Abstract
Motivation: Experimental and predicted data concerning gene transcriptional regulation are distributed among many heterogeneous sources. However, there are no resources to integrate these data automatically or to provide a ‘one-stop shop’ experience for users seeking information essential for deciphering and modeling gene regulatory networks.
Results: IntegromeDB, a semantic graph-based ‘deep-web’ data integration system that automatically captures, integrates and manages publicly available data concerning transcriptional regulation, as well as other relevant biological information, is proposed in this article. The problems associated with data integration are addressed by ontology-driven data mapping, multiple data annotation and heterogeneous data querying, also enabling integration of the user's data. IntegromeDB integrates over 100 experimental and computational data sources relating to genomics, transcriptomics, genetics, and functional and interaction data concerning gene transcriptional regulation in eukaryotes and prokaryotes.
Availability: IntegromeDB is accessible through the integrated research environment BiologicalNetworks at http://www.BiologicalNetworks.org
Contact: baitaluk@sdsc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
A large number of databases and datasets that annotate transcriptional regulatory elements in general or from niche areas of gene regulation have been developed; see the Nucleic Acids Research list of databases on transcriptional regulator sites and transcription factors (TFs) (Cochrane and Galperin, 2010). TRANSFAC (Matys et al., 2006), for example, contains data on TFs, TF binding sites (TFBSs), target genes, promoters and TF classification in several model eukaryotic organisms; while FlyBase (Grumbling and Strelets, 2006) and Arabidopsis gene regulatory information server (AGRIS; Davuluri et al., 2003) are species-centered resources. Resources providing curated information, such as ORegAnno (Griffith et al., 2008) and Transcription Regulatory Regions Database (TRRD; Kolchanov et al., 2002), co-exist with resources that provide computationally derived data, such as TrsDB (Hermoso et al., 2004) and DBD (Kummerfeld and Teichmann, 2006). In addition, there are general biological resources that contain among other information data related to transcriptional regulation. For example, PDB (Berman et al., 2000) and NDB (Berman et al., 2002) contain structures of TFs and their complexes with DNA; Pfam (Finn et al., 2008) and PROSITE (Hulo et al., 2006) contain sequence patterns of TFs. Currently, information concerning transcriptional regulation is dispersed among various resources, many of which are not organized into databases but separate files posted on the web. To fully use and navigate these data, integrated systems are required.
The first data integration systems in molecular biology emerged to bring together internal databases and analysis tools in order to extract novel biological knowledge; examples include GeneExpress (Kolchanov et al., 1999), which is specific to the domain of gene transcriptional regulation, and FlyBase (Drysdale, 2008), which is species-specific. Early systems integrated external databases predominantly by means of URL links. Well-known link-based integrating systems, aka portals or navigators, include Entrez (Sayers et al., 2009), Ensembl (Hubbard et al., 2009), ISYS (Siepel et al., 2001), the Biology Workbench (Subramaniam, 1998), SRS (Etzold and Argos, 1993), Integr8 (Pruess et al., 2005), Galaxy (Giardine et al., 2005) and BioMart (Haider et al., 2009). Such systems serve for index information, allow querying and maintain relationships among the entities from various databases.
With the development of biological ontologies, automatic integration of heterogeneous data sources into data warehouses via integrative data models became feasible. Data warehouses can be separated into two groups. The first group comprises systems that cover particular domains of biological knowledge including cPath (Cerami et al., 2006) and PathSys (Baitaluk et al., 2006a, b), which concern biological pathways; ONDEX (Kohler et al., 2006), which stores data from gene expression microarray experiments; Ensembl Regulatory Build (Hubbard et al., 2009), comprising annotations of potential regulatory regions within the human genome; ChlamyCyc (May et al., 2009), which stores data on Chlamydomonas reinhardtii; SNPnexus (Chelala et al., 2009), comprising functional annotations of SNPs in public databases; and RefDIC (Hijikata et al., 2007), containing cross-reference information from the transcriptome and proteome of immune cells. The second group comprises systems that aim to address general problems of integration of heterogeneous biological data and include Atlas (Shah et al., 2005), BioExtract (www.bioextract.org), Biochemical Network Database (BNDB; Kuntzer et al., 2007), BIOZON (Birkland and Yona, 2006), GUS (Davidson et al., 2001) and InterMine (www.intermine.org).
Data integrity, consistency, redundancy, connectivity, updatability, expandability and complex and ‘fuzzy’ queries are the problems associated with data integration (Birkland and Yona, 2006), which arise from the nature of heterogeneous data and the lack of unified ontology. Therefore, there is a need for integration systems that are able to recognize different ontologies and semantics of the data. In addition, since different databases have various update cycles that can lead to format changes and the discontinuance or addition of data, the integration systems need to automatically and regularly scan databases for updates, recognize format changes and update mapping and data exchange procedures in order to maintain consistency of data. Yet the integration systems should also provide an environment that allows users to integrate their own data and customize the system. An ‘ideal’ integration system should provide ad hoc queries that are broad enough and, at the same time, domain-specific and user-friendly.
This study addresses the aforementioned problems of integrating heterogeneous data in the domain of transcriptional regulation. There are systems that integrate various data concerning transcriptional regulation such as, BNDB (Kuntzer et al., 2007) and SNPnexus (Chelala et al., 2009), which include data from TRANSFAC and CoryneRegNet (Baumbach, 2007). However, there is no system that integrates the full spectra of data concerning transcriptional regulation, together with other relevant biological information, which is currently available in upwards of 60 databases listed in the Nucleic Acids Research depository (Cochrane and Galperin, 2010) under the category ‘transcriptional regulator sites and TFs’. The two major projects in the domain, Ensembl Regulatory Build (Hubbard et al., 2009) and ORegAnno (Griffith et al., 2008), do not represent data warehouses per se, and their aims are distinct from data integration. Ensembl Regulatory Build (Hubbard et al., 2009) provides raw data concerning maps of open chromatin created by DNase I hypersensitivity mapping, covalent modifications of histone protein tails assayed by chromatin immunoprecipitation and annotations of potential regulatory regions within the human genome based on these data, obtained from the ENCODE project (Birney et al., 2007). ORegAnno (Griffith et al., 2008) is an open-source open-access database and literature curation system for community-based annotation of experimentally identified DNA regulatory regions, TFBSs and regulatory variants; it is integrated with Ensembl, PubMed and dbSNP via curated cross references.
