Abstract
Background and Aims
Under conditions of low iron availability, rice plants induce genes involved in iron uptake and utilization. The iron deficiency-responsive cis-acting element binding factors 1 and 2 (IDEF1 and IDEF2) regulate transcriptional response to iron deficiency in rice roots. Clarification of the functions of IDEF1 and IDEF2 could uncover the gene regulation mechanism.
Methods
Spatial patterns of IDEF1 and IDEF2 expression were analysed by histochemical staining of IDEF1 and IDEF2 promoter-GUS transgenic rice lines. Expression patterns of the target genes of IDEF1 and IDEF2 were analysed using transformants with induced or repressed expression of IDEF1 or IDEF2 grown in iron-rich or in iron-deficient solutions for 1 d.
Key Results
IDEF1 and IDEF2 were highly expressed in the basal parts of the lateral roots and vascular bundles. IDEF1 and IDEF2 expression was dominant in leaf mesophyll and vascular cells, respectively. These expression patterns were similar under both iron-deficient and iron-sufficient conditions. IDEF1 was strongly expressed in pollen, ovaries, the aleurone layer and embryo. IDEF2 was expressed in pollen, ovaries and the dorsal vascular region of the endosperm. During seed germination, IDEF1 and IDEF2 were expressed in the endosperm and embryo. Expression of IDEF1 target genes was regulated in iron-rich roots similar to early iron-deficiency stages. In addition, the expression patterns of IDEF2 target genes were similar between iron-rich conditions and early or subsequent iron deficiency.
Conclusions
IDEF1 and IDEF2 are constitutively expressed during both vegetative and reproductive stages. The spatial expression patterns of IDEF1 and IDEF2 overlap with their target genes in restricted cell types, but not in all cells. The spatial expression patterns and gene regulation of IDEF1 and IDEF2 in roots are generally conserved under conditions of iron sufficiency and deficiency, suggesting complicated interactions with unknown factors for sensing and transmitting iron-deficiency signals.
Keywords: IDE, IDEF, iron deficiency, Oryza sativa, reproductive organs, seeds, transcriptional regulation, vegetative organs
INTRODUCTION
All plants require iron (Fe) for their growth and reproduction. Although abundant in mineral soils, Fe is only slightly soluble under aerobic conditions at high soil pH, especially in calcareous soils, which account for about 30 % of the world's cultivated soils. Fe deficiency is a widespread agricultural problem that reduces plant growth and crop yield (Marschner, 1995). Because plants are the primary food source for humans, the nutritional state of plants is of central importance to human health (Grusak and Dellapenna, 1999). Because excess free Fe is toxic to living cells (Guerinot and Yi, 1994), plants tightly control the level of Fe uptake, utilization and storage in response to Fe availability in the environment.
To take up and utilize Fe from the rhizosphere, higher plants have evolved two major strategies (Marschner et al., 1986): reduction (Strategy I) and chelation (Strategy II). The Strategy II mechanism, which is specific to graminaceous plants, is mediated by natural Fe chelators, the mugineic acid family phytosiderophores (MAs). Graminaceous plants synthesize and secrete MAs from their roots to solubilize Fe(III) in the rhizosphere (Takagi, 1976), and the resulting Fe(III)–MAs complexes are taken up by the roots through Yellow Stripe 1 (YS1)/YS1-Like (YSl) transporters in the plasma membrane (Curie et al., 2001, 2008). MAs are biosynthesized from methionine (Mori and Nishizawa, 1987) via four sequential enzymatic reaction steps (Shojima et al., 1990; Bashir et al., 2006). Rice (Oryza sativa) synthesizes 2′-deoxymugineic acid (DMA) and takes up Fe from the rhizosphere using an Fe(III)–DMA transporter such as OsYSL15 (Inoue et al., 2009). Rice also takes up ferrous Fe using the OsIRT1 transporter (Ishimaru et al., 2006).
Under conditions of low Fe availability, rice induces various genes involved in Fe acquisition and utilization (Kobayashi et al., 2005; Kobayashi and Nishizawa, 2008). The genes showing induction under Fe deficiency include those responsible for DMA biosynthesis (nicotianamine synthase genes OsNAS1–3, nicotianamine aminotransferase gene OsNAAT1, DMA synthase gene OsDMAS1; Higuchi et al., 2001; Inoue et al., 2003, 2008; Bashir et al., 2006); the Fe(III)–DMA transporter gene OsYSL15 (Inoue et al., 2009); the ferrous Fe transporter gene OsIRT1 (Bughio et al., 2002; Ishimaru et al., 2006); the Fe(II)- and manganese(II)-nicotianamine transporter gene OsYSL2, which is thought to be involved in the internal transport of Fe within the plant body (Koike et al., 2004); and the basic helix–loop–helix-type transcription factor gene OsIRO2 (Ogo et al., 2006). OsIRO2 positively regulates various Fe deficiency-induced genes related to DMA-based Fe acquisition, including OsNAS1, OsNAS2, OsNAAT1, OsDMAS1 and OsYSL15 (Ogo et al., 2007). Over-expression of OsIRO2, using the cauliflower mosaic virus 35S promoter, results in higher induction of these target genes and enhanced tolerance to prolonged low Fe availability (Ogo et al., 2007, 2009; Y. Ogo et al., unpubl.res.)
