Abstract
Background/Aims
The aim of our study was to define the potential role of virologic response at 12 months of treatment (VR12) in predicting subsequent virologic and clinical outcomes in adefovir (ADV)-treated lamivudine-resistant chronic hepatitis B.
Methods
Two hundred and four patients with lamivudine-resistant chronic hepatitis B virus (HBV) treated with ADV monotherapy were included. Serum HBV DNA was quantified by real-time polymerase chain reactions. VR12 was defined as a HBV DNA level of less than 4 log10 copies/mL after 12 months of ADV treatment.
Results
VR12 was observed in 110 of the 204 patients (54%). The mean HBV DNA reductions from baseline after 12 months of ADV treatment were 3.8 and 1.9 log10 copies/mL in patients with and without VR12, respectively (p<0.001). The hepatitis B "e" antigen (HBeAg) seroconversion rates in patients with and without VR12 were 32% and 14% at 12 months treatment, respectively (p=0.018), and 40% and 27% at 24 months of treatment (p=0.032). The genotypic mutation rates to ADV in patients with and without VR12 were 0% and 6% at 12 months of treatment, respectively (p=0.033), and 21% and 42% at 24 months (p=0.012). The rates of viral breakthrough in patients with and without VR12 were 0% and 7% at 12 months of treatment, respectively (p=0.072), and 9% and 25% at 24 months (p=0.006).
Conclusions
Patients without VR12 may need to switch to or add on other potent antiviral drugs in their medical regimens.
Keywords: Adefovir dipivoxil, Drug resistance, Virologic response
INTRODUCTION
Adefovir dipivoxil (ADV) is a nucleotide analogue that selectively inhibits viral polymerases and reverse transcriptases and it has broad-spectrum antiviral activity against both wild-type hepatitis B virus (HBV) and lamivudine (LMV)-resistant HBV mutants in vitro and in vivo.1,2 The benefits of ADV therapy, as compared to LMV therapy, for the patients with chronic HBV are delayed and infrequent selection of drug-resistant viruses in nucleos(t)ide naïve patients.3,4
Recently, monitoring of the serum HBV DNA levels has become necessary to assess the initial response to the antiviral drug regimen and to determine whether the antiviral effect is maintained during therapy. This monitoring allows predicting of biochemical remission, the subsequent hepatitis B "e" antigen (HBeAg) seroconversion, and the occurrence of drug resistance.5,6 For patients with LMV-treated chronic HBV infection, a greater reduction of the serum HBV DNA levels early in treatment is associated with better clinical outcomes and lower resistance rates.7-9
The aims of our study were to characterize the potential role of the virologic response at the time of 12 months of treatment for predicting the subsequent virologic and clinical outcomes and to determine the factors that are associated with the virologic response at the time of 12 months of treatment for patients with ADV-treated LMV-resistant chronic hepatitis B.
MATERIALS AND METHODS
1. Patients
All the adult patients with LMV-resistant, chronic HBV infection and who were referred to the Liver Clinic of Korea University Guro Hospital in Seoul, Korea, from December 2002 to March 2006 and who had received ADV for at least 6 months were included in this study. The data was retrospectively collected from 204 of these patients. Informed consent was obtained from each patient, and the experimental protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected by the prior approval of the Korea University Guro Hospital Human Research Committee.
2. Biochemical and virologic monitoring
A routine complete blood count and liver biochemical tests to measure the serum levels of aspartate transaminase (AST), alanine transaminase (ALT), total bilirubin, albumin, blood urea nitrogen and creatinine, and a measurement of the prothrombin time were performed for each patient. Hepatitis B surface antigen (HBsAg) was measured using commercially available radioimmunoassay kits (Abbot Laboratories, North Chicago, IL, USA). HBeAg and antibody to HBeAg levels were measured using radioimmunoassay kits (B.J.INEM.Co., Beijing, China). Tests for assessing the biochemical liver function and viral replication were done every 2-3 months during the ADV treatment.
3. HBV DNA quantification by real-time PCR
Quantitative analysis of the serum HBV DNA in the stored serum samples was retrospectively performed. For the real-time polymerase chain reaction (PCR) quantification, the viral DNA was extracted using a Qiagen Blood Kit (Qiagen, Chatworth, CA, USA) according to the manufacturer's instructions. Real-time PCR was performed with an Applied Biosystems 7300 Real Time PCR System® (Applied Biosystems, Foster City, CA, USA) as described previously.10 This method is sufficiently sensitive to detect as few as 300 copies of HBV genome per milliliter of serum.