The present study proposes an approach for integrating all publicly available genomic, transcriptomic, genetic and functional data relevant to transcriptional regulation in eukaryotes and prokaryotes. The resulting integration system, IntegromeDB, has been implemented and is available within the BiologicalNetworks integrated research environment at http:/www.BiologicalNetworks.org (Baitaluk et al., 2006b). Information relating to integrated data can be searched by category and data source, and includes quick searches of genes/proteins, data statistics and data inconsistencies in public data sources (http://www.integromedb.org). Data integration and mapping to the internal database is fully automated and based on Semantic Web technologies such as the Resource Description Framework (RDF; http://www.w3.org/RDF/) and Web Ontology Language (OWL; http://www.w3.org/TR/owl-ref/). The IntegromeDB ontology developed by the authors is presented here, together with the system architecture. The current version of IntegromeDB integrates in excess of 100 000 different data types and features from more than 100 data sources concerning sequences and structures of TFs, their orthologs and binding sites, promoters and other gene regulatory regions, orthologs of target genes, disease relationships, mutations and SNPs, gene expression data, gene function, pathways, protein–protein interactions and other related information. IntegromeDB enables researchers to integrate their own data into the system and query them together with data extracted from other resources.
2 SYSTEM OVERVIEW
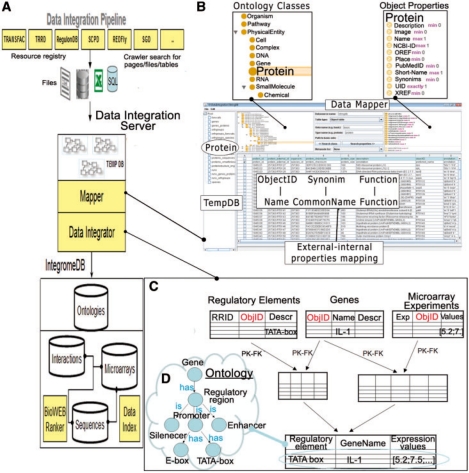
The architecture of the IntegromeDB system is presented in Figure 1. The data integration pipeline contains the following main blocks (Fig. 1A):
Web crawler that automatically searches a list of web sites for data to be integrated.
Data Integration Server that does the following: (i) accepts external data from the web crawler and stores them in the temporary database TempDB; (ii) maps external data to the IntegromeDB database schema, using the IntegromeDB Ontology (Fig. 1B); and (iii) injects data from external tables into the database (Fig. 1C).
The internal database (also called IntegromeDB in Fig. 1A) stores the integrated data according to the IntegromeDB Ontology.
Fig. 1.
The architecture of IntegromeDB. (A) Data integration pipeline, containing the main architectural blocks. (B) Data mapper, administered internally and allowing mapping external data to internal database schema through ontologies. (C) Data integrator logic schema, shown on the example of three different data types, regulatory elements (e.g. TATA box), genes (e.g. IL-1 gene) and microarray experiments, that share the common ObjectID (e.g. GeneID) and are joined through PK–FK relationships depicted in the ontology (D) into a single integrated table.
2.1 Data integration and mapping
The data integration and mapping procedure is fully automated and does not require human intervention at any step, including data collection by traversing external web sites and mapping external data to the internal database schema.
To traverse web sites of interest, the SmartCrawler web crawler is utilized (http://sourceforge.net/projects/smartcrawler/) to crawl the links to a depth of 12. The 12-deep crawl provides a sufficiently broad coverage and retrieves web pages that predominantly contain information relevant to transcriptional regulation. Web crawler searches for web pages, tables and relational databases that can be accessed in any of the following ways: (i) directly by querying an SQL database; (ii) through a HTTP GET operation executed against a database; and (iii) invoking a web service provided by the database. The data source to be integrated is assumed to be either a relational (tab-delimited, Excel, SQL), XML or RDF file with a binding pattern for every relationship disclosed.
The web crawler stores external data in the temporary database TempDB (Fig. 1A) before they are mapped to the IntegromeDB database schema, using IntegromeDB Ontology, and are finally transferred into the database by DataIntegrator.
To map the data, the following four kinds of mapping relations are considered:
‘OntologyClass’ mapping, which describes the type of objects to be integrated. It maps data values from an external source to an ontological term in IntegromeDB.
‘Attributes_for’ mapping, which specifies the attributes for classes that must be integrated. It is a joinable relation that links attributes to integrated objects that are mapped through ontology to an internal OntologyClass.
‘MetaNode_for’ mapping applies to meta-graphs, such as pathways and organisms, and describes which OntologyClass is a meta-graph of another OntologyClass, for example, a protein being part of a particular pathway.
‘Relations_between’ mapping applies to the relationships between objects, such as interactions, co-expression and co-occurrence, and provides OntologyClasses between which the relationship is integrated.
Data Mapper maps external biological data to the IntegromeDB internal RDF-compatible (RDF; http://www.w3.org/RDF/) database schema, transforming biological data into an RDF-compatible format. To transform biological data into an RDF-compatible format and create integrated views of data sources, the data integration procedure includes automatic determination of Node IDs (primary, graph and connector nodes) such as names and synonyms of biological entities. Several algorithms have been implemented to support relevant data ingestion using ‘ontology to data’ mapping, primary key–foreign key (PK–FK) constraints, and ontological data joins that are based on concept IDs rather than actual data. Figure 1C illustrates how integrated data that are mapped to two different ontological concepts, such as ‘TATA box’ and ‘Gene Expression’, can be linked through the PK–FK constraint at the source. Local views can be joined on the basis of ObjectIDs extracted from the ontological source.
In the absence of clear evidence of/reference to a class from the ontology, an automatic procedure that statistically evaluates the content of the integrated table and assigns a term from the IntegromeDB ontology to it is applied. For each distinct word and word combination that is present in the table to be integrated, which are terms in the IntegromeDB ontology, the statistical significance of the term's occurrence (P-value) is calculated using Fisher's exact test. The most significant term is assigned to the table.
2.2 Database schema
The IntegromeDB internal database schema is RDF-compatible (RDF; http://www.w3.org/RDF/); i.e. it stores biological data in an RDF-compatible format, the standard format of the Semantic Web (Good and Wilkinson, 2006). The database architecture and database schema are provided at http://www.BiologicalNetworks.net/Database/tut0.php.
IntegromeDB's internal database is a PostgreSQL database that has been modeled as a node- and edge-typed labeled meta-graph (Hu et al., 2007), where the labels are described by their own schema. The data model has been introduced and described in detail (Baitaluk et al., 2006a, b); therefore, a brief description is provided herein. Objects such as proteins, ligands, molecular complexes and genes, are represented by nodes; the relationships between objects such as up/down-regulation, molecular transport, molecular synthesis and enzymatic activity are represented by edges. The types of nodes and edges are designated in the standard ontologies described further. A label of a node or edge provides specific details about it. For example, for a node ‘gene’, the gene's name, ORF, chromosome, coordinates and other physical, genetic and functional properties, are specified in the label of the node. An edge ‘regulation’ between a protein and a gene could be labeled by the nature of regulation such as activation and the mechanism of regulation, for example, phosphorylation.