Previous study used a promoter analysis of the barley MAs biosynthetic gene IDS2 to identify Fe deficiency-responsive cis-acting elements 1 and 2 (IDE1 and IDE2; Kobayashi et al., 2003). IDE1 and IDE2 are also functional in rice roots and leaves (Kobayashi et al., 2004). Two rice transcription factors, IDEF1 (IDE-binding factor 1) and IDEF2, which specifically bind to IDE1 and IDE2, respectively, were recently identified (Kobayashi et al., 2007; Ogo et al., 2008). IDEF1 and IDEF2 belong to the plant-specific transcription factor families ABI3/VP1 and NAC, respectively. IDEF1 recognizes the CATGC sequence within IDE1, whereas IDEF2 predominantly recognizes CA[A/C]G[T/C][T/C/A][T/C/A] within IDE2 as the core binding site. Both IDEF1 and IDEF2 transcripts are constitutively expressed in rice roots and leaves. Transgenic rice plants with induced expression of IDEF1 under the control of the barley IDS2 promoter exhibited slower progression of leaf chlorosis in Fe-free hydroponic culture and also showed better early growth when germinated on calcareous soil (Kobayashi et al., 2007). Conversely, IDEF1-knockdown lines generated by the RNA interference (RNAi) technique exhibited hypersensitivity in Fe-free hydroponic culture (Kobayashi et al., 2009). Expression levels of IDEF1 also positively affect the expression of various Fe deficiency-responsive genes. On day 7 of Fe deficiency treatment in hydroponic culture, IDEF1 positively regulates OsIRO2, OsIRT1 and OsNAS3, and genes encoding late embryogenesis-abundant proteins (Kobayashi et al., 2007, 2009). Moreover, a majority of the known Fe acquisition/utilization-related genes, including OsIRO2, OsIRT1, OsYSL15, OsYSL2, OsNAS1, OsNAS2, OsNAS3 and OsDMAS1, are positively regulated by IDEF1 during the earlier stages of Fe deficiency (Kobayashi et al., 2009). On the other hand, IDEF2 also regulates Fe homeostasis by inducing another subset of Fe deficiency-responsive genes (Ogo et al., 2008). RNAi-mediated IDEF2-knockdown lines exhibit aberrant Fe distribution between the roots and shoots and are defective in inducing Fe deficiency-responsive genes, including OsYSL2, on day 7 of Fe deficiency treatment in hydroponic culture (Ogo et al., 2008). Earlier responses to Fe deficiency in the IDEF2-knockdown lines have not been investigated.
The present report aims to clarify the basal mechanism and properties of gene regulation mediated by IDEF1 and IDEF2. The spatial patterns of IDEF1 and IDEF2 expression were analysed by histochemical observation of promoter-β-glucuronidase (GUS) transformants. Expression of the target genes of IDEF1 and IDEF2 was also analysed by using transformants with altered expression of IDEF1 or IDEF2 under Fe-rich conditions or during an early stage of Fe deficiency. The results indicate that both IDEF1 and IDEF2 are constitutively expressed during both the vegetative and the reproductive stages and that both function under conditions of Fe sufficiency and deficiency.
MATERIALS AND METHODS
Production of promoter-GUS rice lines
To construct the IDEF1 promoter-GUS vector, PCR was used to amplify the 1994-bp 5′-upstream region of the IDEF1 gene from the translation start site (containing 1832 bp upstream from the transcription start site), using genomic DNA as a template. The primers used were 5′-CTCGAGTTAACCAGGAGACTGACTGG-3′ (forward) and 5′-TCTAGAGTTGCCCTGTTCGCTCGCT-3′ (reverse). The amplified and verified fragment was excised using XhoI and XbaI (underlined) and placed in the pIG121Hm-Xho vector (Kobayashi et al., 2004). For the IDEF2 promoter-GUS vector, the 3464-bp 5′-upstream region of the IDEF2 gene from the translation start site (containing 2000 bp upstream from the transcription start site) was amplified by PCR using genomic DNA and the primers 5′-TCTAGACCTAGGTACAACTTGAGGTAGCGGGACA-3′ (forward) and 5′-TCTAGACACTGCAGTGTATATCTTGCC-3′ (reverse). The amplified and verified fragment was excised using XbaI (underlined) and placed in the pIG121Hm-Xho vector.
The resultant IDEF1 promoter-GUS or IDEF2 promoter-GUS vector was transformed into rice (Oryza sativa L. ‘Tsukinohikari’) using an Agrobacterium-mediated method, as previously described (Hiei et al., 1994; Higuchi et al., 2001). T1, T2 or T3 seeds were used in the analysis. For the IDS2 promoter-GUS experiments, T1 lines of the previously obtained transformants (I2 lines; Kobayashi et al., 2004) were used.
Growth conditions and histochemical observation of promoter-GUS rice lines
For experiments in the vegetative stage, T1, T2 or T3 seeds were germinated on Murashige and Skoog (MS) medium with hygromycin B (50 mg L−1) and transferred to hydroponic culture (Kobayashi et al., 2005). For Fe deficiency treatments, plants that were 25–31 d old were transferred to a nutrient solution without Fe(III)-EDTA for 1–7 d. For analysis during the flowering and maturing periods, the promoter-GUS lines were transplanted to Fe-replete soil with fertilizer, and developing seeds were progressively sampled. Whole roots, transverse sections of roots and leaves, and longitudinally divided flowers and seeds were subjected to histochemical staining as described by Inoue et al. (2003) and Nozoye et al. (2007). Similar staining patterns were observed in at least three independent lines.
Plant materials and growth conditions for expression analysis of downstream genes of IDEF1 and/or IDEF2
Non-transgenic (NT) and transgenic rice (‘Tsukinohikari’) lines with induced or repressed expression of IDEF1 or IDEF2 were used for the expression assays. T1, T2 or T3 seeds of IDS2 promoter-IDEF1 transformants (I2pro-IDEF1; I2p-IDEF1 in Kobayashi et al., 2007), IDEF1-RNAi transformants (Kobayashi et al., 2009) and IDEF2-RNAi transformants (Ogo et al., 2008) were germinated on MS medium with hygromycin B (50 mg L−1) and transferred to hydroponic culture (Kobayashi et al., 2005). NT seeds were germinated on MS medium lacking hygromycin. For the Fe-rich treatment, plants that were 22–25 d old were transferred to a nutrient solution [containing 100 µm Fe(III)-EDTA] supplemented with 15·8 mg L−1 Tetsuriki-TypeX fertilizer (containing approx. 19 µm Fe2+; Aichi Steel, Aichi, Japan; Matsuyama et al., 2008). For Fe-deficiency treatments, plants that were 22–23 d old were transferred to a nutrient solution lacking Fe(III)-EDTA. After 24 h of treatment, roots were harvested from at least three plants of each line to reduce biological variance and were used for expression analysis.
