Abstract
In Drosophila, neurons of the central nervous system are grouped into units called lineages. Each lineage contains cells derived from a single neuroblast. Due to its clonal nature, the Drosophila brain is a valuable model system to study neuron development and circuit formation. To better understand the mechanisms underlying brain development, genetic manipulation tools can be utilized within lineages to visualize, knock down, or over-express proteins. Here, we will introduce the formation and development of lineages, discuss how one can utilize this model system, offer a comprehensive list of known lineages and their respective markers, and then briefly review studies that have utilized Drosophila neural lineages with a look at how this model system can benefit future endeavors.
Keywords: Drosophila, Brain, Lineage, Neuroblast, Axon
Introduction
In the late 1800s, Cajal’s Golgi stains provided the first evidence that the primary functional unit of the nervous system is the neuron (Cajal 1899). Since these initial observations, neuroanatomists and developmental neurobiologists have asked the question: how do neurons arrange themselves into a functional circuit?
The advent of imaging methods such as functional magnetic resonance imaging or diffusion tensor imaging (reviewed in Huppi and Dubois 2006) has paved the way for dramatic progress in understanding the functional centers of the human brain. Research teams are now beginning to decipher how individual processing hubs form a neural network that transmits sensory information to areas that dictate behavioral output. One elegant example of progress in this area is the association of frontal cortico-basal ganglia macrocircuitry with the complex process of decision making and risk assessment (for a thorough review, refer to Fareri et al. 2008). Despite the growing number of reports linking brain centers to behavior, the precise neuronal circuits and the genetic mechanisms putting these circuits together during development are still largely unknown.
Because genes govern the position, morphology, and activity of neurons, one might assume a strong genetic contribution to the establishment of neuronal networks. To address the genetic control of neural circuitry, a system that allows genetic manipulation in a manageable number of neurons is advantageous. The Drosophila brain offers a unique model in which sibling neurons of a parent neuroblast are bundled together in groups of clonal units, called lineages. By combining many neurons into a single larger unit, Drosophila lineages give the resolution necessary to study key aspects of neuron development, such as branch formation, axon guidance, and circuit formation, in an in vivo setting. This is in contrast to the mammalian brain in which ample lineage information is unavailable. Bota and Swanson review a potential online tool, the Brain Knowledge Management System, that aims to piece together a neuron classification system for the mammalian brain (Bota and Swanson 2007); however, it is still unclear if there exists a set of mammalian neurons that organize into distinct units based upon lineage restrictions.
The observation that a neuroblast’s progeny coalesce into a distinct unit was first recognized in the Drosophila mushroom body (Ito et al. 1997). This key realization was preceded by a long line of investigation into insect neurogenesis. Some of the earliest studies on the insect nervous system characterized the stereotypical neuroblasts of the segmental ganglia in grasshopper, locust, and tobacco hornworm (Bate 1976; Booker and Truman 1987). Even the more primitive, wingless insect silverfish presents a highly conserved number and position of neuroblasts in the thoracic ganglia (Truman and Ball 1998).
Almost 20 years after the neuroblasts of the grasshopper segmental ganglia were characterized, Zacharias et al. (1993) questioned if the major parts of the grasshopper brain were organized in a similar manner. Indeed, Zacharias found 130 neuroblasts that were arranged in a stereotyped pattern in the protocerebrum, deuterocerebrum, and tritocerebrum of the grasshopper brain (Zacharias et al. 1993).
These reports highlighted the similarity of nervous system organization among insect species and paved the way for genetic analysis of neurogenesis in the genetically tractable model, Drosophila. Like other insects, Drosophila have a stereotyped neuroblast deposition in the segmental ganglia and brain (Doe 1992; Younossi-Hartenstein et al. 1996; Urbach and Technau 2003), but it was not until clonal technology allowed direct visualization of the bundled progeny of a single neuroblast that the significance of the neural lineage as a unit was recongnized (Ito et al. 1997).
The Drosophila central brain contains a stereotyped pattern of approximately 100 lineages (a relatively small number compared to the estimated 100 billion neurons of the human brain), allowing for the creation of maps detailing tract projections, compartment boundaries, and ultimately circuit formation. Most studies in the Drosophila brain have so far utilized only a small number of available lineages, in particular the four identical lineages forming the mushroom body (Ito et al. 1997), and more recently some of the antennal lobe lineages (Lai et al. 2008), limiting the spectrum of information that can be obtained from this versatile model. This is due mostly to the fact that until recently, little information was available for the majority of brain.