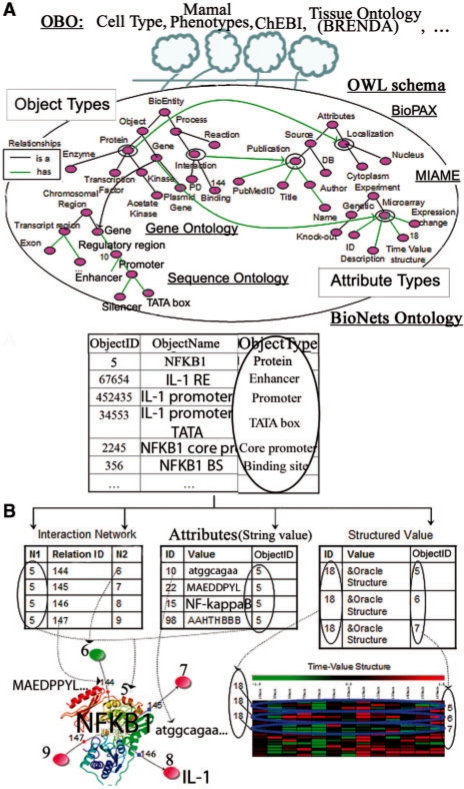
To represent a wide variety of biological data, the IntegromeDB internal database employs a graph-based model that dynamically incorporates (Fig. 1C) new sets of nodes, edges or node/edge labels into the database, and integrates the following four orthogonal data types (Fig. 2B):
Graphs that represent molecular interactions and ontologies; for example, the protein–protein interaction network of NFκB-1 factor with other four proteins, denoted ObjectID 6, 7, 8 and 9 (Fig. 2B). Relations between them could be as follows: 144, 145, 146 and 147.
Histograms that represent time–value structures including gene expression data and metabolite concentrations; for example, microarray expression data obtained in an experiment with ID 18 (structured value in Fig. 2B) can be associated with the genes with ObjectID 5, 6 and 7, coding for NFκB-1 and its interacting proteins.
Trees that represent classifications/ontologies and phylogenies.
Sequences that represent protein/DNA/RNA sequences and protein structures; for example, in Figure 2B the object NFκB-1 protein (ObjectID 5) has such attributes as the name of the object (NFκB-1, ID 15), gene coding sequence (ID 10), DNA-binding motif (ID 22), and a secondary structure of the DNA-binding domain (ID 98).
Fig. 2.
IntegromeDB internal database. (A) IntegromeDB Ontology integrating basic BioNets ontology with 34 OBO ontologies, (B) Heterogeneous data of the database and data structures. The color figure at the bottom left (generated in BiologicalNetworks) shows the structure of NFκB-1 protein (ObjectID 5), fragments of NFκB-1 gene and protein sequences (AttributeID 10, 22), as well as genes and proteins (ObjectID 6, 7, 8, 9) interacting with NFκB-1 that are schematically represented by circles. The color figure at the bottom right shows an image from a microarray experiment (ID 18) that involved NFκB-1 gene and its interactors (ObjectID 6, 7).
The internal database of IntegromeDB internal database contains specialized indexes that allow quick access to ancestor/descendant relationships for transitive relationships, such as ‘subclass-of’ and ‘part-of’. To support ontological queries, IntegromeDB contains a specialized query processing engine described further.
2.3 Ontology model
Databases use different ontologies and some do not use standard ontologies. Therefore, to integrate heterogeneous resources, an ‘integrated’ ontology, IntegromeDB Ontology, which is available as an OWL file at www.integromedb.org has been developed.
IntegromeDB Ontology (Fig. 2A) was developed by manual selection of 34 ontologies that reflect current knowledge of transcriptional regulation, from approximately 100 ontologies provided by the OBO consortium (www.bioontology.org). The selected ontologies include Sequence Ontology, GeneOntology, BioPAX, Disease Ontology, Chemical Ontology, the Functional Genomics Ontology, Phenotype and Trait Ontology and various others provided by the OBO consortium (www.bioontology.org). The selected ontologies were mapped to the BioNets ontology (Baitaluk et al., 2006a) in order to accommodate terms and inter-term relationships relevant to transcriptional regulation. The resulting IntegromeDB Ontology complies with the formal OWL, the World Wide Web Consortium standard (http://www.w3.org/TR/owl-ref/). IntegromeDB Ontology is a graph structure that is automatically generated by Protégé (http://protege.stanford.edu) and represents a semantic part of the IntegromeDB internal database, in which ontological classes form entities, and properties form attributes. IntegromeDB Ontology is a graph, on which graph-like operations such as finding k-neighbors, ancestors or descendants can be performed.
In OWL ontology, terms or classes are represented in hierarchies of ‘is a’ relationships, but the actual definitions are constructed from attributes assigned to each term/class. The OWL schema contains classes that are represented as Object Types in the schema of the IntegromeDB database (Fig. 2A). For example, the ontology class ‘TF’ is a subClassOf ‘Protein’. The general-purpose OWL schema (Fig. 2A) serves as a glue to hold different areas of biological knowledge together and is implemented so that any individual ontology describing another type of biological knowledge, for example, pharmacogenomics, can be introduced and modified with minimum impact on the rest of the system.
Each ontology class has a number of attributes in the form of ‘restriction onProperty value’: cardinality (which is the number of possible values for a property) and negation. For example, NFκB TF is associated with at least one (cardinality) binding site, yet NFκB has no (negation) transmembrane region. In this instance, the computer differentiates between, NFκB TFs and, for example, the ABCA4 transporter, which does not bind DNA and has transmembrane regions. This description logic definition matches accumulated knowledge, enabling automatic classification and management of heterogeneous data.
2.4 Data query
The search engine layer transforms the user query into actual search instructions and contains the following components:
Query processor, which manipulates the user's keyword queries into an internal form (query processor structure and internal query language will be published elsewhere);
Index manager, which uses the Apache Lucene indexing engine to create direct and inverted indexes of all integrated data sources and contains the methods required to create, update and access the indexes.
When the query results are ready, the module developed by the authors, called the BioWEB ranker, calculates the ‘importance’ of every returned object or ontology class. The ‘importance’ is measured as a weighted number of links an object or ontology class has to other objects and ontology classes. BioWEB implements the modified version of Google PageRank algorithm (Page et al., 1999) to sort results in terms of the ‘importance’ score.
The web page www.integromedb.org allows integrated data to be inspected without loading the BiologicalNetworks application. It provides the following querying possibilities: (i) simple keyword/ID search; (ii) wildcard search; (iii) multiple word structured search, such as ‘obesity AND/OR diabetes’, ‘obesity AND diabetes’ or ‘obesity OR diabetes’. Examples of queries are available at http://www.integromedb.org/tut0.php.