4. ADV- and LMV-resistant mutation analysis by mass spectrometry analysis of the oligonucleotide fragment
Detection of the ADV- and LMV-resistant mutations by mass spectrometry analysis of the oligonucleotide fragment via restriction fragment mass polymorphism (RFMP) was performed as previously described.10
Briefly, two microliters of viral DNA was used for the PCR reaction. For the RFMP genotyping, PCR was performed in an 18-µL reaction mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.2 mM of each dNTP, 10 pmol of each primer, and 0.4 units of Platinum® Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA). The amplification condition included an initial denaturation step at 94℃ for 2 minutes, followed by 35 cycles of denaturation at 94℃ for 30 seconds, annealing at 55℃ for 30 seconds, and extension at 72℃ for 30 seconds. Restriction enzyme digestion of the PCR products was performed. The mass spectra were acquired on a linear MALDI-TOF MS (Bruker Daltonics Biflex IV, Billerica, MA, USA) work station.
5. Definitions
Virologic response at 12 months (VR12) was defined as HBV DNA less than 4 log10 copies/mL at 12 months after starting treatment in the patients who had HBV DNA, over 4 log10 copies/mL at treatment baseline. Virologic breakthrough (VBT) was defined as a greater than or equal to a 1 log10 increase in the serum HBV DNA level from the nadir on two consecutive occasions after an initial virologic response. Genotypic mutation was defined as the detection of mutation in the HBV polymerase gene using mass spectrometry analysis of the oligonucleotide fragment.
6. Statistical analysis
Statistical testing was performed using SPSS version 13.0. The values are expressed as means±standard error of the mean (SEM). The HBV DNA levels were logarithmically transformed for analysis. Continuous variables were compared using the 2-tailed Student's t-test. The categorical data were compared using the two-tailed χ2 test or Fisher's exact test. The factors associated with VR12 were analyzed by univariate analysis and multivariate logistic regression analysis. The cumulative probability of ADV-resistant mutations and the HBeAg seroconversion rate was estimated by Kaplan-Meier analysis. p-values<0.05 were considered statistically significant.
RESULTS
1. Baseline characteristics of the patients prior to adefovir treatment
Two hundred four patients were included in the study. Fifty-five patients who were not followed up for longer than 12 months after starting ADV were excluded from the final analysis. Table 1 summarizes the baseline characteristics of the 149 LMV-resistant patients who were treated with ADV. One hundred twenty (81%) patients were men and 20 were women, and the mean age of the total patients was 45 years. One hundred eight (72%) patients were HBeAg-positive with a mean serum ALT level of 186 IU/L. The average level of serum HBV DNA was 7.24 log10 copies/mL. Sixty-one patients had compensated liver cirrhosis. The mean duration of LMV treatment prior to beginning treatment with ADV was 31 months, and the mean duration of ADV treatment was 18.2 months.
Table 1.
Baseline Characteristics of 149 Lamivudine-resistant ADV-treated Chronic Hepatitis B Patients
Virologic response at 12 month (VR12) was defined as hepatitis B virus (HBV) DNA less than 4 log10 copies/mL after 12 months of adefovir (ADV) treatment. Values are represented as Mean±Standard error of the mean.
2. Comparison of the HBV reduction in the patients with or without VR12
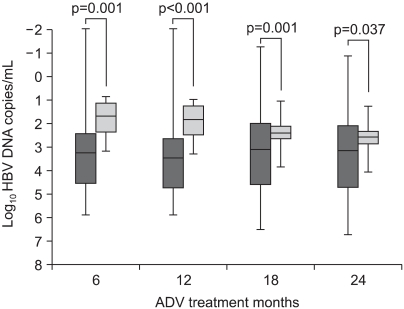
VR12 was achieved in 81 of the 149 patients (54%). Compared with the patients who did not achieve VR12, those who did had a greater reduction of their serum HBV DNA from baseline to 12 months of treatment (3.8 log10 copies/mL vs 1.9 log10 copies/mL, p<0.001). At 24 months treatment, the mean reduction of the serum HBV DNA levels was 3.5 log10 copies/mL vs 2.4 log10 copies/mL in the patients who did and did not achieve VR12, respectively (p=0.037) (Fig. 1).
Fig. 1.