In the following review, we will (1) discuss how Drosophila lineages develop, (2) describe how one can visualize and manipulate lineages in an in vivo setting, and (3) briefly introduce lineages that are already in use as model systems to study neuron development.
Lineages are clonal units
The Drosophila brain begins as a cluster of approximately one hundred neuroblasts that appear at embryonic stage 9-11 (Younossi-Hartenstein et al. 1996; Urbach and Technau 2003). During embryonic stages, neuroblasts divide in a stem cell-like manner to produce two hemispheres of roughly 1,500 primary neurons each. The majority of neuroblast divisions generate a ganglion mother cell (GMC) and another neuroblast. The GMC then divides again to produce two neurons. In specific cases, the neuroblast also generates a glial cell or a transient-amplifying GMC (Boone and Doe 2008); however, these additional activities are beyond the scope of this review.
Over the course of embryonic development, the primary neurons project primary axon tracts (PAT) into the center of the brain (Nassif et al. 1998; Younossi-Hartenstein et al. 2006; Larsen et al. 2009), creating a central neuropile surrounded by a cortex of cell bodies. The clonal unit containing a mother neuroblast and its embryonically produced progeny is referred to as a “primary lineage.” After a brief stage of quiescence, the same neuroblasts again divide during larval stages to produce a new set of secondary neurons (“secondary lineage”). The secondary neurons emit secondary axon tracts (SATs) along the existing PATs. By the end of larval development, the full lineage consists of primary neurons, secondary neurons, and the mother neuroblast (Fig. 1, left panel). Lineages mature during metamorphosis and persist in the adult brain.
Fig. 1.
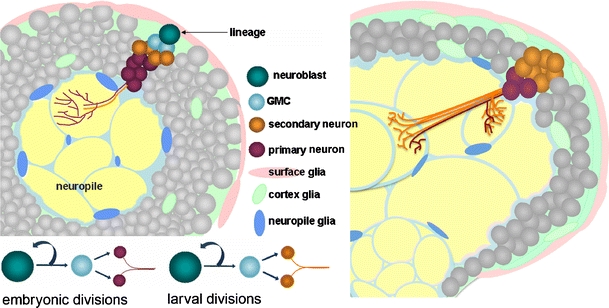
Lineage development from larva to adult. Cartoon diagram highlighting a single neural lineage in the third instar brain (left) and in the adult brain (right). Note the decreased contribution of primary neurons in the adult brain compared to the third instar brain. GMC ganglion mother cell
Lineages as a primary functional unit
Each lineage undergoes a series of changes between the late embryonic and adult stage. By embryonic stage 16, short neurites have emerged from the neurons of the primary lineages to form the initial connectivity of a central neuropile (Younossi-Hartenstein et al. 2006; Larsen et al. 2009). Most lineages project into a defined region of the neuropile, producing the initial compartments of the larval neuropile that are later surrounded by glia (Younossi-Hartenstein et al. 2003).
Larsen et al. (2009) have followed the lineages into later stages and have found that the spatial relationship of primary and secondary neurons within a lineage is dictated by the initial projection of the PAT. The primary lineages establish the initial neuropile compartments, and the secondary lineages then use these primary scaffolds (Larsen et al. 2009), as well as glial boundaries (Spindler et al. 2009), in their own arborization patterns during pupal stages. Hence, the secondary neurons follow the primary neuron schematic to generate neural circuits for the adult behaviors.
Lineage branching patterns
Unlike the bipolar neurons of the mammalian brain, Drosophila neurons are unipolar and contain a long neurite (conventionally called “axon”) that emits collateral branches (Fig. 2a). All primary and secondary neurons in a given lineage tend to abide by a specific pattern for that lineage (Larsen et al. 2009). The various branching patterns of lineages can be subdivided into three types: type PD lineages have distinct proximal arbors and distal terminal arbors, type C lineages have continuous branch patterns along the entire axon, and type D lineages have only distal terminal branching (Larsen et al. 2009; Fig. 2b).
Fig. 2.