More extensive querying functionality is available in the BiologicalNetworks application, which can be downloaded at http://www.BiologicalNetworks.org. The specially designed query interface supports structured advanced queries to allow querying of any logical combination of bioentities, bioprocesses/relations and their properties. For example, the context query:
(geneID in (:like(NuclSequence, ANY)) ):gene.geneID, gene.enchancer
retrieves the set of objects (genes) that have attributes containing the specified query phrase ‘sequences of enhancers.’
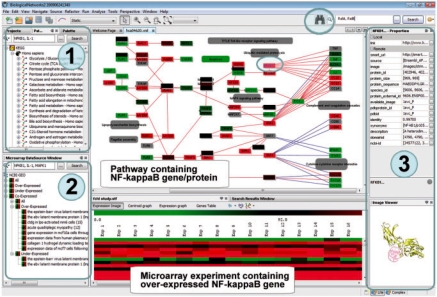
Other examples of context queries and queries by attributes and databases/datasets, can be found in Supplementary Material 1. The examples of queries provided are internal queries generated in response to queries constructed using the tool in the BiologicalNetworks application called ‘Comprehensive search by attributes’ (it is located in the upper right corner of the program and depicted by a binocular; see Fig. 4).
Fig. 4.
Screenshot of the BiologicalNetworks application providing access to IntegromeDB on the example of NFκB-1 query (complex layout). (1) Pathways window, showing the result of search of pathways containing NFκB-1 and IL-1 genes in human. To speed up the search, the user can first select a species, clicking the upper left look-up button. One of the selected pathways, Toll-like receptor signaling pathway, is displayed in the middle top window after selecting it using the right mouse button. (2) Microarray data window, showing the result of search in the GEO database of experiments that involved NFκB-1, IL-1, and MAPK1 genes. One of the selected experiments in which NFκB1, IL-1 and MAPK1 genes are over-expressed and co-expressed (Pearson's correlation >0.75) simultaneously is displayed in the middle bottom window. To do so, the user needs to select ‘Open’ in the menu, which is opened using the right mouse button. On the displayed pathway, the genes are automatically colored according to the expression at a particular time point or experiment (the user needs to select that in the pop-up menu, which appears after clicking the coloring tool on the upper tab above the microarray data window): red, up-regulated genes; green, down-regulated; gray, not present in the selected experiment. (3) Property window, displaying the selected object's (NFκB-1) properties collected from all integrated data sources.
Searching by sequence is under development. Section 3 ‘Integration of sequences with meta-graph data’ presents the proposed approach to the problem of querying the database by sequence.
2.5 Data provenance, reconciliation and consistency
Data are integrated into the system having been automatically collected from databases listed in the NAR repository (Cochrane and Galperin, 2010), using web crawling technologies. The full list of databases that have been integrated so far is provided at http://www.BiologicalNetworks.net/Database/tut5.php.
Data from databases that have been already integrated in the system are updated monthly. In addition, the web crawlers are continuously searching for and adding data from databases that have not yet been integrated into the system. Web crawlers are guided by the NAR Database depository (Cochrane and Galperin, 2010), which currently lists more than 1200 databases; 102 of which have been integrated in IntegromeDB on 02/10/2010, including all databases in the category ‘transcriptional regulator sites and TFs’. Mass integration of databases from other categories is a subject of data storage availability (the current size of IntegromeDB exceeds 5TB). Current statistics concerning integrated data by category are provided at www.integromedb.org. Also, statistics calculated by contribution of integrated data sources can be found at http://www.integromedb.org/stat.php.
To address the problem of data cleaning and conflict resolution, reconciliation procedures that identify controversies or inconsistencies in data have been developed. Examples of data inconsistencies include, but are not limited to, the following: (i) two different genes being assigned to the same synonym; (ii) two genes with the same name pointing to different chromosomal locations; (iii) two genes with different names pointing to the same chromosomal location; and (iv) different objects having names with a common string; for example, p53, p53(361–393), p53(modified:Thr:212) and pCMX-mutant-p53. These inconsistencies should be resolved by a curator, but owing to limited human resources and fully automated data integration, human intervention does not occur; details on retrieved properties by data sources can be viewed for each gene/protein by clicking ‘Details by Data Sources’ on the query result page at http://www.integromedb.org.
To evaluate the quality of integrated data, inconsistencies in the databases that were integrated in IntegromeDB were estimated. Since calculation of all inconsistencies among gene/protein IDs only, including their synonyms, would require more than 1013 all-against-all comparisons, the number of inconsistencies was estimated. Four databases, namely GeneCards, String, BIND and Uniprobe, which are the largest contributors of mammalian gene/protein IDs being integrated, were selected. All genes/proteins from the 10 largest genomes were selected from these databases, and for each gene/protein from the final dataset, comprising ∼400 000 IDs, bidirectional occurrences of names and synonyms in the databases were calculated. Inconsistencies were found in ∼3% of genes/proteins (∼12 000) and are documented at http://www.integromedb.org. This level of inconsistency nearly corresponds to that expected from manually curated data; users should expect ∼3% of retrieved data to be inconsistent when querying IntegromeDB.
3 INTEGRATION OF SEQUENCES WITH META-GRAPH DATA
Data represented by graphs, histograms and trees—interaction networks, 3D structures of node proteins, expression values of mRNA products and molecular interaction types—can be integrated into the labeled meta-graph database in a straightforward manner (Fig. 2B; see also Fig. 2 in Baitaluk et al., 2006b). However, integrating meta-graph data and sequence data requires superimposing meta-data on genomic sequence elements to create multiple annotations for the genomic sequences. This is not a trivial task owing to the orthogonal nature of integrated data that are represented by sequences, graphs and time/value dependencies.
Herein is a description of an approach to the problem of integrating sequences with meta-graph data. Specifically, Relational Interval (RI)-tree structures that are used for navigation through sequences, including scroll upstream/downstream, get_next gene/operon/chromosome, and annotation of multiple overlapping sequences are described. However, a description of the suffix tree structures used for sequence searches is beyond the scope of this article. It should be noted that features described in this section are currently implemented at the database level and are available as binaries upon request; their implementation at the level of the user interface will be described elsewhere.