The median hepatitis B virus (HBV) DNA reduction from baseline after 12 months of treatment with adefovir (ADV) in patients with (█) or without (▒) a virologic response at 12 months (VR12). VR12 was defined as HBV DNA less than 4 log10 copies/mL after 12 months of treatment in patients with HBV DNA exceeding 4 log10 copies/mL at the treatment baseline. The median and quartile values are represented as a horizontal bar and square, respectively.
3. HBeAg seroconversion and the biochemical response in the patients with or without VR12
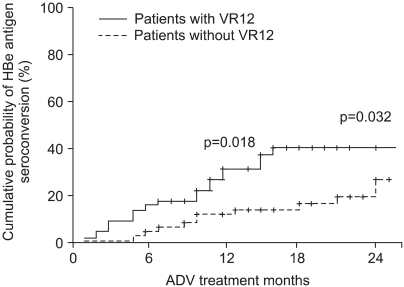
Compared with the patients who did not achieve VR12, those who did had a higher cumulative HBeAg seroconversion rate at 12 months treatmet (32% vs 14%, p=0.018) and at 24 months treatment (40% vs 27%, p=0.032) (Fig. 2). The ALT normalization at 6 months treatment was 88% vs. 69% for each group, respectively (p<0.05). At 12 and 24 months after starting treatment, there was no significant difference of ALT normalization between the two groups (p=NS).
Fig. 2.
Cumulative incidence of HBeAg seroconversion in patients with or without a virologic response at 12 months (VR12).
4. Genotypic mutation to ADV in the patients with or without VR12
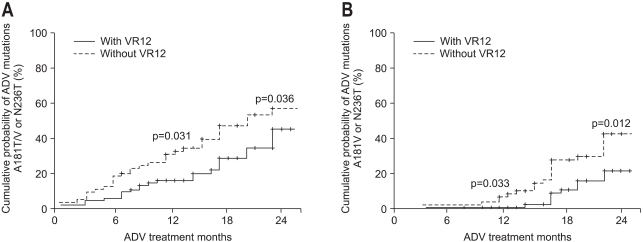
The ADV-resistant mutations rtA181V/T or rtN236T were detected in 57 of the 149 patients (38.3%) during the course of ADV treatment. Compared with the patients who did not achieve VR12, those who did had a significantly lower incidence of developing genotypic mutation to ADV at 12 months treatment (46% vs 18%, p=0.034). At 24 months treatment, the proportion of patients who developed genotypic mutation to ADV was 55% and 44%, in each group, respectively (p=0.036) (Fig. 3A).
Fig. 3.
The cumulative incidences of genotypic resistance -including A181V, N236T, and A181T (A), and A181V and N236T (B)- to adefovir (ADV) in patients with or without virologic response at 12 months (VR12) among the lamivudine-resistant chronic hepatitits B patients who were treated with ADV.
Locarnini et al.11 reported that the rtA181T mutation induces no antiviral resistance to ADV in vitro. When we analyzed the patients who carried only rtA181V or rtN236T, mutations were detected in 33 of 149 (22.1%) during the course of ADV treatment. Compared with patients who did not achieve VR12, those who did showed a higher rate of genotypic mutation to ADV at 12 months treatment (0% vs 6% p=0.033). This difference in the incidence of genotypic mutation between the two groups was still present at 24 months treatment (21% vs 42%, p=0.012) (Fig. 3B).
5. Viral breakthrough in the patients with or without VR12
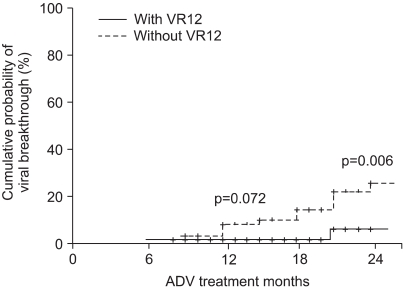
Viral breakthrough was more common in the patients without VR12 as compared to that of the patients with VR12. At 12 months of ADV treatment, viral breakthrough had developed in 0% and 7% of the patients with and without VR12, respectively (p=0.072). At 24 months treatment, viral breakthrough was found in 9% and 25% of the patients in each group, respectively (p=0.006) (Fig. 4).
Fig. 4.
Cumulative incidence of a virologic breakthrough in patients with or without virologic response at 12 months (VR12).