Quantitative RT-PCR analysis
Total RNA was extracted from rice roots using the NucleoSpin RNA Plant Mini Kit (Macherey-Nagel, Düren, Germany). First-strand cDNA was synthesized using ReverTra Ace reverse transcriptase (TOYOBO, Tokyo, Japan) by priming with oligo-d(T)17. Quantitative RT-PCR was performed with a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA) and SYBR Premix Ex TaqTM (Perfect Real Time) reagent (TaKaRa, Tokyo, Japan). The gene-specific primers used for PCR are listed in Supplementary Data Table S1 (available online). To exclude the detection of over-expressed RNAi trigger transcripts, the DNA-binding B3 region for IDEF1 and the non-NAC region for IDEF2 were quantified. Transcript abundance was normalized against rice Actin transcript levels and is expressed as a ratio relative to the levels in NT under Fe-rich conditions.
RESULTS
Spatial patterns of IDEF1 expression
Cell-type specificity of IDEF1 expression was investigated by histochemical observation of transgenic rice plants with the introduced IDEF1 promoter (2·0 kb)–GUS construct (Fig. 1). Expression of IDEF1 was observed in every organ investigated. In roots, IDEF1 expression was relatively dominant in the basal part of the lateral roots and near the root tips (Fig. 1A, B). Observation of transverse sections revealed that IDEF1 expression in the primary roots was mainly limited to the phloem cells (Fig. 1C–F). In leaf blades, IDEF1 expression was dominant in mesophyll cells and weaker in small vascular bundles (Fig. 1G–I). The phloem companion cells showed the most staining in the vascular bundles (Fig. 1I), whereas the main vascular bundle was not stained (data not shown). In the basal part of the leaf sheath, mesophyll and small vascular cells of the inner layers showed high IDEF1 expression (Fig. 1J, K), whereas the outer layers showed little expression (Fig. 1J). These expression patterns in vegetative tissues were similar for Fe sufficiency and early or subsequent Fe deficiency (Fig. 1A–K; data not shown).
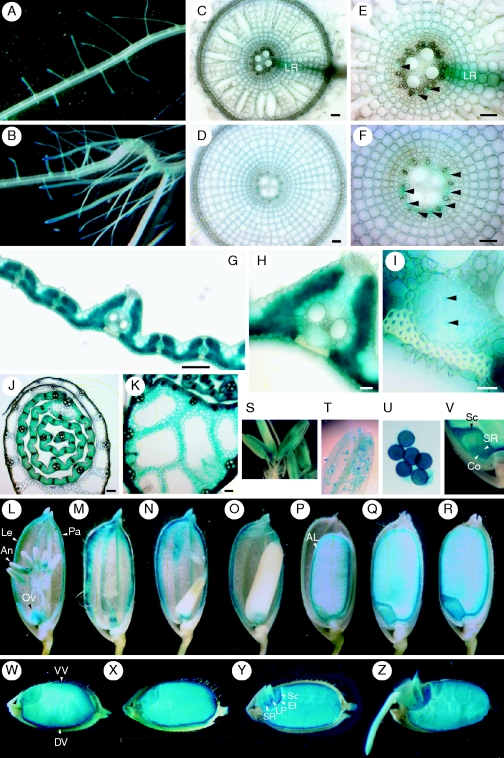
Fig. 1.
Cellular localization of IDEF1 expression as observed by histochemical staining of IDEF1 promoter-GUS expression in transgenic rice plants. (A) Fe-sufficient roots. (B) Fe-deficient roots. (C) Transverse section of Fe-sufficient roots. (D) Transverse section of Fe-deficient roots. (E, F) Enlarged parts of vascular tissues shown in (C) and (D). Arrowheads: phloem cells. (G–I) Transverse sections of Fe-deficient leaf blades. (I) An enlarged part of the phloem cells in (H). Arrowheads: phloem companion cells. (J, K) Transverse sections of the basal parts of Fe-deficient (J) or Fe-sufficient (K) leaf sheaths. (L–V) Flowers and maturing seeds. (L, S–U) Before anthesis. (M–R, V) After anthesis. Just after fertilization (M), and 3 d (N), 5 d (O), 10 d (P), 15 d (Q) and 30 d (R) after fertilization. (S) An enlarged part of the anther. (T) Pollen sac. (U) Pollen. (V) An enlarged part of the embryo 30 d after fertilization. (W–Z) Germinating seeds. Just before sowing (W), and 1 d (X), 2 d (Y) and 3 d (Z) after sowing. Abbreviations: AL, aleurone layer; An, anther; Co, coleorhiza; DV, dorsal vascular bundle; Et, epithelium; Le, lemma; LP, leaf primodium; LR, lateral root; Ov, ovary; Pa, palea; Sc, scutellum; SR, seminal root; VV, ventral vascular bundle. Scale bars = 50 µm for (C–F); 100 µm for (G, J, K); and 10 µm for (H, I).
