Abstract
Summary: Dynamic programming (DP) is a general optimization strategy that is successfully used across various disciplines of science. In bioinformatics, it is widely applied in calculating the optimal alignment between pairs of protein or DNA sequences. These alignments form the basis of new, verifiable biological hypothesis. Despite its importance, there are no interactive tools available for training and education on understanding the DP algorithm. Here, we introduce an interactive computer application with a graphical interface, for the purpose of educating students about DP. The program displays the DP scoring matrix and the resulting optimal alignment(s), while allowing the user to modify key parameters such as the values in the similarity matrix, the sequence alignment algorithm version and the gap opening/extension penalties.
We hope that this software will be useful to teachers and students of bioinformatics courses, as well as researchers who implement the DP algorithm for diverse applications.
Availability and Implementation: The software is freely available at: http:/melolab.org/sat. The software is written in the Java computer language, thus it runs on all major platforms and operating systems including Windows, Mac OS X and LINUX.
Contact: All inquiries or comments about this software should be directed to Francisco Melo at fmelo@bio.puc.cl
1 INTRODUCTION
Dynamic programming (DP) algorithm was first formalized by the mathematician Richard Bellman in the early 1950s (Bellman, 1952). Since then, DP has been adapted to solve many optimization problems in different areas of science (Bellman, 1966). DP, in various forms, constitutes the core of bioinformatics. It is central to several software applications, such as those comparing sequences and structures of proteins, RNA and DNA (Sankoff, 2000). The pivotal role of DP algorithm in Bioinformatics has led it to be considered as an essential topic of any undergraduate and graduate course in bioinformatics. Despite the simplicity of its formulation, DP algorithm is a subject that is not easy for the students to learn. Extensive practice is essential for students to fully appreciate it workings.
Given its importance, and its mandatory presence in bioinformatics courses, there are not interactive computer tools designed to help students to understand the specific details of the DP algorithm. We have addressed this need by developing Sequence Alignment Teacher (SAT), a simple software application, which interactively calculates the DP matrix in real time. SAT also visually illustrates the consequences of tweaking the various parameters of the algorithm. We hope this simple software tool will not only be useful for teachers and students of bioinformatics courses but also to future bioinformatics developers as an error check tool when they start implementing this algorithm for the first time.
2 SOFTWARE DESCRIPTION
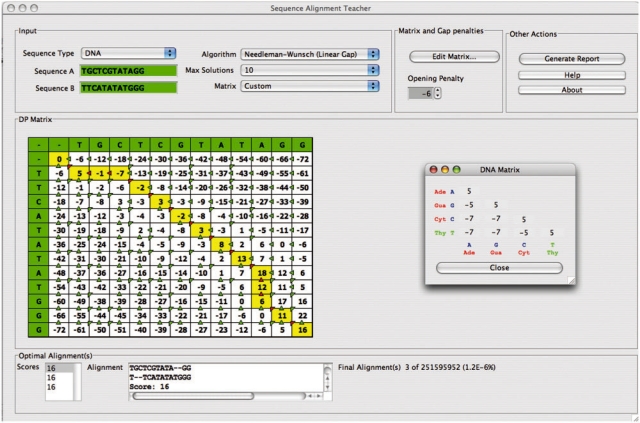
SAT requires that the user provide the input sequence data and select the different alignment parameters. This can be done on the graphics window that is launched on starting SAT (Fig. 1). The graphic window display contains five major sections:
Fig. 1.
Screen snapshot of SAT graphical interface.
Input Sequences, DP algorithm and Similarity Matrix (Fig. 1 top left): It contains two text fields to input the DNA or protein sequences that will be aligned, and three combo boxes to select the version of DP algorithm, the similarity matrix and the maximum number of optimal solutions that should be reported.
Similarity Matrix and Gap penalties (Fig. 1 top middle): The user can visualize and modify the values in the similarity matrix and the gap opening/extension penalties. Default similarity matrices include identity and transition/transversion for DNA and BLOSUM50, BLOSUM62, PAM100, PAM250 for proteins. Users can create their own custom matrices by inputting substitution values of their choice. Gap penalties can be similarly modified. Upon changing any of these values, the system will update the results of the calculations in real time.
Other Actions (Fig. 1 top right): The user can generate a report of the current data and results in PDF format. Brief help guidelines about how to use the software and contact information for authors are also provided.
DP Matrix (Fig. 1 middle): It shows the filled DP matrix along with the backtrace path of the currently selected optimal alignment. The user can click on any position of the matrix, to generate optimal sub-alignments starting from the 5′ end or the amino terminus of the sequence up to the selected position in the DP matrix. The user can re-establish the original full optimal alignment by clicking a refresh button.
Optimal Alignment(s) (Fig. 1 bottom): It displays the overall alignment score and the corresponding optimal alignment. The total number of optimal alignments displayed depends on the value specified by the user. This section also displays the total number of optimal alignments, the total number of possible alignments and the percentage of optimal solutions. By clicking on any score on the left, the corresponding optimal alignment will be shown in the window on the right. In response to this user action, the DP matrix and optimal backtrace path will also be immediately updated.
Three different DP algorithm versions were implemented here (Durbin et al., 1998), a global Needleman–Wunsch algorithm with linear and affine gap penalties and a local Smith–Waterman with linear gap penalties.
It is important to note that this software is neither intended to teach the theory of DP nor constitute another sequence alignment tool, but to provide an interactive exploring/testing framework that supports the learning process of this important algorithm. For this reason, the current version of the software is limited to short sequences to facilitate the display in laptop computers and LCD projectors in the classroom. There is plenty of bibliographic material available in the literature dedicated to the theory of DP. We recommend as a general and brief primer to DP algorithm and sequence alignment the excellent vignette by Sean Eddy (Eddy, 2004). A nice source containing a detailed description of DP algorithms for sequence alignment is the book by Durbin et al. (1998).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Javier Castellanos, Alex Slater and Tomás Norambuena for helpful discussions. We are also grateful to Drs Manfred Sippl, Andrej Sali, M.S. Madhusudhan and their research groups for testing the software and giving us important feedback.
Funding: Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) from Chile (1080158).
Conflicts of Interest: none declared.
REFERENCES
- Bellman R. On the theory of dynamic programming. Proc. Natl Acad. Sci. USA. 1952;38:716–719. doi: 10.1073/pnas.38.8.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellman R. Dynamic programming. Science. 1966;153:34–37. doi: 10.1126/science.153.3731.34. [DOI] [PubMed] [Google Scholar]
- Durbin R, et al. Biological Sequence Analysis. 1st. Vol. 2. Cambridge: Cambridge University Press; 1998. pp. 12–32. [Google Scholar]
- Eddy SR. What is dynamic programming? Nat. Biotechnol. 2004;22:909–910. doi: 10.1038/nbt0704-909. [DOI] [PubMed] [Google Scholar]
- Sankoff D. The early introduction of dynamic programming into computational biology. Bioinformatics. 2000;16:41–47. doi: 10.1093/bioinformatics/16.1.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.