Abstract
Do general principles govern the genetic causes of phenotypic evolution? One promising idea is that mutations in cis-regulatory regions play a predominant role in phenotypic evolution because they can alter gene activity without causing pleiotropic effects. Recent evidence that revealed the genetic basis of pigmentation pattern evolution in Drosophila santomea supports this notion. Multiple mutations that disrupt an abdominal enhancer of the pleiotropic gene tan partly explain the reduced pigmentation observed in this species.
Which mutations underlie phenotypic evolution?
Do developmental mechanisms influence genetic evolution? That is, can we predict what types of mutations underlie phenotypic evolution? The simplest model predicts that mutations with the least pleiotropic effects are most likely to contribute to phenotypic variation and divergence. The rationale for this model comes from a synthesis of Fisher’s geometrical model of phenotypic evolution [1] with modern developmental biology. Fisher conceived of mutational effects as randomly oriented with respect to phenotypes. Given that phenotypes are complex and multidimensional, he assumed that mutations affect many traits at the same time. Fisher’s model thus implies that most mutations generate pleiotropic effects. Such pleiotropic mutations will typically degrade fitness, because it is difficult to improve fitness if each mutation simultaneously affects many aspects of a phenotype. Modern developmental biology has provided an escape from this conundrum by demonstrating that cis-regulatory DNA is often functionally modular: genetically separable elements are responsible for discrete phases and patterns of expression [2–5]. Thus, although nearly all genes perform pleiotropic roles in development, cis-regulatory regions ensure that not all mutations have pleiotropic effects [3]. Accordingly, evolution is expected to favor mutations that cause fewer pleiotropic effects [6,7], such as mutations in cis-regulatory regions. Is this model correct? The decisive data—the mutations themselves—have been hard to identify. Although mapping natural variation to chromosomal regions is now easy, the final step of identifying the causal genes and mutations remains a massive challenge. Now Jeong et al. [8] have added an important piece to the puzzle by demonstrating that the evolution of Drosophila santomea pigmentation patterning has been driven by mutations at a cis-regulatory element of the pleiotropic enzyme tan, not once, but three separate times (Figure 1).
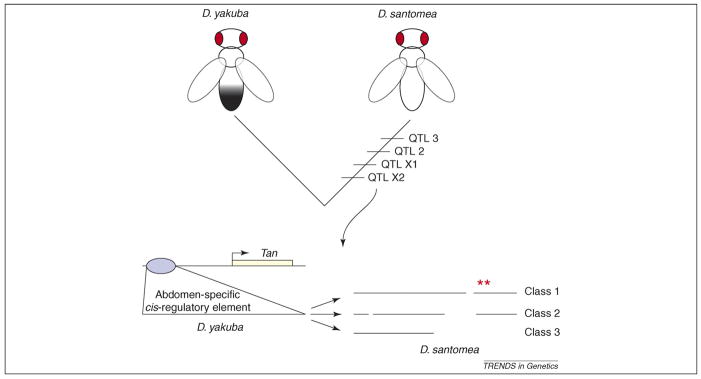
Figure 1.
Genetic causes of abdominal pigmentation evolution. The two closely related species, Drosophila yakuba and Drosophila santomea, shown schematically at the top, display different levels of abdominal pigmentation. This difference results from changes in at least four quantitative trait loci (QTLs) that all occurred on the lineage leading to D. santomea [9]. Two QTLs reside on the X chromosome (QTL X1 and QTL X2), one on chromosome II (QTL 2) and one on chromosome III (QTL 3). The order of evolved QTLs is shown for illustration purposes only. The true order of QTL evolution is unknown. Jeong et al. [8] investigated the genetic changes underlying QTL X2. They found that changes in an abdomen-specific cis-regulatory element of the tan gene, shown below, had evolved in D. santomea. They identified three classes of alleles carrying different mutations that all incapacitate the cis-regulatory element. The cis-regulatory enhancer sequences are represented as horizontal lines at the bottom. Point mutations that alter function are shown as asterisks above the D. santomea sequences. Deletions are shown as gaps in the sequence.
Pale abdomens lack tan
Drosophila abdominal pigmentation patterns vary among species. D. santomea, endemic to São Tomé, an island off the west coast of Africa, lost pigmentation in the posterior abdominal segments since divergence from their darkly pigmented sister species D. yakuba. Linkage mapping studies implicated a locus of large effect on the X chromosome as one of four loci contributing to the pigmentation difference between these species [9]. The linkage peak falls near the candidate gene tan, an enzyme required for pigmentation as well as for vision [10]. In D. yakuba, tan mRNA is expressed in the abdominal epidermal cells that produce darkly pigmented cuticle. In D. santomea, tan mRNA is undetectable in the epidermal cells. In both species, however, tan is expressed in the cells that produce pigmented bristle cells. Thus, the absence of tan expression in epidermal cells likely resulted from altered regulation of tan and not a complete loss-of-function tan allele.