6. The factors associated with VR12
When comparing the baseline characteristics between the patients with or without VR12, there was no significant difference in age, gender, and the previous duration of LMV treatment. The patients who achieved VR12 were less likely to be HBeAg-positive (62% vs 85%, p=0.001), and they had higher ALT levels (352 IU/L vs 212 IU/L, p=0.002) and lower serum HBV DNA levels (6.9 log10 copies/mL vs 7.5 log10 copies/mL, p=0.002) compared to those of the patients who did not have VR12.
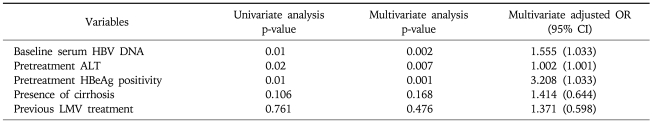
When we analyzed the factors associated with VR12 by multivariate analysis, the baseline serum HBV DNA levels, the pretreatment ALT levels, and the presence or absence of HBeAg were the independent factors with statistical significance (Table 2).
Table 2.
Factors Associated with VR12
Virologic response at 12 month (VR12) was defined as a hepatitis B virus (HBV) DNA level less than 4 log10 copies/mL at 12 months treatment.
OR, odds ratio; CI, confidence interval; HBeAg, hepatitis B "e" antigen; LMV, lamivudine.
DISCUSSION
It has been suggested that the antiviral response at key time points of treatment may be a valuable predictor of future treatment outcomes. In previous study for patients with chronic hepatitis C virus (HCV) infection, the reduction of the serum HCV RNA levels at 1 or 3 months treatment had a strong power to predict a sustained virologic response and the total duration of treatment.12,13 However, in patients with a chronic HBV infection, an antiviral response as an indicator of future treatment outcomes has not yet been fully determined. Previous studies suggested that the HBV genotype,14 the ALT level,9,15 the degree of the decline of the HBV DNA level during therapy.16,17 the presence of HBeAg,15,18 the presence of core promoter mutations,19 and HBV core-related antigens20 were factors that could predict the emergence of YMDD mutants.
We performed this study to characterize the potential role of the virologic response at 12 months treatment to predict subsequent virologic and clinical outcomes and to determine the factors associated with a virologic response at 12 months treatment in ADV-treated LMV-resistant chronic HBV patients.
In our study, the extent of the serum HBV DNA reduction and the incidence of HBeAg seroconversion and ALT normalization in the ADV-treated LMV-resistant chronic HBV-infected patients were similar to those previously reported.21-23 In a Korean study on ADV treatment for lamivudine-resistant hepatitis B virus genotype C infection, Kim et al.24 reported that the incidence of serum HBV DNA loss and serum HBeAg seroconversion were 88.2% and 14.3% at 12 months of treatment, respectively. This was also similar to our results.
The virologic response at 12 months treatment (VR12) was defined as the reduction of the serum HBV DNA level below the level of 4 log10 copies/mL at 12 months of treatment in patients who had serum HBV DNA levels over 4 log10 copies/mL at baseline. VR12 was achieved in 54% of the ADV-treated LMV-resistant patients. The mean serum HBV DNA reduction from baseline to 12 and 24 months treatment was greater in the patients with VR12 compared with that of the patients without VR12. In the patients with VR12, HBeAg seroconversion was more common.
In our study, only 54% of the LMV-resistant patients treated with ADV achieved VR12. Low antiviral activity or inefficient conversion from the prodrug (adefovir dipivoxil) to the active compound (adefovir diphosphate) or previous treatment experience with nucleoside might account for this suboptimal response to ADV. Other recent studies have shown that the currently approved dose of ADV was suboptimal and this has been further supported by recent studies showing its inferiority to tenofovir 300 mg.25 In our study, most of the patients who failed to achieve VR12 did not have further viral reduction despite continued ADV treatment.
The genotypic mutations to ADV more frequently occurred in the patients who did not achieve VR12 than in those who did. In our study, the ADV resistant mutations rtA181V/T or rtN236T were detected in 57 of 149 patients (38.3%). Since we detected the genotypic mutation to ADV using a sensitive and specific RFMP analysis, which could detect 1% mutants of the total viral population, these factors might explain the higher detection rate of the ADV mutations in our cases compared to that in the previous reports.3,4,26 Also, pre-existing LMV-resistant mutations and patient factors might have influenced the risk of ADV resistance. Similar to what occurs in LMV experienced ADV treated patients, it has been reported that an entecavir-resistant mutation developed more frequently in patients carrying LMV mutations than it did in nucleoside-naïve patients.27
Locarinini et al.11 reported that rtA181T did not show resistance to ADV in their in vitro phenotypic analysis. In recent papers, however, it became apparent that rtA181T has a resistance to ADV likewise rtA181V.27,28 When we analyzed the patients only carrying rtA181V or rtN236T, mutations were detected in 22.1% during two years of ADV treatment, which was similar to the results in other reports. Even after excluding the patients carrying the A181T mutation, the incidence of genotypic mutation to ADV (rtA181V or rtN236T) was higher in the patients who didn't have VR12 than that in those patients with VR12.