Drosophila lineage properties. Cartoon diagram contrasting a bipolar vertebrate neuron (a) and a unipolar Drosophila neuron (b). Dendritic arbors are shown in orange and axonal terminals are colored green. Dendrites are segregated from axons in neurons with proximal-distal branching (PD), while pre-synaptic and post-synaptic branches are mixed in continuous (c) and distal (d) branching patterns. mcd8GFP expression (c, e) versus proximal localization of the dendritic marker DSCAM1.1:GFP (d, f) in adult preparations of two lineages: BLD5 visualized with atonal-GAL4 (c, d) and DALv2 visualized with EB1-GAL4 (e, f). Axonal visualization, or lack thereof, is labeled with a white arrow
While the embryonic-born primary lineages establish a general schematic for the secondary neuron siblings, the primary neurons have a generally wider and more dispersed branching pattern than the secondary axon tracts (Fig. 3). In the brain, arbors of single dopaminergic primary neurons innervate much of the brain, while the secondary neurons from the parent lineage retain a more selective innervation pattern (L. Cruz, unpublished data). The mechanism, however, by which secondary neurons focus their innervation pattern, is currently unknown.
Fig. 3.
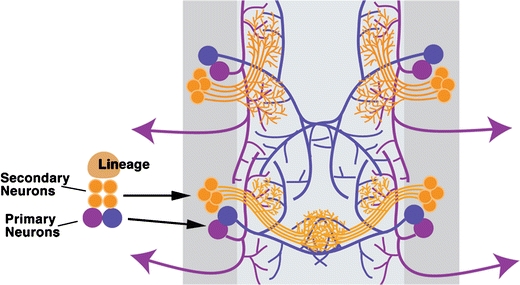
Secondary versus primary arbor patterning. A schematic diagram representing the widely dispersed projections of the embryonic-born primary neurons (blue/purple) versus the localized branching of larval-born secondary neurons (orange) in the ventral nerve segments
Currently, the definition of primary versus secondary neuron is based on embryonic versus larval birth date. Recent evidence suggests that there is a special population of late embryonic-born neurons that remain arrested in an undifferentiated state until larval stages (Zhou et al. 2009). Because the temporal differentiation of these late-embryonic born neurons is in parallel with the larval born neurons, perhaps a more appropriate basis for the classification of primary versus secondary exists in the timing of differentiation rather than birth stage.
Regardless of classification, the branching pattern of both primary and secondary neurons establishes the flow of information through tightly positioned pre-synaptic and post-synaptic inputs. While it is generally thought that proximal arbors are dendritic the exact distinction of pre-synaptic vs. post-synaptic domains of Drosophila lineages of each type is unclear (for a review of Drosophila synapse biology, please refer to (Prokop and Meinertzhagen 2006). Due to the presence of lineages with only one area of arborization, it is likely that both pre- and post-synaptic terminals co-exist along a shared axon tract in these lineages. In the Musca domestica, for example, detailed descriptions of mixed pre- and post-synaptic terminals in both proximal and terminal arbors have already been described (Strausfeld 1976).
Using type PD lineages, markers for Drosophila dendritic versus axonal terminals have been identified such as DSCAM containing exon 17.1 (Wang et al. 2004) and nodlacZ (Clark et al. 1997; Wang et al. 2004). The localization of tau-lacZ (Callahan and Thomas 1994) and nSyb (Srahna et al. 2006) labels axons. In each case, the dendritic markers tend to localize to the proximal arborizations, and the axonal markers are segregated into the distal domain. Shown here, the adult DALv2 (Fig. 2c,d) and the pupal BLD5 (Fig. 2e,f) lineages localize the dendritic marker DSCAM17.1:GFP to the proximal domain of the SAT as compared to uniform distribution of mcd8GFP into both the proximal and distal domains. Whether or not a distinct dendritic versus axonal domain exists for all lineage types has not been addressed. Further studies defining the pre/post-synaptic domains within each lineage using the above markers should help clarify this ambiguity.
Visualization and manipulation of lineages in vivo
In previous studies, lineages were labeled with antibodies against proteins that segregate to specialized regions of the neuron, such as synapses or adhesion sites (Nassif et al. 1998; Dumstrei et al. 2003a; Nassif et al. 2003; Pereanu and Hartenstein 2006). For example, N-cadherin (a cell-adhesion molecule) labels all axon tracts from embryonic stages onward; in addition, this molecule is concentrated in synapses, and therefore serves as a suitable marker for neuropile compartments at larval, pupal, and adult stages. Antibodies against fasciclin II and III or against Futsch (Hybridoma 22C10) label-specific axon bundles. Neurotactin (a surface glycoprotein) is transiently expressed in all neurons. It labels primary axons and neuropile of embryos; in the early larva, it is downregulated in primary neurons and reappears in secondary neurons. During mid-to-late larval periods, as well as in the early pupa, anti-neurotactin is the best marker for secondary axon tracts. In the larval, pupal, and adult brains, antibodies against the cell-surface trans-membrane protein neuroglian mark the long axon tracts that connect neuropile compartments.