IDEF1 expression was also present in flowers and developing seeds throughout all stages (Fig. 1L–R). Prior to anthesis, IDEF1 expression was dominant in the anthers (Fig. 1L, S), among which extremely strong staining was observed in pollen (Fig. 1T, U). Strong expression was also observed in the ovaries and the vascular bundles of the lemma and palea (Fig. 1L). After fertilization, the ovary showed high IDEF1 expression (Fig. 1M). The vascular bundles of the lemma and palea maintained high expression during the flowering and grain-filling stages (Fig. 1M–P). There was strong staining of the embryo and the aleurone layer in the late stages of maturation (Fig. 1P–R). In embryos, the scutellum, seminal root and coleorhiza were densely stained (Fig. 1V).
In germinating seeds, strong IDEF1 expression was observed in both the endosperm and the embryo (Fig. 1W–Z). Expression in the endosperm was especially strong on the outer surface, including the aleurone layer and dorsal and ventral vascular bundles, whereas expression in the embryo was especially strong in the epithelium, scutellum, and vascular bundles of the leaf primordium and seminal roots.
Spatial patterns of IDEF2 expression
Cell-type specificity of IDEF2 expression was similarly investigated using IDEF2 promoter (3·5 kb)–GUS transformants (Fig. 2). In roots, IDEF2 expression was dominant in the vascular bundles of the lateral roots (Fig. 2A–D). Expression of IDEF2 in the primary roots was mainly restricted to the epidermis, exodermis and vascular cells (Fig. 2E–G). In the leaf blades, IDEF2 expression was prominent in small and large vascular bundles but was very weak in mesophyll cells (Fig. 2H–J). Similar to IDEF1, the expression patterns in vegetative tissues were not altered by Fe availability (data not shown).
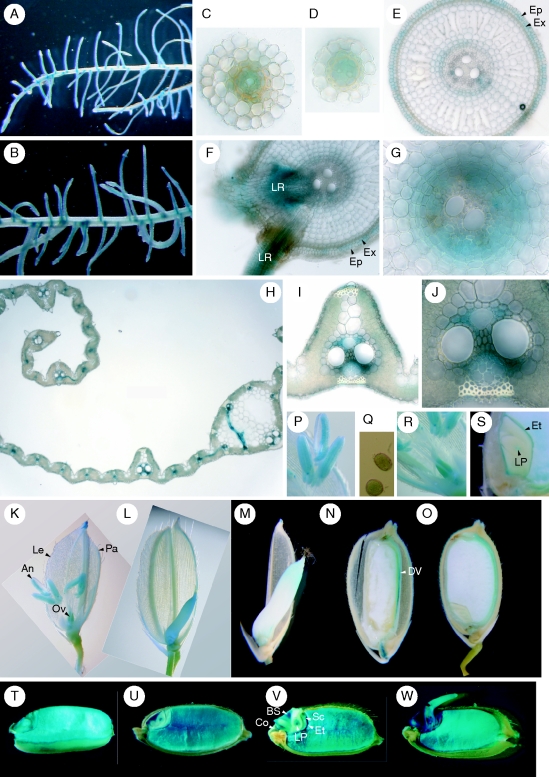
Fig. 2.
Cellular localization of IDEF2 expression as observed by histochemical staining of IDEF2 promoter-GUS expression in transgenic rice plants. (A) Fe-sufficient roots. (B) Fe-deficient roots. (C, D) Transverse sections of Fe-sufficient secondary roots. (E–G) Transverse sections of Fe-deficient main roots. (G) Enlarged parts of vascular tissues. (H–J) Transverse sections of Fe-deficient leaf blades. (J) An enlarged part of the phloem cells in (I). (K–S) Flowers and maturing seeds. (K, P–R) Before anthesis. (P) An enlarged part of the anther. (Q) Pollen. (R) Ovary. (L–O, S) After anthesis: 3 d (L), 5 d (M), 10 d (N) and 30 d (O) after fertilization. (S) An enlarged part of the embryo 30 d after fertilization. (T–W) Germinating seeds. Just before sowing (T), and 1 d (U), 2 d (V) and 3 d (W) after sowing. Abbreviations: An, anther; BS, bud scale; Co, coleorhiza; DV, dorsal vascular bundle; Ep, epidermis; Et, epithelium; Ex, exodermis; Le, lemma; LP, leaf primodium; LR, lateral root; Ov, ovary; Pa, palea; Sc, scutellum.
IDEF2 was also expressed in flowers and developing seeds (Fig. 2K–S). Before anthesis, IDEF2 expression was observed in anthers, including pollen, ovaries, and vascular bundles of the lemma and palea (Fig. 2K, P–R). After fertilization, the ovaries were still stained (Fig. 2L, M). In the late maturation stages, IDEF2 expression was dominant in the dorsal vascular tissues (Fig. 2N, O). In embryos, the epithelium and leaf primordium were mainly stained (Fig. 2S).
In germinating seeds, IDEF2 expression was strong in both the endosperm and the embryo (Fig. 2T–W). Strong expression was observed in the whole endosperm. In embryos, expression was dominant in the leaf primordium, scutellum, epithelium, and vascular bundles of the scutellum, bud scales and coleorhiza.
Spatial patterns of barley IDS2 promoter-GUS expression in germinating seeds
The spatial expression patterns of IDEF1 and IDEF2 were compared with their main target genes (Supplementary Data Table S2, available online). Among them, the barley IDS2 gene promoter is assumed to be a putative heterologous target of IDEF1 and IDEF2 when introduced into rice, because it contains the canonical IDE1 and IDE2 elements (Kobayashi et al., 2003, 2004). As the cell-type expression pattern of the IDS2 promoter in rice has been investigated only in vegetative tissues (Kobayashi et al., 2004), histochemical analysis of germinating seeds of IDS2 promoter-GUS transformants was carried out (Fig. 3). In contrast to IDEF1 and IDEF2, expression driven by the IDS2 promoter was absent in the endosperm (Fig. 3A–D). In embryos, IDS2 promoter activity was dominant in the outer layer of the basal seminal root and was also detected in the bud scales and coleorhiza.