In the patients without VR12, viral breakthrough occurred at a significantly higher rate than it did in the patients with VR12. In our multivariate analysis, the factors associated with VR12 were the baseline serum HBV DNA levels, the pretreatment ALT levels, and the presence or absence of HBeAg. Recently, Buti et al. reported that the female gender, HBeAg negativity and low baseline serum HBV DNA levels were associated with the response to ADV treatment, which was similar to our findings.29,30
Although our study has several limitations, such as a relatively small number of patients, a relatively short-term follow-up period, and a potential referral bias, the virologic response at 12 months of ADV treatment may predict the subsequent virologic and clinical outcomes. Patients who do not achieve VR12 may need to switch to or add other potent antiviral drugs to their medical regimens.
References
- 1.Schiff ER, Lai CL, Hadziyannis S, et al. Adefovir dipivoxil therapy for lamivudine-resistant hepatitis B in pre- and post-liver transplantation patients. Hepatology. 2003;38:1419–1427. doi: 10.1016/j.hep.2003.09.040. [DOI] [PubMed] [Google Scholar]
- 2.Peters MG, Hann Hw H, Martin P, et al. Adefovir dipivoxil alone or in combination with lamivudine in patients with lamivudine-resistant chronic hepatitis B. Gastroenterology. 2004;126:91–101. doi: 10.1053/j.gastro.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 3.Villeneuve JP, Durantel D, Durantel S, et al. Selection of a hepatitis B virus strain resistant to adefovir in a liver transplantation patient. J Hepatol. 2003;39:1085–1089. doi: 10.1016/j.jhep.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Angus P, Vaughan R, Xiong S, et al. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003;125:292–297. doi: 10.1016/s0016-5085(03)00939-9. [DOI] [PubMed] [Google Scholar]
- 5.Mommeja-Marin H, Mondou E, Blum MR, Rousseau F. Serum HBV DNA as a marker of efficacy during therapy for chronic HBV infection: analysis and review of the literature. Hepatology. 2003;37:1309–1319. doi: 10.1053/jhep.2003.50208. [DOI] [PubMed] [Google Scholar]
- 6.Gauthier J, Bourne EJ, Lutz MW, et al. Quantitation of hepatitis B viremia and emergence of YMDD variants in patients with chronic hepatitis B treated with lamivudine. J Infect Dis. 1999;180:1757–1762. doi: 10.1086/315147. [DOI] [PubMed] [Google Scholar]
- 7.Lai CL, Dienstag J, Schiff E, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687–696. doi: 10.1086/368083. [DOI] [PubMed] [Google Scholar]
- 8.Buti M, Sanchez F, Cotrina M, et al. Quantitative hepatitis B virus DNA testing for the early prediction of the maintenance of response during lamivudine therapy in patients with chronic hepatitis B. J Infect Dis. 2001;183:1277–1280. doi: 10.1086/319677. [DOI] [PubMed] [Google Scholar]
- 9.Yuen MF, Sablon E, Hui CK, Yuan HJ, Decraemer H, Lai CL. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology. 2001;34:785–791. doi: 10.1053/jhep.2001.27563. [DOI] [PubMed] [Google Scholar]
- 10.Yeon JE, Yoo W, Hong SP, et al. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006;55:1488–1495. doi: 10.1136/gut.2005.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locarnini S QX, Arterburn S, Snow A, Brosgart CL, Currie G. Incidence and predictors of emergence of adefovir resistant HBV during four years of adefovir dipivoxil (ADV) therapy for patients with chronic hepatitits B (CHB) J Hepatol. 2005;42(Suppl 2):17. [Google Scholar]
- 12.Zeuzem S, Pawlotsky JM, Lukasiewicz E, et al. International, multicenter, randomized, controlled study comparing dynamically individualized versus standard treatment in patients with chronic hepatitis C. J Hepatol. 2005;43:250–257. doi: 10.1016/j.jhep.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Berg T, Sarrazin C, Herrmann E, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37:600–609. doi: 10.1053/jhep.2003.50106. [DOI] [PubMed] [Google Scholar]