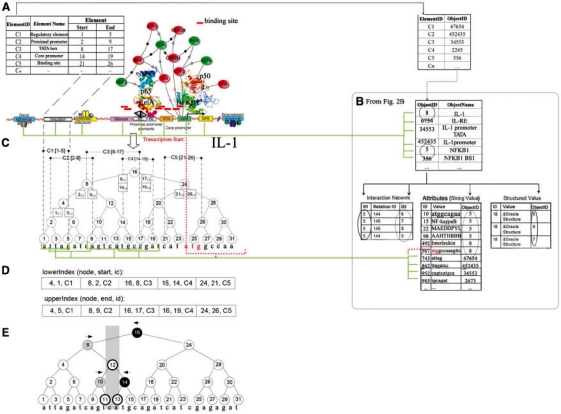
Sequences (genomic/protein) are integrated with meta-graph data using an ElementId–ObjectID connection table (Fig. 3A). Where elements are sequence elements, for example, a core promoter, TATA box or binding site, they are attributed to a particular gene by means of known localization in the gene, according to the GeneBank global position. Internal enumerations in integrated databases such as TRANSFAC provides localization of a regulatory region in respect to the transcription start and are recalculated accordingly. The connection table assigns sequence elements to meta-graph objects, so that sequence elements, represented as a RI-tree structure, become graph objects within the meta-graph database. All heterogeneous data, which are integrated in the meta-graph database, appear to be mapped on genomic intervals and vice versa. As a result, DNA sequences, molecular interaction graphs, 3D protein structures, images of expression, and other meta-data become annotated within the same context.
Fig. 3.
Integration of genomic sequences with meta-graph data. (A) Integrated tables and visualization of the final integrated product on the sequence of IL-1 gene (see Fig. 2). (B) Connection of MetaGraph part of the database (from Fig. 2B) to Sequence part of the database. (C) Five sample intervals (regulatory regions) from the IL-1 upstream region for construction of an example RI-tree. Virtual backbone of the RI-tree and registration positions are depicted. (D) Relational indexes lowerIndex and upperIndex. (E) Query preparation step for the query interval (11, 13) (shaded in gray): leftQueries (8, 10), rightQueries (14, 16) and innerQueries (11, 13). The color figure was generated in BiologicalNetworks.
Figure 3 demonstrates how five sample intervals on the sequence of the IL-1 gene are represented by an RI-tree and how navigation queries are processed for such a tree. Let us consider five intervals: C1(1, 5), C2(2, 9), C3(8, 17), C4(14, 19) and C5(21, 26) (Fig. 3A and B). The virtual backbone with root value 16 covers the data space from 1 to 31 nt (Fig. 3C). The five intervals are registered at the nodes 4, 8, 16 and 24, respectively. The interval (1, 5) is represented by the entries 4, 1 and C1 in the lowerIndex and by 4, 5 and C1 in the upperIndex, as 4 is the registration node and 1 and 5 are the start and end points of the interval, respectively (Fig. 3D).
To process an interval intersection query (start, end) based on the RI-tree, two phases are distinguished, the query preparation phase and the declarative query processing phase. The first phase descends the virtual backbone from the root node down to the start and the end, respectively (Fig. 3E). The traversal is performed arithmetically, and the visited nodes are collected in two different main-memory tables, leftQueries and rightQueries, both obeying the unary relational schema (node). Nodes to the left of the start could contain intervals that overlap the start and are inserted into leftQueries. Nodes to the right of the end could contain intervals that overlap the end and are inserted into rightQueries. Where these nodes are taken from the paths, the set of all nodes between the start and the end belong to the innerQuery, which is represented by a single range query on the node values. All intervals registered at the nodes from the innerQuery are guaranteed to intersect the query and will, therefore, be reported without further comparison. The query preparation phase is performed entirely in the main memory with no I/O operations.
In the second phase, transient tables are joined with relational indexes upperIndex and lowerIndex, as follows:
SELECT id FROM upperIndex AS i JOIN :leftQueries USING (node) WHERE i.end >= :start
UNION ALL
SELECT id FROM lowerIndex AS i JOIN :rightQueries USING (node) WHERE i.start <= :end
UNION ALL
SELECT id FROM lowerIndex // or upperIndex WHERE node BETWEEN :start AND :end
The end point of each interval registered at the nodes in leftQueries is compared with the start, and the start point of each interval in rightQueries is compared with the end. The innerQuery corresponds to a simple range scan over the intervals with the nodes in the interval between the start and the end.
4 DATA ACCESS AND SYSTEM EVALUATION
This section describes how data in IntegromeDB can be accessed and provides several examples of application of the system.
IntegromeDB is accessible through the BiologicalNetworks application, which can be downloaded at http://www.BiologicalNetworks.org. The web page www.integromedb.org has been developed to allow the user a quick inspection of integrated data for specific genes/proteins without loading the BiologicalNetworks application.
4.1 Querying IntegromeDB web page
The web page www.integromedb.org provides keyword/ID, wildcard and multiple word search capabilities, statistics on integrated data by category and database, information relating to retrieved properties by data sources for each gene/protein that can be accessed from the query result page, and data inconsistencies in public data. The web site was designed primarily for the purpose of giving the user an opportunity to look at integrated data rather than to provide complex data analysis capabilities, which are implemented in the BiologicalNetworks application.
The remainder of this section, explores the IntegromeDB web site search capabilities using two example queries: ‘relb AND diabetes AND Alzheimer’ and ‘rela AND diabetes AND Alzheimer’. RelA (p65) and RelB TFs belong to the family of NFκB factors; they can form heterodimers with other NFκB factors, p50 (NFκ B-1), p52 (NFκB-2), c-Rel, and with each other, and RelA can form homodimers. RelA is activated in the classical/canonical NFκB activation pathway that is stimulated by pro-inflammatory cytokines, such as TNF-α and IL-1, and pathogen-associated molecular patterns (Hoffmann et al., 2006). In addition, RelB is released in the alternative/non-canonical pathway that is activated by other cytokines. The canonical and non-canonical pathways have distinct regulatory functions: the canonical pathway is involved in innate immunity and cell survival; the non-canonical pathway is important in adaptive immunity, lymphoid organ development and B-cell maturation (Bonizzi and Karin, 2004). However, inflammatory processes involving both the canonical and non-canonical NFκB activation pathways directly underlie insulin resistance in peripheral tissues and astrocytes in the brain and play an essential role in the etiology of Alzheimer's disease and diabetes mellitus (Granic et al., 2009).
The first query, ‘relb AND diabetes AND Alzheimer’, returns four genes: Tnf, ESR1, CD40 and AhR. In comparison, EB-eye search (http://www.ebi.ac.uk; Jones et al., 2008) for the same query returns no entries from any database and Entrez (Sayers et al., 2009) returns three genes: Tnf, ESR1 and CD40. Aryl hydrocarbon receptor (AhR) protein, which was returned by IntegromeDB but not by Entrez, interacts with RelB, and Ahr:RelB dimers regulate transcription of many genes, functioning as coordinators of inflammatory responses (Vogel et al. 2007; Vogel and Matsumura, 2009). Therefore, AhR relates to RelB, diabetes and Alzheimer's disease. One reason that Entrez did not return the AhR gene is that it searches keywords, publications and gene properties; while IntegromeDB searches relations between publications to the database objects (genes) associated with query words.