Fig. 3.
Cellular localization of IDS2 promoter-driven expression in germinating seeds as observed by histochemical staining of IDS2 promoter-GUS expression in transgenic rice plants. Just before sowing (A), and 1 d (B), 2 d (C) and 3 d (D) after sowing. Arrowheads: outer layer of the basal seminal root.
IDEF1-mediated gene regulation under Fe-rich conditions
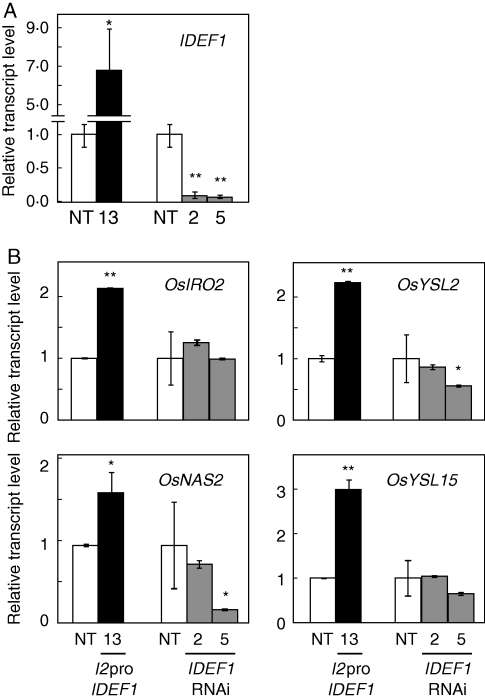
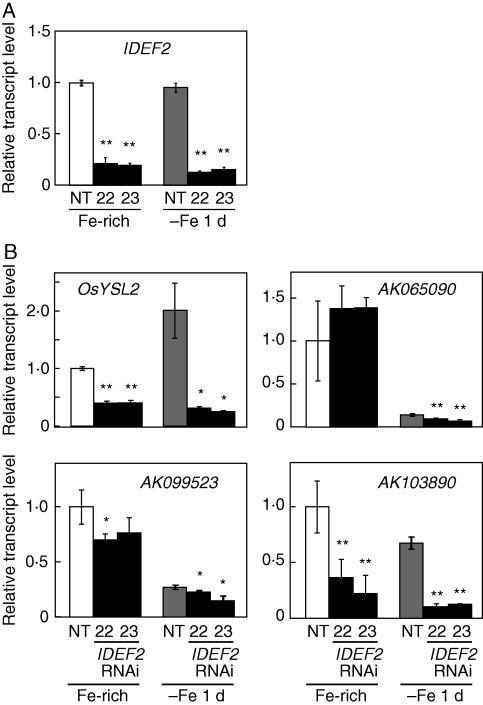
IDEF1 positively regulates various Fe deficiency-responsive genes in Fe-deficient roots and leaves (Kobayashi et al., 2007, 2009). To identify IDEF1 function under conditions of Fe sufficiency, NT or transgenic rice plants with induced or repressed IDEF1 expression were grown hydroponically and supplied with extra Fe fertilizer (Tetsuriki-TypeX; Matsuyama et al., 2008) for 1 d to ensure Fe availability. Roots grown under this Fe-rich condition were subjected to quantitative RT-PCR analysis (Fig. 4). Transgenic lines with induced IDEF1 expression driven by the IDS2 promoter (I2pro-IDEF1; Kobayashi et al., 2007) had IDEF1 levels about seven times higher than that in NT, even under Fe-rich conditions (Fig. 4A), confirming that the IDS2 promoter drives substantial basal expression in Fe-replete roots. IDEF1 expression levels in IDEF1-knockdown lines (IDEF1-RNAi; Kobayashi et al., 2009) were strongly repressed to 0·09 and 0·06 times that in NT in lines 2 and 5, respectively (Fig. 4A). Expression levels of several IDEF1 target genes during early Fe deficiency (Kobayashi et al., 2009) were quantified in these plants (Fig. 4B). OsIRO2, OsYSL2, OsNAS2 and OsYSL15 exhibited higher expression in the roots of the I2pro-IDEF1 line than in NT roots. Expression levels of these genes were similar in NT and IDEF1-RNAi line 2, whereas expression levels of OsYSL2 and OsNAS2 were repressed in IDEF1-RNAi line 5.
Fig. 4.
Quantitative RT-PCR analysis of IDEF1 (A) and its typical target genes OsIRO2, OsYSL2, OsNAS2 and OsYSL15 (B) under Fe-rich conditions in hydroponic culture. I2pro-IDEF1 transformants (line 13, T3) or IDEF1-RNAi transformants (line 2, T3; line 5, T2) were individually cultured with non-transformants (NT) and supplied with 15·8 mg L−1 Tetsuriki-TypeX fertilizer for 24 h. Transcript abundances in the roots were quantified and are expressed as ratios relative to NT levels (mean ± s.d.; n = 3–6). Significant differences from NT, analysed using a t-test (*P < 0·05; **P < 0·01), are shown.