- 14.Zollner B, Petersen J, Puchhammer-Stockl E, et al. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology. 2004;39:42–50. doi: 10.1002/hep.20016. [DOI] [PubMed] [Google Scholar]
- 15.Nafa S, Ahmed S, Tavan D, et al. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in patients treated by lamivudine for chronic hepatitis B. Hepatology. 2000;32:1078–1088. doi: 10.1053/jhep.2000.19619. [DOI] [PubMed] [Google Scholar]
- 16.Puchhammer-Stockl E, Mandl CW, Kletzmayr J, et al. Monitoring the virus load can predict the emergence of drug-resistant hepatitis B virus strains in renal transplantation patients during lamivudine therapy. J Infect Dis. 2000;181:2063–2066. doi: 10.1086/315519. [DOI] [PubMed] [Google Scholar]
- 17.Zollner B, Schafer P, Feucht HH, Schroter M, Petersen J, Laufs R. Correlation of hepatitis B virus load with loss of e antigen and emerging drug-resistant variants during lamivudine therapy. J Med Virol. 2001;65:659–663. doi: 10.1002/jmv.2087. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki F, Tsubota A, Arase Y, et al. Efficacy of lamivudine therapy and factors associated with emergence of resistance in chronic hepatitis B virus infection in Japan. Intervirology. 2003;46:182–189. doi: 10.1159/000071460. [DOI] [PubMed] [Google Scholar]
- 19.Lok AS, Hussain M, Cursano C, et al. Evolution of hepatitis B virus polymerase gene mutations in hepatitis B e antigen-negative patients receiving lamivudine therapy. Hepatology. 2000;32:1145–1153. doi: 10.1053/jhep.2000.19622. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka E, Matsumoto A, Suzuki F, et al. Measurement of hepatitis B virus core-related antigen is valuable for identifying patients who are at low risk of lamivudine resistance. Liver Int. 2006;26:90–96. doi: 10.1111/j.1478-3231.2005.01200.x. [DOI] [PubMed] [Google Scholar]
- 21.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, et al. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B. N Engl J Med. 2005;352:2673–2681. doi: 10.1056/NEJMoa042957. [DOI] [PubMed] [Google Scholar]
- 22.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, et al. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology. 2006;131:1743–1751. doi: 10.1053/j.gastro.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-negative chronic hepatitis B. N Engl J Med. 2003;348:800–807. doi: 10.1056/NEJMoa021812. [DOI] [PubMed] [Google Scholar]
- 24.Kim do Y, Kim HJ, Lee CK, et al. Efficacy of adefovir dipivoxil in the treatment of lamivudine-resistant hepatitis B virus genotype C infection. Liver Int. 2007;27:47–53. doi: 10.1111/j.1478-3231.2006.01407.x. [DOI] [PubMed] [Google Scholar]
- 25.van Bommel F, Zollner B, Sarrazin C, et al. Tenofovir for patients with lamivudine-resistant hepatitis B virus (HBV) infection and high HBV DNA level during adefovir therapy. Hepatology. 2006;44:318–325. doi: 10.1002/hep.21253. [DOI] [PubMed] [Google Scholar]
- 26.Westland CE, Yang H, Delaney WE, 4th, et al. Week 48 resistance surveillance in two phase 3 clinical studies of adefovir dipivoxil for chronic hepatitis B. Hepatology. 2003;38:96–103. doi: 10.1053/jhep.2003.50288. [DOI] [PubMed] [Google Scholar]
- 27.European Association for the Study of the Liver. EASL clinical practice guidelines: management of chronic hepatitis B. J Hepatol. 2009;50:227–242. doi: 10.1016/j.jhep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. doi: 10.1053/j.gastro.2009.08.063. e1-2. [DOI] [PubMed] [Google Scholar]
- 29.Tenney DJ, Rose RE, Baldick CJ, et al. Two-year assessment of entecavir resistance in Lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present. Antimicrob Agents Chemother. 2007;51:902–911. doi: 10.1128/AAC.00833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buti M, Elefsiniotis I, Jardi R, et al. Viral genotype and baseline load predict the response to adefovir treatment in lamivudine-resistant chronic hepatitis B patients. J Hepatol. 2007;47:366–372. doi: 10.1016/j.jhep.2007.04.011. [DOI] [PubMed] [Google Scholar]