in human liver. Pharmacol. Toxicol. 1998;182:209–217. doi: 10.1111/j.1600-0773.1998.tb01427.x. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Ren B. Genome-wide location analysis: insights on transcriptional regulation. Hum. Mol. Genet. 2006;15(Spec No 1):R1–R7. doi: 10.1093/hmg/ddl043. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoforms to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JC. Epigenetics in development. Dev. Dyn. 2007;236:1144–1156. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr. Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. Constitutive mRNA expression of various glutathione S-transferase isoforms in different tissues of mice. Toxicol. Sci. 2007;100:513–524. doi: 10.1093/toxsci/kfm233. [DOI] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. Induction of hepatic glutathione S-transferases in male mice by prototypes of various classes of microsomal enzyme inducers. Toxicol. Sci. 2008;106:329–338. doi: 10.1093/toxsci/kfn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Hartley DP, Cherrington NJ, Klaassen CD. Tissue expression, ontogeny, and inducibility of rat organic anion transporting polypeptide 4. J. Pharmacol. Exp. Ther. 2002;301:551–560. doi: 10.1124/jpet.301.2.551. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Hassett C, Costa P. Developmental expression and in situ localization of the phenobarbital-inducible rat hepatic mRNAs for cytochromes CYP2B1, CYP2B2, CYP2C6, and CYP3A1. Mol. Pharmacol. 1990;38:462–470. [PubMed] [Google Scholar]
- Pineau T, Daujat M, Pichard L, Girard F, Angevain J, Bonfils C, Maurel P. Developmental expression of rabbit cytochrome P450 CYP1A1, CYP1A2 and CYP3A6 genes. Effect of weaning and rifampicin. Eur. J. Biochem. 1991;197:145–153. doi: 10.1111/j.1432-1033.1991.tb15893.x. [DOI] [PubMed] [Google Scholar]
- Raijmakers MTM, Steegers EAP, Peters WHM. Glutathione-S-transferases and thiol concentrations in embryonic and early fetal tissues. Hum. Reprod. 2001;16:2445–2450. doi: 10.1093/humrep/16.11.2445. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Muramatsu M. Regulation of glutathione transferase P: a tumor marker of hepatocarcinogenesis. Biochem Biophys Res Commun. 2007;357:575–578. doi: 10.1016/j.bbrc.2007.03.174. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- Satoh K, Itoh K, Yamamoto M, Tanaka M, Hayakari M, Ookawa K, Yamazaki T, Sato T, Tsuchida S, Hatayama I. Nrf2 transactivator-independent GSTP1-1 expression in “GSTP1-1 positive” single cells inducible in female mouse liver by DEN: a preneoplastic character of possible initiated cells. Carcinogenesis. 2002;23:457–462. doi: 10.1093/carcin/23.3.457. [DOI] [PubMed] [Google Scholar]
- Slitt AL, Cherrington NJ, Hartley DP, Leazer TM, Klaassen CD. Tissue distribution and renal developmental changes in rat organic cation transporter mRNA levels. Drug Metab. Dispos. 2002;30:212–219. doi: 10.1124/dmd.30.2.212. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR) Proc. Natl. Acad. Sci. U.S.A. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee LB, Gilmore KS, Meyer DJ, Ketterer B, Yves Vandenberghe Y, Yeoh GC. Expression of glutathione S-transferase during rat liver development. Biochem. J. 1992;282:209–218. doi: 10.1042/bj2820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM, Tew KD, He L, King JB, Hanigan MH. Role of glutathione S-transferase Pi in cisplatin-induced nephrotoxicity. Biomed. Pharmacother. 2009;63:79–85. doi: 10.1016/j.biopha.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol. Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yijia L, Danyan Z, Yue D, Meiyuan G, Xin H, Kuifen M, Yu Y. MAPEG expression in mouse embryonic stem cell-derived hepatic tissue system. Stem Cells Dev. 2008;17:775–784. doi: 10.1089/scd.2007.0241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.