IDEF2-mediated gene regulation under Fe-rich conditions and during an early stage of Fe deficiency
The target genes of IDEF2 were previously identified during a 7-d Fe deficiency treatment (Ogo et al., 2008). In the present study, expression of these IDEF2 target genes under Fe-rich or early Fe-deficient conditions was analysed. IDEF2 knockdown lines (IDEF2-RNAi; Ogo et al., 2008) or NT plants were hydroponically grown with supplemented Tetsuriki-TypeX fertilizer or transplanted to Fe-free solution for 1 d, and the roots were subjected to quantitative RT-PCR (Fig. 5). IDEF2 expression was effectively repressed to 0·13–0·21 times that in NT in IDEF2-RNAi lines 22 and 23 under both Fe-rich and early Fe-deficient conditions (Fig. 5A). Among the typical IDEF2 target genes under prolonged Fe deficiency, OsYSL2, AK099523 (function unknown) and AK103890 (ubiquitin gene) showed repressed expression in IDEF2-RNAi lines under both Fe-rich and early Fe-deficient conditions (Fig. 5B). Expression of AK065090 (heme peroxidase gene) was repressed in IDEF2-RNAi lines under Fe-deficient conditions but not in Fe-rich conditions (Fig. 5B). Although these four genes are induced after 7 d of Fe deficiency (Ogo et al., 2008), only OsYSL2 showed induction in NT roots after 1 d of Fe deficiency; expression of AK065090, AK099523 and AK103890 was repressed (Fig. 5B).
Fig. 5.
Quantitative RT-PCR analysis of IDEF2 (A) and its typical target genes OsYSL2, AK065090, AK099523 and AK103890 (B) under Fe-rich and early Fe-deficient conditions in hydroponic culture. IDEF2-RNAi transformants (lines 22 and 23, T1) and NT were either supplied with 15·8 mg L−1 Tetsuriki-TypeX fertilizer (Fe-rich) or cultured in Fe-free solution (–Fe 1 d) for 24 h. Transcript abundances in roots were quantified and are expressed as ratios relative to NT levels in Fe-rich conditions (mean ± s.d.; n = 3–6). Significant differences from NT, analysed using a t-test (*P < 0·05; **P < 0·01), are shown.
DISCUSSION
IDEF1 and IDEF2 are expressed throughout the life cycle
Plants manage to maintain Fe homeostasis under conditions of low Fe availability through the induction of Fe uptake- and utilization-related genes, although this Fe deficiency response has been investigated almost exclusively in vegetative tissues, predominantly roots (Kobayashi and Nishizawa, 2008). Recent studies have revealed that IDEF1 and IDEF2 play crucial roles in the regulation of Fe deficiency-responsive genes in rice roots and leaves (Kobayashi et al., 2007, 2009; Ogo et al., 2008). The present study utilized the IDEF1 and IDEF2 promoters containing 2·0 and 3·5 kb, respectively, from the translation start site for promoter-GUS analysis, because these promoter fragments contain the whole 5′-unstranslated regions and about 2·0 kb of the upstream regions from the transcription start sites. Constitutive expression derived from both the IDEF1 and the IDEF2 promoters was observed not only in vegetative tissues but also in reproductive tissues (Figs 1 and 2), consistent with previous Northern blot and microarray analyses (Kobayashi et al., 2007; Ogo et al., 2008; K. Usuda et al., University of Tokyo, Tokyo, unpubl. res.). This suggests that these promoter fragments contain all the essential elements for expression. Notably, the dominant Fe deficiency-responsive genes involved in Fe uptake and/or utilization are also expressed in reproductive tissues grown in Fe-replete soil or media (Table S2, available online; Koike et al., 2004; Nozoye et al., 2007; Inoue et al., 2009; Ogo et al., 2009). The presence of IDEF1 and IDEF2 expression in reproductive tissues suggests the regulation of their target genes, including those related to Fe homeostasis, in these tissues.
IDEF1 belongs to the ABI3/VP1 family of transcription factors, which regulate seed-specific gene expression through recognition of RY elements (CATGCA; Suzuki et al., 1997; Suzuki and McCarty, 2008). IDEF1 is an exceptional member in this family in that it is able to efficiently recognize a shorter sequence (CATGC) and, thus, the Fe deficiency-responsive IDE1 element (Kobayashi et al., 2007). However, IDEF1 also recognizes RY elements and regulates late embryogenesis-abundant genes in Fe-deficient roots and leaves (Kobayashi et al., 2009). Therefore, IDEF1 might have a dual function in reproductive tissues: CATGC-mediated regulation of Fe utilization-related genes and RY element-mediated regulation of seed maturation-related genes.
Overlapping expression of IDEF1 and IDEF2 with their target genes is achieved in restricted cell types
Previous promoter-GUS analyses revealed that expression patterns of the dominant Fe deficiency-responsive genes involved in MA-based Fe acquisition, including OsNAS1, OsNAS2, OsNAAT1, OsDMAS1 and OsYSL15, are very similar in roots and leaves (Table S2; Inoue et al., 2003, 2008, 2009; Bashir et al., 2006; Ishimaru et al., 2006). Expression of these genes is observed mainly in phloem tissues of Fe-sufficient roots and leaves and is strongly induced throughout all tissues in the roots and leaves in response to Fe deficiency. Expression of these genes is positively regulated by the Fe deficiency-inducible transcription factor OsIRO2 (Ogo et al., 2007). The expression pattern of OsIRO2 is also highly similar to that of these Fe uptake/utilization-related genes (Y. Ogo et al., unpubl. res.), suggesting that OsIRO2 directly regulates these target genes in the same cells in which it is expressed.