While markers that label all lineages are convenient for mapping and gaining an overall idea about lineage position in the brain, it is difficult to study a single lineage in detail using immunostaining alone. To highlight individual lineages, one can utilize the GAL4/upstream activating sequence (UAS) system (Brand and Perrimon 1993) as well as genetic techniques such as flip-out clones (Harrison and Perrimon 1993) and mosaic analysis with a repressible cell marker (MARCM; Lee and Luo 2001). During late embryogenesis, primary neural clusters develop unique patterns of gene expression. Sprecher and colleagues (2007) have mapped 18 key developmental control genes in 72 identified lineages to produce a genetic identity for each lineage based on the combination of genes expressed (Sprecher et al. 2007). Based on this premise, promoters or putative enhancer fragments in the 5′, introns, or 3′ regions of gene X can be inserted before a GAL4 transgene and crossed to a line carrying a UAS-reporter construct, such as GFP or lacZ. Additionally, BACs and knock-ins are sufficient to generate lines that are active in only a small set of neurons (Venken et al. 2006; Kurtovic et al. 2007). A list is presented in Table 1 with publicly available GAL4 lines exhibiting a lineage-specific pattern. The number of available lines is sure to expand based on a large collection of GAL4 constructs currently being screened for neuron and lineage specific patterns (Pfeiffer et al. 2008).
Table 1.
Lineage trajectories, type, and known markers
| Family | 3 L 1o Proximal projections | 3 L 1o Distal projections | 3 L 2o Terminations | Adult 2o proximal projection | Adult 2o distal projection | Branch type | Marker |
|---|---|---|---|---|---|---|---|
| BAmv1 | BPM | LAL | FSB, SMP | Type PD | Period-GAL4 | ||
| BAmv3 | AL | calyx, LH | Type D | GH146-GAL4, sine-oculis-GAL4 | |||
| BAla1 | BA | CPL | CPL | AL | LH | Type PD | Period-GAL4 |
| BAla3 | BPM | BPM | LMC, VLP | Engrailed-GAL4 | |||
| BAlc | BA, iACT | AL | calyx, LH | Type PD | GH146-GAL4 | ||
| BAlp 1-3 | BPM, BLP | Period-GAL4 | |||||
| DALcl1 | OT | LT | Type PD | STAT92E10XGFP* | |||
| DALv2 | BC | CPM | LT, IMP, LAL | EB | Type PD | Engrailed-GAL4, engrailed-lacZ | |
| DALv3 | CPM | LT | EB | Type PD | Period-GAL4, EB1-GAL4* | ||
| DPLam | CPL, CPI, DA | CPL | Type C | Engrailed-GAL4 | |||
| DPLl | CPLd | Sine-oculis-GAL4 | |||||
| BLD5 | IOL | COL | Type PD | Atonal-GAL4 | |||
| BLVa1 | CPLd | Sine-oculis-GAL4 | |||||
| MB | Calyx | MB medial and dorsal lobe | MB medial and dorsal lobe | Calyx | MB medial and dorsal lobe | Type PD | OK107-GAL4, GH146-GAL4, 201Y-GAL4 |
Type PD lineages have distinct proximal arbors and distal terminal arbors. Type C lineages have continuous branch patterns along the entire axon. Type D lineages have only distal terminal branching (Larsen et al. 2009)
BA baso-anterior, BC baso-central, BPM baso-posterior-medial, CPL(d) centro-posterior-lateral (dorsal), CPI centro-posterior-intermediate, DA dorso-anterior, MB mushroom body, iACT inner antennal cerebral tract, BLP baso-lateral-posterior, CPM centro-posterior-medial, LAL lateral accessory lobe, EB ellipsoid body, AL antennal lobe, COL contralateral optic lobe, IOL ipsilateral optic lobe, OT optic tubercle, LT lateral triangle, IMP inferior-medial-protocerebrum, FSB fan-shaped body, SMP superior-medial protocerebrum, LH lateral horn, VMC ventro-medial cerebrum, VLP ventro-lateral protocerebrum
In most cases where Gal4 drivers show any association with lineages (rather than complex and essentially uninterpretable populations of scattered neurons), more than one lineage is labeled (see Table 1). In these cases, flip-out clones are employed to visualize only a single lineage. Flip-out clones utilize a flippase recognition target (FRT)-flanked termination sequence that is incorporated at the interface between the target promoter and the GAL4 transgene, inhibiting the expression of GAL4 protein. Upon the induction of flipase protein, via heat shock-driven expression, FRT site recombination removes the termination sequence. Subsequently, GAL4 protein is expressed and a visualization marker can be driven under control of an upstream-activating sequence (Fig. 4). Only neuroblasts that successfully remove the termination sequence generate a labeled lineage. With a specifically timed heat shock (taking into account developmental stage and length of heat shock), clones can be limited to only one lineage of the brain.