The second query, ‘rela AND diabetes AND Alzheimer’, returns six genes: Tnf, K60 (IL-8), PTGS2, BAX, STMY3 and Mlana. Entrez returns nine genes: PTGS2, Tnf (mouse), IL8, TNF, TP53, SIRT1, PRKCD, ESR1 and PRKACA. The query results of Entrez and IntegromeDB only have three common genes, but this can be explained by the fact that Entrez is more up to date. However, IntegromeDB returned three genes that were not found in Entrez: BAX, STMY3, and Mlana. Therefore, we investigated these three genes, searching the query result pages for the query words.
BAX (Bcl2-associated protein X) gene expression is regulated by NFκB factors, specifically RelA (Grimm et al., 2005). It was demonstrated that patients with diabetes (Varo et al., 2003) and those with Alzheimer's disease (Ait-ghezala et al., 2008) have a pro-inflammatory state indicated by elevated levels of plasma sCD40L. In addition, BAX mRNA levels are altered in peripheral blood mononuclear cells from individuals with mild cognitive impairment and Alzheimer's patients (Gatta et al., 2009).
STMY3 (Stromelysin-3 precursor) gene expression is associated with the expression of p53 in various cancers (Sharma et al., 2004). p53 is directly regulated by RelA (Jeong et al., 2004). Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE, were reported in brain from subjects with amnestic mild cognitive impairment and Alzheimer's disease (Cenini et al., 2008). In addition, polymorphisms in p53 are known to be associated with diabetes (Szoke et al., 2009).
Mlana (Melan-A protein, MART-1) stimulates T-cells to increase secretion of TNF-α (Elluru et al., 2008), which is a direct target of RelA (Shakhov et al., 1990). Expression of TNF-α increases in both diabetes (Gordin et al., 2008) and Alzheimer's disease (Baranowska-Bik et al., 2008).
Therefore, the three genes considered above are directly or indirectly associated with RelA, Alzheimer's disease and diabetes. The fact that IntegromeDB found these genes, while Entrez did not supports the aforementioned statement that IntegromeDB approaches integration of data and searches differently from Entrez. In particular, IntegromeDB integrates data objects (genes) and performs searches by object properties rather than searching keywords in publications. The examples considered clearly demonstrate the power of the proposed approach: novel knowledge concerning gene-disease associations was obtained using IntegromeDB in a matter of minutes, and no other system could reveal those associations.
4.2 BiologicalNetworks application
BiologicalNetworks serves as an environment for navigating, visualizing and analyzing integrated data. Within the application, the user can search pathways [Fig. 4(1)], microarray experiments [Fig. 4(2)] and data on transcriptional regulation [Fig. 4(3)]. Figure 4 demonstrates how these three types of search can be integrated and visualized: first, select the Toll-like receptor signaling pathway that contains NFκB1 and IL-1 genes [Fig. 4(1)]; second, select a microarray experiment that involved NFκB1, IL-1 and MAPK1 genes, and color the genes on the pathway according to their color on the microarray image [this was done using the tool on the top panel of the window that displays the microarray image, Fig. 4(2)]; third, select the NFκB1 gene on the pathway and explore the properties of this gene/protein that have been collected from all integrated databases [Fig. 4(3)]. The properties of any gene/protein available in the integrated resources can be explored without examining pathways or microarray data; the user can perform a simple search in the search window or a complex search by attributes using the tool ‘Search by attribute value’ located in the top right corner of the application.
The rest of this section explains how to use BiologicalNetworks to find murine genes that are common targets of c-Rel and RelA TFs and contain experimentally identified binding sites. In addition, the types of cells expressing the genes, the signals to which the genes respond and co-expression of the genes can be identified. There are many ways to use the application in order to find the aforementioned data, and highlighted below is one such method.
First, the TFs of interest must be identified. In the top right corner of the application select ‘Mus Musculus’ using a look-up menu on the left of the search window, type ‘rela, c-rel’ (comma means ‘OR’, ‘c-Rel OR RelA’) in the search window and click ‘Search’. The search results are shown in the middle bottom ‘Search Result Window’, populating the IntegromeDB folder.
Second, target genes must be identified. In the result search window, select the proteins with the names Rela and Rel (c-Rel) (use Ctrl+left mouse to make the selection) and using the right mouse button select ‘Build Pathway’ in the pop-up window to open the ‘Build Pathway Wizard’. In the wizard, select the option ‘Find targets for selected nodes’. The network of nodes appears in the pathway window. The network can be zoomed and moved using the mouse. In addition, in this window each node and its attributes can be explored using the tool for node selection on the above panel, depicted by an arrow. Select, using the tool, all nodes that are common for c-Rel and RelA (when selected, the nodes are colored blue).
Third, identify from the common targets (nodes selected at the previous step) those with regulatory regions containing experimentally identified binding sites. When the pathway window is active, click on the image of the binocular in the right-hand corner to launch the tool ‘Search by attribute value’. In the tool, expand (do not select) ‘entity’, then ‘organism’, then ‘physicalEntity’ and select ‘protein’. The tool will load attribute types for selected entities (time of loading depends on how many attributes the selected entity has). Attribute types can be sorted by name. Select, by clicking on the right box depicting the plus sign, the attributes starting with ‘bs’, such as ‘bs_name’ (at the moment of writing this text, nine such attributes appeared), ‘tf’, such as ‘tfbs_name’ (four attributes), and the attribute ‘the_sequence_of_the_binding_site’. Expand ‘relation’ then ‘interaction’, and select ‘interaction_transcription’ to load attributes for this entity. Select all attributes starting with ‘binding’, such as ‘binding_motif’ and ‘binding_seq_cis_elements’. Altogether, we selected 18 attribute types, applied to them the common expression ‘not empty/exist’ and common operator ‘OR’. Click ‘OK’ in the tool. The search gives the following result: among nine common targets of c-Rel and RelA, two genes, recombination activating gene 2 (Rag2) and interleukin 12b (IL-12b) have information on TFBSs that has been integrated in IntegromeDB. Rag2 and IL-12b are known NFκB targets (Murphy et al. 1995; Verkoczy et al., 2005).
Fourth, identify microarray experiments in which Rag2 and IL-12b are co-expressed. Type in the search window for microarray experiments [Fig. 4(2)] ‘rag2, il-12b’, select ‘Mus Musculus’ in the look-up window on the left of the search window, select an option to search for co-regulated genes only on the right of the search window. At the time of writing, the search gave 16 GEO profiles to browse through.