In contrast to these synchronous expression patterns, the present results revealed that the regions of IDEF1 and IDEF2 expression in roots and leaves are more restricted than their target genes (Figs 1 and 2; Table S2). The predominant expression of IDEF1 and IDEF2 in the basal parts of the lateral roots and phloem cells (Figs 1A–F, 2A–G) is consistent with the preferred expression of the Fe utilization-related genes OsYSL2, OsNAS1, OsNAS2, OsDMAS1, OsYSL15 and OsIRT1 in these cells (Inoue et al., 2003, 2009; Koike et al., 2004; Bashir et al., 2006; Ishimaru et al., 2006). This observation suggests that IDEF1 and IDEF2 induce Fe deficiency-responsive genes in these cells and that this may play an important role in Fe utilization. However, lack of substantial expression of IDEF1 in the epidermis/exodermis and cortex of the main roots (Fig. 1C–F) is not consistent with induction of its main target genes throughout the Fe-deficient roots (Table S2; Inoue et al., 2003, 2009; Bashir et al., 2006; Ishimaru et al., 2006). In addition, IDEF1 expression in leaves is stronger in mesophyll cells than in vascular cells (Fig. 1G–I), opposite to the predominant expression of Fe utilization-related genes in vascular tissues (Table S2; Inoue et al., 2003, 2009; Koike et al., 2004; Bashir et al., 2006). If shoot-borne long-distance signal which regulates Fe deficiency-induced genes in roots might exist, as suggested by Enomoto et al. (2007, 2009), the predominant expression of IDEF1 in root vascular tissues might be favourable for receiving such signals. However, induction of Fe uptake-related genes in the epidermis/exodermis of the main roots, where IDEF1 expression is absent, would require the radial movement of IDEF1 proteins and/or related factors from vascular cells. Horizontal signal transfer has also been suggested in Arabidopsis sulfur deficiency response from the vasculature-expressed key transcription factor SULFUR LIMITATION 1 (SLIM1) toward its epidermis-expressed target gene SULTR1;2 (Maruyama-Nakashita et al., 2006). Radial movement of transcription factors was reported for SHOOT-ROOT (SHR) from Arabidopsis stele to adjacent cells in root cell patterning (Nakajima et al., 2001). Alternatively, Fe deficiency-induced expression in the root epidermis/exodermis might be mediated by a distinct mechanism from IDEF1-dependent transactivation in vascular cells.
Expression of IDEF1 and IDEF2 in reproductive organs tended to be more widely distributed than their target genes in vegetative organs (Table S2). In germinating seeds, the expression patterns of IDEF1, IDEF2 and OsNAS1 were similar; all were expressed in the embryo and endosperm (Figs 1W–Z, 2T–W, Table S2; Nozoye et al., 2007). In contrast, other Fe utilization-related genes were not expressed in the endosperm (Table S2; Nozoye et al., 2007; Inoue et al., 2009). In addition, expression of the heterologously introduced barley IDS2 gene promoter in germinating seeds was restricted to the embryo and was absent in the endosperm (Fig. 3), even though the IDS2 promoter contains a functional IDE1 and IDE2 set (Kobayashi et al., 2003, 2004). These observations indicate that the expression of IDEF1 and IDEF2 is not necessarily sufficient for the induction of the vegetative target genes in germinating seeds, suggesting the presence of unknown factors mediating IDEF-based gene expression in seeds. Also, the target genes of IDEF1 and IDEF2 might not be identical between reproductive and vegetative organs.
In floral tissues, IDEF1 expression was especially high in pollen (Fig. 1T, U). IDEF2 was also expressed in pollen but to a lesser extent (Fig. 2P, Q). Interestingly, an Fe(III)-DMA transporter gene, OsYSL18, is expressed in pollen grains (Aoyama et al., 2009). Although the expression of OsYSL18 is neither induced under Fe deficiency nor regulated by IDEF1 or IDEF2 in vegetative tissues (Aoyama et al., 2009; T. Kobayashi et al. and Y. Ogo et al., unpubl. res.), IDEF1 and IDEF2 might regulate OsYSL18 or other genes involved in pollen Fe acquisition.
IDEF1 and IDEF2 regulate gene expression in both Fe-sufficient and Fe-deficient roots
The constitutive expression of IDEF1 and IDEF2 suggests a role in sensing Fe deficiency signals and triggering a transcriptional cascade. In this respect, it is important to know whether IDEF-mediated transactivation occurs even under conditions of Fe sufficiency. Recent analysis suggested that IDEF1 regulates the expression of its target genes even when grown in standard hydroponic culture, similar to early Fe-deficiency stages (Kobayashi et al., 2009). However, it was not clear whether this transactivation is due to the basal activity of IDEF1 under Fe sufficiency or, alternatively, due to a response to physiological Fe deficiency that might occur even in standard ‘Fe-sufficient’ culture. To discriminate among these possibilities, IDEF-mediated transactivation under Fe-rich conditions was confirmed (Figs 4 and 5). By using Tetsuriki fertilizer (Matsuyama et al., 2008), the basal expression of sensitive Fe deficiency-inducible genes was repressed to lower levels than in standard ‘Fe-sufficient’ conditions (data not shown). Even under these ‘Fe-rich’ conditions, the I2pro-IDEF1 transformants induced substantially higher IDEF1 expression (Fig. 4A), indicating the presence of basal activity of the IDS2 promoter that is possibly conferred by IDE1 and IDE2, and thus IDEF1 and IDEF2. Furthermore, the transcript levels of IDEF1 were positively correlated with those of their target genes as under early Fe deficiency (Fig. 4B). Similarly, the gene expression patterns of IDEF2 knockdown lines (Fig. 5) indicated that the expression of many IDEF2 target genes on day 7 of Fe deficiency was also regulated by IDEF2 under Fe-rich conditions as well as during early Fe deficiency. In contrast to IDEF1, IDEF2 appears to cause no remarkable alteration in the gene regulation patterns between early and subsequent stages of Fe deficiency (Fig. 5; Ogo et al., 2008). These results indicate that IDEF1 and IDEF2 function to activate the expression of Fe homeostasis-related genes even under Fe sufficiency. Thus, IDEF1 and IDEF2 may already be present before the onset of Fe deficiency in their active forms. At present, other factors that mediate the IDEF functions are not known.