Fig. 4.
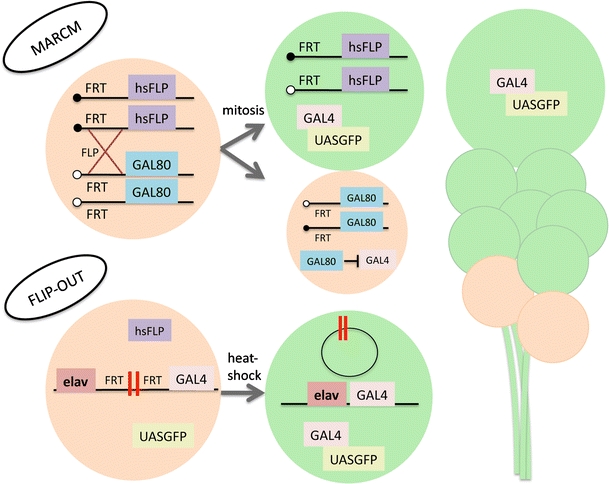
Lineage-based clonal analysis. Cartoon diagram explaining the genetic technique of mosaic analysis with a repressible cell marker (MARCM; top) and flip-out induced clones (bottom). MARCM clones are generated by inducing recombination at FRT sites during the G2 phase of mitosis through conditional expression of flipase under the control of the hsp-promoter. Upon division, recombined chromosomes are segregated so that one daughter cell contains both copies of GAL80 and the other lacks inhibition of the GAL4 system. If the neuroblast loses the GAL80 transgene, all subsequent divisions generate neurons that express GAL4-driven GFP expression. Flip-out clones are generated by flanking a termination sequence (double red bar) with FRT sites between the promoter/enhancer and the GAL4 transgene. Upon conditional expression of flipase via a brief period of heat shock, the termination sequence is extracted by recombination of the FRT sites, and GAL4 protein is produced, allowing for marker expression under control of an upstream activating sequence (UAS). If the termination sequence is extracted from the neuroblast, all further progeny from that neuroblast will express the marker protein
While lineage-specific GAL4 lines and flip-out clones help visualize wild-type lineage morphology, studying null phenotypes is more difficult due to potential embryonic lethality from the genetic mutation. MARCM is used to generate a mosaic of homozygous loss-of-function clones containing a visualization marker. MARCM clones result from the removal of GAL80 repression of the UAS-GAL4 system by conditional mitotic recombination in the neuroblast (Lee and Luo 2001; Fig. 4). By visualizing MARCM clones with pan-neuronal drivers, homozygous mutant lineages are compared to wild-type lineages throughout the brain. Single-cell clones are marked if recombination occurs in the GMC rather than the neuroblast. For a narrower approach, lineage-specific GAL4 lines can be used to ease the identification of lineages visualized using MARCM.
Lineages and neuropile compartments are invariant
The position and trajectory of lineages is stereotypical between individual Drosophila brains. Each lineage is categorized by the position of the cell bodies within the cortex of the brain and their respective projection patterns (Truman et al. 2004; Pereanu and Hartenstein 2006). Embryonic and larval maps have been generated for the neuroblasts (Urbach and Technau 2004; Younossi-Hartenstein et al. 2006), pioneer tracts (Nassif et al. 1998), PATs (Younossi-Hartenstein et al. 2006; Sprecher et al. 2007), SATs (Dumstrei et al. 2003b; Pereanu and Hartenstein 2006), and neuropile compartments (Younossi-Hartenstein et al. 2003) of the larval brain. All lineages belong to a specific family based on position within the brain. To date, approximately 100 lineages belonging to 12 families have been defined and mapped (Fig. 5). At third instar stage, each family will have developed a set of stereotyped-projection patterns that innervate only a specific set of neuropile compartments (Pereanu and Hartenstein 2006). Individual lineages and lineage families that have been characterized by lineage-specific GAL4 lines are listed here (Table 1) along with the primary and secondary lineage projection pattern, branching type, and the marker line. For a comprehensive map of all secondary lineage projections, please see Pereanu and Hartenstein (2006).
