Abstract
Summary: We present a suite of Unix shell programs for processing any number of phylogenetic trees of any size. They perform frequently-used tree operations without requiring user interaction. They also allow tree drawing as scalable vector graphics (SVG), suitable for high-quality presentations and further editing, and as ASCII graphics for command-line inspection. As an example we include an implementation of bootscanning, a procedure for finding recombination breakpoints in viral genomes.
Availability: C source code, Python bindings and executables for various platforms are available from http://cegg.unige.ch/newick_utils. The distribution includes a manual and example data. The package is distributed under the BSD License.
Contact: thomas.junier@unige.ch
1 INTRODUCTION
Phylogenetic trees are a fundamental component of evolutionary biology, and methods for computing them are an active area of research. Once computed, a tree may be further processed in various ways (Table 1). Small datasets consisting of a few trees of moderate size can be processed with interactive GUI programs. As datasets grow, however, interactivity becomes a burden and a source of errors, and it becomes impractical to process large datasets of hundreds of trees and/or very large trees without automation.
Table 1.
Selected Newick utilities programs and their functions
| Program | Function |
|---|---|
| nw_clade | Extracts clades (subtrees), specified by labels |
| nw_distance | Extracts branch lengths in various ways (from root, from parent, as matrix, etc.) |
| nw_display | Draws trees as ASCII or SVG (suitable for further editing for presentations or publications), several options |
| nw_match | Reports matches of a tree in a larger tree |
| nw_order | Orders tree nodes, without altering topology |
| nw_rename | Changes node labels |
| nw_reroot | Reroots trees on an outgroup, specified by labels |
| nw_trim | Trims a tree at a specified depth |
| nw_topology | Retains topological information |
SVG, Scalable vector graphics.
Automation is facilitated if the programs that constitute an analysis pipeline can easily communicate data with each other. One way of doing this in the Unix shell environment is to make them capable of reading from standard input and writing to standard output—such programs are called filters.
Although there are many automatable programs for computing trees [e.g. PhyML (Guindon and Gascuel, 2003), PHYLIP (Felsenstein, 1989)], programs for processing trees [e.g. TreeView (Page, 2002), iTOL (Letunic and Bork, 2007)] are typically interactive. Here, we present the Newick utilities, a set of automatable filters that implement the most frequent tree-processing operations.
2 RESULTS
The Newick utilities have the following features:
no user interaction is required;
input is read from a file or from standard input; output is written to standard output;
all options are passed on the command line (no control files);
the input format is Newick (Archie et al., 1986);
the output is in plain text (Newick, ASCII graphics or SVG);
there are no limits to the number or size of the input trees;
each program performs one function, with some variants; and
the programs are self-documenting (option -h).
2.1 Example: Bootscanning
Bootscanning (Salminen, 1995) locates recombination breakpoints by identifying (locally) closest relatives of a reference sequence. An example implementation is as follows:
produce a multiple alignment of all sequences, including the reference;
divide the alignment into equidistant windows of constant size (e.g. 300 bp every 50 bp);
compute a maximum-likelihood tree for each window;
root the trees on the appropriate outgroup (not the reference);
from each tree, extract the distance (along the tree) from the reference to each of the other sequences; and
plot the result (Fig. 1).
Fig. 1.
Bootscanning using PhyML, EMBOSS, Muscle, Newick utilities, GNUPlot and standard Unix shell programs. The species with the lowest distance is the reference's nearest neighbor (by distance along tree branches). A recombination breakpoint is predicted near position 450, as the nearest neighbor changes abruptly.
The distribution includes a Bash script, bootscan.sh, that performs the procedure with Muscle (Edgar, 2004) (Step 1), EMBOSS (Rice et al., 2000) (Step 2), PhyML (Step 3), GNUPlot (Step 6) and Newick utilities for Steps 4 and 5. This method was used to detect breakpoints in human enterovirus (Tapparel et al., 2007).
3 DISCUSSION
The Newick utilities add tree-processing capabilities to a shell user's toolkit. Since they have no hard-coded limits, they can handle large amounts of data; since they are non-interactive, they are easy to automate into pipelines, and since they are filters, they can easily work with other shell tools.
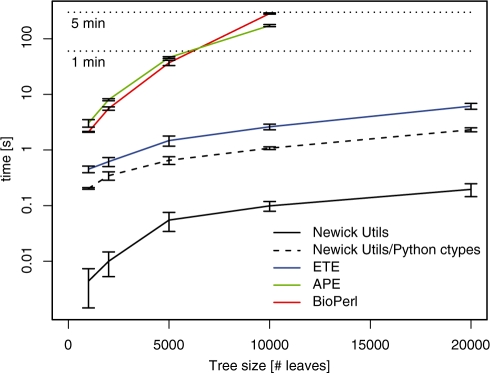
Tree processing may also be programmed using a specialized package [e.g. BioPerl (Stajich et al., 2002), APE (Paradis et al., 2004) or ETE (Huerta-Cepas et al., 2010)], but this implies knowledge of the package, and such programs tend to be slower and use more resources than their C equivalents. The difference is particularly apparent for large trees (Fig. 2).
Fig. 2.
Average run times (10 samples) of rerooting tasks on various tree sizes in different implementations. The task involved reading, rerooting and printing out the tree as Newick. Runs of the BioPerl and APE implementation on the 20 000-leaf tree did not complete. Error bars show 1 SD. Computer: 3 GHz 64 bit Intel Core 2 Duo, 1 GB RAM, Linux 2.6. Made with R (R Development Core Team, 2008).
3.1 Python bindings
To combine the advantages of a high-level, object-oriented language for the application logic with a C library for fast data manipulation, one can use the Newick utilities through Python's ctypes module. This allows one to code a rerooting program in 25 lines of Python while retaining good performance (Fig. 2). A detailed example is included in the documentation.
Some users will feel more at ease working in the shell or with shell scripts, using existing bioinformatics tools; others will prefer to code their own tools in a scripting language. The Newick utilities are designed to meet the requirements of both.
ACKNOWLEDGEMENTS
We wish to thank the members of the E.Z. group for feedback and beta testing.
Funding: The Infectigen Foundation; Swiss National Science Foundation (grant 3100A0-112588 to E.Z.).
Conflict of Interest: none declared.
REFERENCES
- Archie J, et al. 1986. http://evolution.genetics.washington.edu/phylip/newicktree.html. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Page R. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0602s01. Chapter 6, Unit 6.2. [DOI] [PubMed] [Google Scholar]
- Paradis E, et al. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Rice P, et al. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0. [Google Scholar]
- Salminen M. Identification of breakpoints in intergenotypic recombinants of HIV type I by bootscanning. AIDS Res. Hum. Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C, et al. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics. 2007;8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]