Jeong et al. [8] performed simple crosses to demonstrate that the absence of tan expression in D. santomea results from X-linked factors and not from the evolution of transacting factors located on other chromosomes. Together with an earlier study [9] showing tight linkage of pigmentation pattern with tan, these results suggested that the change in tan expression patterns might have resulted from a change in the tan cis-regulatory region. As this cis-regulatory region was previously uncharacterized, Jeong et al. [8] made a series of reporter constructs using D. melanogaster DNA and found a cis-regulatory region 3–4 kb upstream from the transcription start site, in the intergenic region between two other genes, that drives expression in the same abdominal pattern observed for endogenous D. melanogaster tan.
Jeong et al. [8] found that this cis-regulatory region could drive tan cDNA expression and rescue abdominal pigmentation in D. melanogaster tan mutants. This transgene also partially restored pigmentation in D. santomea, implying that altered tan activity is indeed a major cause of the reduced pigmentation in D. santomea abdomens.
Additional observations indicate that changes in this cis-regulatory region have caused pigmentation evolution. First, the D. yakuba and D. santomea tan genes encode identical proteins, ruling out any effects caused by coding changes. Second, the D. yakuba abdominal cis-regulatory element drives tan expression in the posterior abdomen, whereas the same region from D. santomea fails to do so. Jeong et al. [8] dissected this regulatory element further and identified two single nucleotide substitutions that together cause the loss of enhancer activity.
Thus, mutations that alter tan abdominal expression, without affecting the gene’s other roles, underlie a portion of the pigmentation pattern divergence observed in D. santomea. Surprisingly, a survey of the tan locus from multiple D. santomea natural isolates uncovered three separate alleles that independently eliminate tan abdominal enhancer activity [8]. A second allele carries a 30-bp deletion in the tan abdominal cis-regulatory element, and a third harbors a 212-bp deletion in the same region. No tan alleles with a functional abdominal cis-regulatory element were found in the D. santomea isolates. Remarkably, the three nonfunctional regulatory sequences arose independently from a functional allele, based on the sequences inferred for their ancestors.
The mutations that cause these three evolutionarily independent alleles all occur within a few bases of one another in the tan abdominal cis-regulatory element. These results are consistent with a model wherein the pleiotropic roles of tan bias evolutionarily relevant mutations toward the abdominal cis-regulatory module and away from gene regions that might alter other tan functions.
Concluding remarks
These new results provide the latest example of phenotypic evolution via parallel mutations at a single cis-regulatory element. In humans, for example, a change in temporal regulation of lactase expression has arisen multiple times through mutations in a small cis-regulatory element [11]. In this case, expression later in development has been added without altering the structure of the enzyme or its early expression.
The cis-regulatory hypothesis for evolutionary change does not require that all genetic changes occur at cis-regulatory sites. Rather, it predicts that natural selection will often favor the least pleiotropic route to phenotypic evolution. When evolution requires spatially or temporally regulated gene expression, the least pleiotropic mutations are likely to be found in cis-regulatory regions. At other times, natural selection will favor a change in protein sequence. By example, when host immunity gene products physically interact with pathogens we expect that protein sequence evolution will occur in response to changes in pathogen populations [12]. However, even in such cases, pathogen attack can sometimes be foiled by specifically altering the regulation of a target protein. In human populations from West Africa, a cis-regulatory mutation eliminates DARC (Duffy blood group, chemokine receptor) expression in red blood cells, and blocks malaria infection, without altering DARC expression in other cells types [13]. Of course, evolution will always use the available molecular variation. However, given the choice, natural selection will favor mutations that cause the fewest pleiotropic effects.
The new work from Jeong et al. [8] on tan evolution provides yet one more clearly documented case for the importance of cis-regulatory evolution. Such examples have been slow in coming, largely because it is more technically demanding to identify functional cis-regulatory changes than functional changes in proteins. As the field develops new approaches to fine mapping natural variation, we expect to gain deeper understanding of the rules that influence when evolution favors cis-regulatory mutations. For now, the basic conceptual model remains healthy. Evolution tinkers where the tinkering is good. For traits that require regulated gene expression, cis-regulatory DNA is the place.
References
- 1.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; 1930. [Google Scholar]
- 2.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution Int J Org Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 4.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 5.Davidson EH. The Regulatory Genome: Gene Regulatory Networks Indevelopment and Evolution. Academic; 2006. [Google Scholar]
- 6.Orr HA. Adaptation and the cost of complexity. Evolution Int J Org Evolution. 2000;54:13–20. doi: 10.1111/j.0014-3820.2000.tb00002.x. [DOI] [PubMed] [Google Scholar]
- 7.Otto SP. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc Biol Sci. 2004;271:705–714. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong S, et al. The evolution of gene regulation underlies the morphological divergence of two closely related Drosophila species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Carbone MA, et al. Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea. Genetics. 2005;171:211–225. doi: 10.1534/genetics.105.044412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.True JR, et al. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchison A. Partitioning of genetic variation between regulatory and coding gene segments: the predominance of software variation in genes encoding introvert proteins. Immunogenetics. 1997;46:46–52. doi: 10.1007/s002510050241. [DOI] [PubMed] [Google Scholar]
- 13.Tournamille C, et al. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]