Fig. 5.
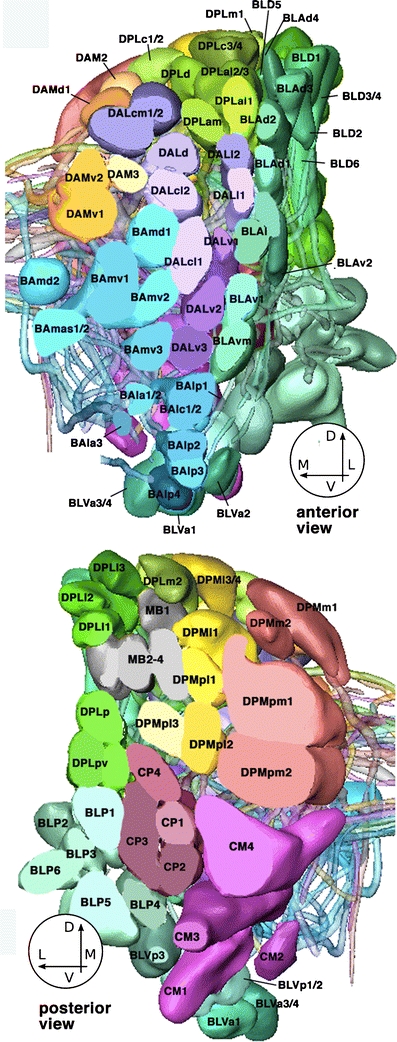
3D digital models of all lineages in one brain hemisphere. Top anterior view, bottom posterior view. For both models, the superficial region of the cortex was “sliced off” to allow for clearer view of lineages. Lineages are annotated according to Pereanu and Hartenstein (2006)
Each lineage has a unique morphology and the progression of its development can be traced from embryo through adult stages. Shown here is a global marker for larval lineages (Fig. 6a), a volume rendering of GFP expression under the control of a multiple-lineage GAL4 line in the adult brain (Fig. 6b), a lineage-specific lacZ line (Fig. 6c), and a MARCM clone in a single lineage at third instar and adult (Fig. 6d). These images are shown to highlight the wide range of tools available to the Drosophila community to label multiple lineage types throughout development.
Fig. 6.

Assortment of lineage markers. a Right hemisphere of a third instar brain labeled with anti-Neurotactin (BP106, green) and anti-DNCadherin (magenta). b Volume rendering of an adult brain with GFP expression under the control of GH146-GAL4. Visualized lineages include BAla1, BAmv3, and mushroom body lineages. c Right hemisphere of a third instar brain with lacZ expression in the DALv3 lineage under the control of the engrailed promoter. d MARCM induced clone in the Adult BAla1 lineage (inset shows third instar clone), visualized under the control of the period-GAL4 driver. All neuropiles are labeled with anti-DNcadherin (magenta). In all panels, medial is to the left and dorsal is up. SAT secondary axon tract, AL antennal lobe, LH lateral horn, MB mushroom body
While over 100 lineages have been identified in the larval brain, only a few of these lineages have been utilized in previous studies. Common model lineages that have been used include the mushroom body lineages, projection neurons of the BA family, ellipsoid body lineages of the DAL family, and the dorsal cluster neurons (DCNs) of the BLD5 lineage. The mushroom body lineages demonstrate great versatility having been used to: screen mutants and gain-of-function constructs for neural defects (Nicolai et al. 2003; Pereanu and Hartenstein 2006), screen small molecules that rescue disease-related mutations (Chang et al. 2008), study the function of specific genes in aspects of neuron maturation such as axon guidance and/or branching (Ng et al. 2002; Wang et al. 2002; Zhan et al. 2004; Martini and Davis 2005; Boyle et al. 2006; Kurusu and Zinn 2008), study pruning mechanisms (Watts et al. 2003; Awasaki and Ito 2004; Watts et al. 2004), establish additional functions of human disease-related genes (Liu et al. 2000; Tessier and Broadie 2008), and relate neural circuits to Drosophila learning and memory behaviors (reviewed in (Davis 1993). The projection neuron containing BA lineages and the DCN containing BLD5 lineage have been used as model systems with clonal analysis to identify proteins that affect neural morphology leading to incorrect circuit formation (Srahna et al. 2006; Komiyama and Luo 2007; Spletter et al. 2007; Lichtneckert et al. 2008). Finally, the few reports that utilize more than one or two lineages present evidence that each lineage develops using unique mechanisms with differential uses for highly conserved proteins such as DE-cadherin (Fung et al. 2009) or surrounding cell types such as glia (Spindler et al. 2009).