To the best of the authors' knowledge, there is no system in the public domain that allows a similar type of search to be executed; that is, to find TF targets, regulators or interacting partners associated with specific attributes gathered from more than a hundred databases. To improve searching and navigation through all integrated data, a new, scenario-oriented navigation interface in BiologicalNetworks is currently under development.
5 DISCUSSION
As far as we are aware, no current integration solution addresses the overlapping nature of integrated data. The majority of existing solutions achieve horizontal integration; data sources are treated as complementary to one another, and issues associated with data aggregation are ignored. The approach proposed here and implemented in the IntegromeDB system allows an integrated warehouse of data to be created from various databases and files in different formats, including web pages.
Unlike traditional warehouses such as Atlas (Shah et al., 2005), BNDB (Kuntzer et al., 2007) and GUS (Davidson et al., 2001) that employ star and snowflake models over relational data, IntegromeDB employs a graph-based model (Baitaluk et al., 2006a) that has been developed for integrating interaction networks. The graph-based model allows natural integration of genomic sequences, which are represented as RI-trees, with graph-structured data such as gene interaction graphs, ontologies, taxonomies and protein classifications. The data model means that IntegromeDB is scalable in respect to the number of integrated data, allowing more resources to be integrated than other systems such as cPath (Cerami et al., 2006), ONDEX (Kohler et al., 2006), BIOZON (Birkland and Yona, 2006) and BNDB (Kuntzer et al., 2007) (Table 1).
Table 1.
Comparison of IntegromeDB with integration systems: cPath (Cerami et al., 2006), ONDEX (Kohler et al., 2006), BIOZON (Birkland and Yona, 2006) and BNDB (Kuntzer et al., 2007)
| cPath | ONDEX | BIOZON | BNDB | IntegromeDB | |
|---|---|---|---|---|---|
| Scalability to the number of data types | no | yes | *a | no | yes |
| Number of integrated databases | 8 | 25 | 20 | 10 | > 100 |
| Ad hoc queries | *c | no | no | no | yes |
| Integration engine | no | yes | no | no | yes |
| Sequence annotation | no | no | no | no | yes |
| Sequence search | no | no | no | no | yes |
| OBO integration | no | yes | no | no | yes |
| Multidimensional datae | *b | *b | no | no | yes |
| Web search | yes | no | yes | *d | yes |
| Research environment | Cytoscape | Ondex | no | no | BiologicalNetworks |
| Open/easy access | yes | *d | yes | *d | yes |
aOnly 10 object types are presented.
bGene expression data only.
cWeb interface for predefined queries exists, but no interface for ad hoc queries.
dAvailable only after registration and sign up.
eMultidimensional data is represented by Time/Value, Value/Space, Time/Value/Space, etc. dependencies, for example microarray gene expression matrixes, protein abundance data, chemical concentration in the cell, etc.
One of the advantages of the IntegromeDB architecture is that its generic internal data model allows annotation and the querying of genomic sequences as well as other meta-data (this feature is not yet available at the level of the user interface). Four integrations systems, cPath, ONDEX, BIOZON and BNDB, which are conceptually similar to IntegromeDB, do not present sequence annotation and queries by sequence (Table 1). The problem of integrating sequences with meta-graph data was addressed by implementing sequence navigation and annotation using RI-tree structures, and sequence searching using suffix tree structures (Gusfield 1997; Farach-Colton et al. 2000; Giegerich et al., 2003).
Ontology-driven data integration and mapping strengthen the proposed approach. Out of the four integration systems that were compared with IntegromeDB, integration with OBO is only provided in ONDEX (Table 1). The ontology-driven approach adopted in the proposed system provides advantages over traditional databases as it allows data integration processes to be automated. However, several limitations exist including the need for human intervention. These limitations predominantly arise from inconsistencies in ontologies and their periodic changes and revisions, reflecting the current state of scientific knowledge. This means that human intervention is unlikely to be eliminated in the near future.
IntegromeDB can be considered as a mixture of two approaches, Data Integration in its classical sense and the Semantic Web. The Semantic Web technologies, such as the RDF (http://www.w3.org/RDF/) and the OWL (http://www.w3.org/TR/owl-ref/), have the potential to add a new dimension to data integration in systems biology, which is expected to adopt these technologies (Ruttenberg et al., 2007). However, one major problem with the Semantic Web is the lack of semantic content; the majority of biological information is either not semantically codified or is codified with poor axiomatization (Egaña, 2008). This means that using the ‘pure’ Semantic Web approach is still problematic (Good and Wilkinson, 2006). Several mechanisms to address the problems of semantic codification, such as resolving biological identifiers, have been proposed and include OKKAM IDs (http://www.okkam.org/), MIRIAM URIs (Laibe and Le Novere, 2007), LSIDs (http://lsrn.org), URIs (http://bio2rdf.wiki.sourceforge.net/Banff+Manifesto) and shared names (http://neurocommons.org/page/Shared_names). In the IntegromeDB system, biological identifiers are resolved by mapping external identifiers to internal identifiers using IntegromeDB ontology and filtering duplicates; this procedure is maximally automated, obviating the need for significant human intervention.
IntegromeDB is integrated into a research environment and has an open-access web-search interface. Data integrated in IntegromeDB are accessible through the integrated research environment BiologicalNetworks at www.BiologicalNetworks.org and the web-search interface at http://integromedb.org. IntegromeDB will evolve by expanding the scope of data and improving the user interface. The IntegromeDB has a general purpose graph architecture and is data-type neutral, and there is the prospect of further data integration of orthogonal sources of information such as chemical and pharmacological data from PharmGKB, microarray data from ArrayExpress, disease data from OMIM, and others. Further development of the user interface will be focused on implementing sequence searches, navigation and annotation, slick representation of integrated data and more intuitive and scenario-focused navigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Sedova and A. Gupta for valuable discussions and assistance in creation of IntegromeDB and W. Bluhm for critical reading of the manuscript.
Funding: National Institutes of Health (R01GM084881 to M.B., R01GM085325 to J.P.).
Conflict of Interest: none declared.