In contrast to observations in rice, constitutive IDEF1 over-expression in tobacco resulted in IDE1-mediated transactivation in Fe-deficient roots but not in Fe-sufficient roots (Kobayashi et al., 2007), suggesting the requirement of Fe-deficient conditions for IDEF1 function in tobacco plants. Nevertheless, the lack of information on IDEF1 orthologues in tobacco, as well as the distinct tissue-specific response of IDE1 between rice and tobacco (Kobayashi et al., 2003, 2004, 2007), makes it difficult to explain the IDEF1 function from the heterologous tobacco system. Future identification of IDEF-interacting factors, as well as the nature of the Fe deficiency signal, will be required to understand the gene regulation mechanisms mediated by IDEF1 and IDEF2.
In summary, the constitutive expression of IDEF1 and IDEF2 during the rice life cycle and generally conserved gene regulation mediated by IDEF1 and IDEF2 between Fe sufficiency and deficiency suggest complicated interactions with unknown factors in sensing and transmitting Fe deficiency signals to facilitate Fe utilization-related genes.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Satoshi Mori (NPO-WINEP) and Dr Hirohiko Sasamoto (Aichi Steel Co.) for providing us with the Tetsuriki-TypeX fertilizer and Ms Kanako Usuda (The University of Tokyo) for her assistance with rice culture. This research was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
LITERATURE CITED
- Aoyama T, Kobayashi T, Takahashi M, et al. OsYSL18 is a rice iron(III)–deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Molecular Biology. 2009;70:681–692. doi: 10.1007/s11103-009-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K, Inoue H, Nagasaka S, et al. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. Journal of Biological Chemistry. 2006;43:32395–32402. doi: 10.1074/jbc.M604133200. [DOI] [PubMed] [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, et al. Cloning an iron-regulated metal transporter from rice. Journal of Experimental Botany. 2002;53:1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- Curie C, Panavience Z, Loulergue C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, et al. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Annals of Botany. 2008;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y, Hodoshima H, Shimada H, et al. Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta. 2007;227:81–89. doi: 10.1007/s00425-007-0596-x. [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Hashida S, Shoji K, et al. Brown P., editor. Expression of iron uptake genes in roots are affected by long-distance signals both in non-graminaceous and in graminaceous plants. Proceedings of XVI International Plant Nutrition Colloquium. 2009 Paper 1209. [Google Scholar]
- Grusak MA, Dellapenna D. Improving the nutrient composition of plants to enhance human nutrition and health. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:133–161. doi: 10.1146/annurev.arplant.50.1.133. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiology. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, et al. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, et al. Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. The Plant Journal. 2001;25:159–167. doi: 10.1046/j.1365-313x.2001.00951.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, et al. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2 and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. The Plant Journal. 2003;36:366–381. doi: 10.1046/j.1365-313x.2003.01878.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Takahashi M, Kobayashi T, et al. Identification and localisation of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Molecular Biology. 2008;66:193–203. doi: 10.1007/s11103-007-9262-8. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, et al. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. Journal of Biological Chemistry. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+ The Plant Journal. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. Regulation of iron and zinc uptake and translocation in rice. In: Hirano HY, Hirai A, Sano Y, Sasaki T, editors. Biotechnology in agriculture and forestry 62. Rice biology in the genomics era. The Netherlands: Springer; 2008. pp. 321–335. Krips bv, Meppel. [Google Scholar]
- Kobayashi T, Nakayama Y, Itai RN, et al. Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. The Plant Journal. 2003;36:780–793. doi: 10.1046/j.1365-313x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakayama Y, Takahashi M, et al. Construction of artificial promoters highly responsive to iron deficiency. Soil Science and Plant Nutrition. 2004;50:1167–1175. [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, et al. Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. Journal of Experimental Botany. 2005;56:1305–1316. doi: 10.1093/jxb/eri131. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogo Y, Itai RN, et al. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proceedings of the National Academy of Sciences USA. 2007;104:19150–19155. doi: 10.1073/pnas.0707010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Itai RN, Ogo Y, et al. The rice transcription factor IDEF1 is essential for the early response to iron deficiency and induces vegetative expression of late embryogenesis abundant genes. The Plant Journal. 2009;60:948–961. doi: 10.1111/j.1365-313X.2009.04015.x. [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, et al. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. The Plant Journal. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. 2nd edn. London: Academic Press; 1995. [Google Scholar]
- Marschner H, Römheld V, Kissel M. Different strategies in higher plants in mobilization and uptake of iron. Journal of Plant Nutrition. 1986;9:695–713. [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, et al. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. The Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Sasamoto H, Nakazono M, et al. The application of tetsuriki-agri (and/or aqua) to the plants. Proceedings of XIV International Symposium on Iron Nutrition and Interactions in Plants. 2008 69. [Google Scholar]
- Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophores in Graminaceae plants. Plant and Cell Physiology. 1987;28:1081–1092. [Google Scholar]
- Nakajima K, Sena G, Nawy T, et al. Intercellular movement of the putative transcription factor SHR in root pattening. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Nozoye T, Inoue H, Takahashi M, et al. The expression of iron homeostasis-related genes during rice germination. Plant Molecular Biology. 2007;64:35–47. doi: 10.1007/s11103-007-9132-4. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, et al. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. Journal of Experimental Botany. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. The Plant Journal. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Kobayashi T, Itai RN, et al. A novel NAC transcription factor IDEF2 that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. Journal of Biological Chemistry. 2008;283:13407–13417. doi: 10.1074/jbc.M708732200. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Kobayashi T, et al. Brown P., editor. Overexpression of OsIRO2 improves both iron uptake and translocation to seeds in rice. Proceedings of XVI International Plant Nutrition Colloquium. 2009 paper 1204. [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, et al. Biosynthesis of phytosiderophores: in vitro biosynthesis of 2'-deoxymugineic acid from L-methionine and nicotianamine. Plant Physiology. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty D. Functional symmetry of the B3 network controlling seed development. Current Opinion in Plant Biology. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of Viviparous1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Science and Plant Nutrition. 1976;22:423–433. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.