Automation of lineage identification and mapping circuitry
The ability to compare data to a standard model is a desired aspect of most model systems. While a standard model of the Drosophila brain in its entirety has been available for years (Rein et al. 2002); it has not been until recent months that a program allowing automated lineage identification has become accessible (A. Cardona et al., submitted). Using dynamic programming on manually segmented data, this newly developed mapping program enables users to match larval neurons with any of the 100 lineages. Therefore, a user needs very little training or knowledge about lineage identification to gain an immediate developmental history of a neuron of interest. In addition, the common use of such a mapping program will lead to a systematic naming system for all neurons in the Drosophila brain by assigning individual neurons to a given lineage.
The foremost goal for many is to relate gene expression, circuit morphology, and behavioral control. Similar to mammalian brains, specific functions have been attributed to compartments in the Drosophila brain. This is epitomized in studies concerning learning and memory in the mushroom body (reviewed in (Davis 1993, 2005; Masse et al. 2009) and motor control by the adult central complex (Martin et al. 1999). If more compartments in the adult brain are specialized for specific behaviors, the automated lineage identification program can then be used to map neurons of interest to a specific lineage, revealing clues to the function of that particular neuron.
Concluding comments
Drosophila lineages have a well-described developmental timeline, originate from a single mother neuroblast (which enables easy clonal analysis), contain groups of bundled neurons that project in a unified direction, have specific genetic identification, can be easily visualized with antibodies or lineage-specific GAL4 lines, and can be manipulated via genetic techniques widely used throughout the Drosophila community. The larger size of stereotypical Drosophila neuron bundles allow for a unique opportunity to study circuitry at the cellular level with manageable resolution. In addition to the advantages of a lineage model, Drosophila offers a short life cycle, a large number of progeny, and easy access to the brain. While we do not yet have the capability to completely understand the development and workings of our own brains, we can begin to learn about nervous system development by picking the brains of flies.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J Embryol Exp Morphol. 1976;35:107–123. [PubMed] [Google Scholar]
- Booker R, Truman JW. Postembryonic neurogenesis in the CNS of the tobacco hornworm Manduca sexta. I. Neuroblast arrays and the fate of their progeny during metamorphosis. J Comp Neurol. 1987;255:548–559. doi: 10.1002/cne.902550407. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Swanson LW. The neuron classification problem. Brain Res Rev. 2007;56:79–88. doi: 10.1016/j.brainresrev.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M, Nighorn A, Thomas JB. Drosophila Eph receptor guides specific axon branches of mushroom body neurons. Development. 2006;133:1845–1854. doi: 10.1242/dev.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cajal SRy (1899) Comparative study of the sensory areas of the human cortex: Harvard University
- Callahan CA, Thomas JB. Tau-beta-galactosidase, an axon-targeted fusion protein. Proc Natl Acad Sci U S A. 1994;91:5972–5976. doi: 10.1073/pnas.91.13.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124:461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- Davis RL. Mushroom bodies and Drosophila learning. Neuron. 1993;11:1–14. doi: 10.1016/0896-6273(93)90266-T. [DOI] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 2003;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V. Early development of the Drosophila brain: V. Pattern of postembryonic neuronal lineages expressing DE-cadherin. J Comp Neurol. 2003;455:451–462. doi: 10.1002/cne.10484. [DOI] [PubMed] [Google Scholar]
- Fareri DS, Martin LN, Delgado MR. Reward-related processing in the human brain: developmental considerations. Dev Psychopathol. 2008;20:1191–1211. doi: 10.1017/S0954579408000576. [DOI] [PubMed] [Google Scholar]
- Fung S, Wang F, Spindler SR, Hartenstein V. Drosophila E-cadherin and its binding partner Armadillo/ beta-catenin are required for axonal pathway choices in the developing larval brain. Dev Biol. 2009;332:371–382. doi: 10.1016/j.ydbio.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3:424–433. doi: 10.1016/0960-9822(93)90349-S. [DOI] [PubMed] [Google Scholar]