REFERENCES
- Ait-ghezala G, et al. Diagnostic utility of APOE, soluble CD40, CD40L, and Abeta1-40 levels in plasma in Alzheimer's disease. Cytokine. 2008;44:283–287. doi: 10.1016/j.cyto.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Baitaluk M, et al. PathSys: integrating molecular interaction graphs for systems biology. BMC Bioinformatics. 2006a;7:55. doi: 10.1186/1471-2105-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baitaluk M, et al. BiologicalNetworks: visualization and analysis tool for systems biology. Nucleic Acids Res. 2006b;34:W466–W471. doi: 10.1093/nar/gkl308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowska-Bik A, et al. Plasma beta amyloid and cytokine profile in women with Alzheimer's disease. Neuro Endocrinol. Lett. 2008;29:75–79. [PubMed] [Google Scholar]
- Baumbach J. CoryneRegNet 4.0—A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The Nucleic Acid Database. Acta Crystallogr. D Biol. Crystallogr. 2002;58:889–898. doi: 10.1107/s0907444902003487. [DOI] [PubMed] [Google Scholar]
- Birkland A, Yona G. BIOZON: a system for unification, management and analysis of heterogeneous biological data. BMC Bioinformatics. 2006;7:70. doi: 10.1186/1471-2105-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Cenini G, et al. Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE, in brain from subjects with amnestic mild cognitive impairment and Alzheimer's disease. J. Cell. Mol. Med. 2008;12:987–994. doi: 10.1111/j.1582-4934.2008.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, et al. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelala C, et al. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane GR, Galperin MY. The 2010 Nucleic Acids Research Database Issue and online Database Collection: a community of data resources. Nucleic Acids Res. 2010;38:D1–D4. doi: 10.1093/nar/gkp1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SB, et al. K2Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst. J. 2001;40:512–530. [Google Scholar]
- Davuluri RV, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R. FlyBase: a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- Egaña M, et al. In situ migration of handcrafted ontologies to reason-able forms. Data Knowl. Eng. 2008;66:147–162. [Google Scholar]
- Elluru SR, et al. Induction of maturation and activation of human dendritic cells: a mechanism underlying the beneficial effect of Viscum album as complimentary therapy in cancer. BMC Cancer. 2008;8:161. doi: 10.1186/1471-2407-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold T, Argos P. SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- Farach-Colton M, et al. On the sorting-complexity of suffix tree construction. J. Assoc. Comput. Mach. 2000;47:987–1011. [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta L, et al. Peripheral blood mononuclear cells from mild cognitive impairment patients show deregulation of Bax and Sod1 mRNAs. Neurosci. Lett. 2009;453:36–40. doi: 10.1016/j.neulet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegerich R, et al. Efficient implementation of lazy suffix trees. Softw. Pract. Exp. 2003;33:1035–1049. [Google Scholar]
- Good BM, Wilkinson MD. The Life Sciences Semantic Web is full of creeps! Brief. Bioinform. 2006;7:275–286. doi: 10.1093/bib/bbl025. [DOI] [PubMed] [Google Scholar]
- Gordin D, et al. Acute hyperglycaemia induces an inflammatory response in young patients with type 1 diabetes. Ann. Med. 2008;40:627–633. doi: 10.1080/07853890802126547. [DOI] [PubMed] [Google Scholar]
- Granic I, et al. Inflammation and NF-kappaB in Alzheimer's disease and diabetes. J. Alzheimers Dis. 2009;16:809–821. doi: 10.3233/JAD-2009-0976. [DOI] [PubMed] [Google Scholar]
- Griffith OL, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm T, et al. EBV latent membrane protein-1 protects B cells from apoptosis by inhibition of BAX. Blood. 2005;105:3263–3269. doi: 10.1182/blood-2004-07-2752. [DOI] [PubMed] [Google Scholar]
- Grumbling G, Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield D. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press; 1997. [Google Scholar]
- Haider S, et al. BioMart Central Portal—unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoso A, et al. TrSDB: a proteome database of transcription factors. Nucleic Acids Res. 2004;32:D171–D173. doi: 10.1093/nar/gkh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata A, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, et al. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Hu Z, et al. Towards zoomable multidimensional maps of the cell. Nat. Biotechnol. 2007;25:547–554. doi: 10.1038/nbt1304. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SJ, et al. HTLV-I Tax induces a novel interaction between p65/RelA and p53 that results in inhibition of p53 transcriptional activity. Blood. 2004;104:1490–1497. doi: 10.1182/blood-2003-12-4174. [DOI] [PubMed] [Google Scholar]
- Jones P, et al. PRIDE: new developments and new datasets. Nucleic Acids Res. 2008;36:D878–D883. doi: 10.1093/nar/gkm1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J, et al. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics. 2006;22:1383–1390. doi: 10.1093/bioinformatics/btl081. [DOI] [PubMed] [Google Scholar]
- Kolchanov NA, et al. Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res. 2002;30:312–317. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchanov NA, et al. Integrated databases and computer systems for studying eukaryotic gene expression. Bioinformatics. 1999;15:669–686. doi: 10.1093/bioinformatics/15.7.669. [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzer J, et al. BNDB—the Biochemical Network Database. BMC Bioinformatics. 2007;8:367. doi: 10.1186/1471-2105-8-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laibe C, Le Novere N. MIRIAM resources: tools to generate and resolve robust cross-references in systems biology. BMC Syst. Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, et al. ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TL, et al. Regulation of interleukin 12 p40 expression through an NF-kappa B half-site. Mol. Cell. Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L, et al. Technical Report. Stanford InfoLab; 1999. The PageRank citation ranking: bringing order to the web. [Google Scholar]
- Pruess M, et al. The Integr8 project—a resource for genomic and proteomic data. In Silico Biol. 2005;5:179–185. [PubMed] [Google Scholar]
- Ruttenberg A, et al. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8:S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, et al. Atlas—a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhov AN, et al. Structural analysis of the rabbit TNF locus, containing the genes encoding TNF-beta (lymphotoxin) and TNF-alpha (tumor necrosis factor) Gene. 1990;95:215–221. doi: 10.1016/0378-1119(90)90364-w. [DOI] [PubMed] [Google Scholar]
- Sharma R, et al. Prognostic significance of stromelysin-3 and tissue inhibitor of matrix metalloproteinase-2 in esophageal cancer. Oncology. 2004;67:300–309. doi: 10.1159/000081331. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. ISYS: a decentralized, component-based approach to the integration of heterogeneous bioinformatics resources. Bioinformatics. 2001;17:83–94. doi: 10.1093/bioinformatics/17.1.83. [DOI] [PubMed] [Google Scholar]
- Subramaniam S. The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins. 1998;32:1–2. [PubMed] [Google Scholar]
- Szoke D, et al. Polymorphisms of the ApoE, HSD3B1, IL-1beta and p53 genes are associated with the development of early uremic complications in diabetic patients: results of a DNA resequencing array study. Int. J. Mol. Med. 2009;23:217–227. [PubMed] [Google Scholar]
- Varo N, et al. Elevated plasma levels of the atherogenic mediator soluble CD40 ligand in diabetic patients: a novel target of thiazolidinediones. Circulation. 2003;107:2664–2669. doi: 10.1161/01.CIR.0000074043.46437.44. [DOI] [PubMed] [Google Scholar]
- Verkoczy L, et al. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes. Immunity. 2005;22:519–531. doi: 10.1016/j.immuni.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem. Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CF, et al. Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem. Biophys. Res. Commun. 2007;363:722–726. doi: 10.1016/j.bbrc.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.