- Huppi PS, Dubois J. Diffusion tensor imaging of brain development. Semin Fetal Neonatal Med. 2006;11:489–497. doi: 10.1016/j.siny.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Zinn K. Receptor tyrosine phosphatases regulate birth order-dependent axonal fasciculation and midline repulsion during development of the Drosophila mushroom body. Mol Cell Neurosci. 2008;38:53–65. doi: 10.1016/j.mcn.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler SR, Fung S, Pereanu W, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev Biol. 2009;335(2):289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/S0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lichtneckert R, Nobs L, Reichert H. Empty spiracles is required for the development of olfactory projection neuron circuitry in Drosophila. Development. 2008;135:2415–2424. doi: 10.1242/dev.022210. [DOI] [PubMed] [Google Scholar]
- Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat Cell Biol. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- Martin JR, Raabe T, Heisenberg M. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J Comp Physiol [A] 1999;185:277–288. doi: 10.1007/s003590050387. [DOI] [PubMed] [Google Scholar]
- Martini SR, Davis RL. The dachshund gene is required for the proper guidance and branching of mushroom body axons in Drosophila melanogaster. J Neurobiol. 2005;64:133–144. doi: 10.1002/neu.20130. [DOI] [PubMed] [Google Scholar]
- Masse NY, Turner GC, Jefferis GS. Olfactory information processing in Drosophila. Curr Biol. 2009;19:R700–R713. doi: 10.1016/j.cub.2009.06.026. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Embryonic development of the Drosophila brain. I. Pattern of pioneer tracts. J Comp Neurol. 1998;402:10–31. doi: 10.1002/(SICI)1096-9861(19981207)402:1<10::AID-CNE2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Nicolai M, Lasbleiz C, Dura JM. Gain-of-function screen identifies a role of the Src64 oncogene in Drosophila mushroom body development. J Neurobiol. 2003;57:291–302. doi: 10.1002/neu.10277. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Meinertzhagen IA. Development and structure of synaptic contacts in Drosophila. Semin Cell Dev Biol. 2006;17:20–30. doi: 10.1016/j.semcdb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Rein K, Zockler M, Mader MT, Grubel C, Heisenberg M. The Drosophila standard brain. Curr Biol. 2002;12:227–231. doi: 10.1016/S0960-9822(02)00656-5. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V. Drosophila cortex and neuropile glia influence secondary axon tract growth, path finding, and fasciculation in the developing larval brain. Dev Biol. 2009;334(2):355–368. doi: 10.1016/j.ydbio.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Develop. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher SG, Reichert H, Hartenstein V. Gene expression patterns in primary neuronal clusters of the Drosophila embryonic brain. Gene Expr Patterns. 2007;7:584–595. doi: 10.1016/j.modgep.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srahna M, Leyssen M, Choi CM, Fradkin LG, Noordermeer JN, Hassan BA. A signaling network for patterning of neuronal connectivity in the Drosophila brain. PLoS Biol. 2006;4:e348. doi: 10.1371/journal.pbio.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ. Atlas of an insect brain. Berlin: Springer; 1976. [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Ball EE. Patterns of embryonic neurogenesis in a primitive wingless insect, the silverfish, Ctenolepisma longicaudata: comparison with those seen in flying insects. Dev Genes Evol. 1998;208:357–368. doi: 10.1007/s004270050192. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays. 2004;26:739–751. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/S0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/S0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra PM, Hartenstein V. Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455:435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nguyen B, Shy D, Hartenstein V. Embryonic origin of the Drosophila brain neuropile. J Comp Neurol. 2006;497:981–998. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]
- Zacharias D, Williams JLD, Meier T, Heinrich R. Neurogenesis in the insect brain: cellular identification and molecular characterization of brain neuroblasts in the grasshopper embryo. Development. 1993;118:941–955. [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhou B, Williams DW, Altman J, Riddiford LM, Truman JW (2009) Temporal patterns of broad isoform expression during the development of neuronal lineages in Drosophila. Neural Dev 4:39 [DOI] [PMC free article] [PubMed]


