Abstract
Background
Productive agriculture needs a large amount of expensive nitrogenous fertilizers. Improving nitrogen use efficiency (NUE) of crop plants is thus of key importance. NUE definitions differ depending on whether plants are cultivated to produce biomass or grain yields. However, for most plant species, NUE mainly depends on how plants extract inorganic nitrogen from the soil, assimilate nitrate and ammonium, and recycle organic nitrogen. Efforts have been made to study the genetic basis as well as the biochemical and enzymatic mechanisms involved in nitrogen uptake, assimilation, and remobilization in crops and model plants. The detection of the limiting factors that could be manipulated to increase NUE is the major goal of such research.
Scope
An overall examination of the physiological, metabolic, and genetic aspects of nitrogen uptake, assimilation and remobilization is presented in this review. The enzymes and regulatory processes manipulated to improve NUE components are presented. Results obtained from natural variation and quantitative trait loci studies are also discussed.
Conclusions
This review presents the complexity of NUE and supports the idea that the integration of the numerous data coming from transcriptome studies, functional genomics, quantitative genetics, ecophysiology and soil science into explanatory models of whole-plant behaviour will be promising.
Keywords: Nitrogen use efficiency, remobilization, quantitative trait loci, natural variation, nitrate transporter, ammonium assimilation, glutamine synthetase, leaf senescence, nutrient recycling
INTRODUCTION
Plants have a fundamental dependence on inorganic nitrogen and 85–90 million metric tonnes of nitrogenous fertilizers are added to the soil worldwide annually (Good et al., 2004). Nitrogen is one of the most expensive nutrients to supply and commercial fertilizers represent the major cost in plant production. Furthermore, there is serious concern regarding nitrogen loss in the field, giving rise to soil and water pollution. Incomplete capture and poor conversion of nitrogen fertilizer also causes global warming through emissions of nitrous oxide. Lowering fertilizer input and breeding plants with better nitrogen use efficiency (NUE) is one of the main goals of research on plant nutrition (Hirel et al., 2007). The ability of a plant to capture nitrogen from the soil depends on soil type, environment and species. It has been estimated that 50–70 % of the nitrogen provided to the soil is lost (Hodge et al., 2000). Therefore, improving NUE is essential in order to reduce damage due to nitrate leaching, ecosystem saturation and water pollution. NUE in plants is complex and depends on nitrogen availability in the soil and on how plants use nitrogen throughout their life span. As a concept, NUE is expressed as a ratio of output (total plant N, grain N, biomass yield, grain yield) and input (total N, soil N or N-fertilizer applied). Increasing NUE and limiting nitrogen fertilizer use are both important and challenges to preserve the environment and improve a sustainable and productive agriculture.
The use of nitrogen by plants involves several steps, including uptake, assimilation, translocation and, when the plant is ageing, recycling and remobilization. For crops, NUE has been defined as the grain yield per unit of nitrogen available from the soil, including nitrogen fertilizer. For Arabidopsis and other model plants, a physiological NUE index is expressed as the fresh or dry matter produced per nitrogen content in the plant (Good et al., 2004). In both cases, NUE is the product of nitrogen uptake efficiency (NupE) and nitrogen utilization efficiency (NUtE), which is the optimal combination between nitrogen assimilation efficiency (NAE) and nitrogen remobilization efficiency (NRE). Until now, most plant cultivars have been selected under non-limiting nitrogen conditions. Reducing the excessive input of fertilizers without affecting productivity raises questions about the adaptive responses of such cultivars to low nitrogen availability.
The biochemical mechanisms involved in N uptake, assimilation and remobilization have been widely studied to identify the bottlenecks associated with NUE. Quantitative genetics is also conducted using both crops and model plants to evaluate the genetic basis of NUE. An overall examination of the physiological, metabolic and genetic aspects of NUE is presented in this review.
NITROGEN SOURCES AND UPTAKE
Although generally low, soil nitrogen availability can fluctuate greatly in both space and time due to factors such as precipitation, temperature, wind, soil type and pH. Therefore, the preferred form in which N is taken up depends on plant adaptation to soil conditions. Generally, plants adapted to low pH and reducing soils as found in mature forests or arctic tundra tend to take up ammonium or amino acids, whereas plants adapted to higher pH and more aerobic soils prefer nitrate (for a review see Maathuis, 2009).
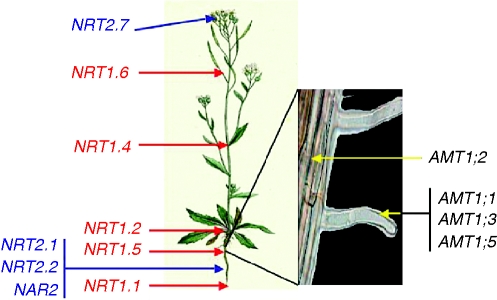
Nitrate uptake occurs at the root level and two nitrate transport systems have been shown to coexist in plants and to act co-ordinately to take up nitrate from the soil solution and distribute it within the whole plant (Fig. 1) (review in Daniel-Vedele et al., 1998; Tsay et al., 2007).
Fig. 1.
Schematic presentation of the known localisation of NRT1, NRT2 and AMT genes in Arabidopsis. Two nitrate transport systems have been shown to coexist in plants and to act co-ordinately to take up nitrate from the soil solution and distribute nitrate within the whole plant. The role of each ammonium transporter has been shown by the study of single, double, triple and quadruple mutants.
It is generally assumed that the NRT1 gene family mediates the root low-affinity transport system (LATS), with the exception of the AtNRT1·1, which is both a dual affinity transporter (Wang et al., 1998; Liu et al., 1999) and a nitrate sensor (Ho et al., 2009). In Arabidopsis, 53 genes belong to the NRT1 family. Among them 51 genes are expressed and exhibit different tissue expression patterns in the whole plant (Tsay et al., 2007), suggesting a specialized and unique function for at least some of them. AtNRT1·1 (formerly Chl1) is the most extensively studied gene and was the first to have been isolated (Tsay et al., 1993). The protein is located on the plasma membrane and the gene is expressed in the epidermis of the root tips and in the cortex and endodermis in the more mature part of the root (Huang et al., 1999). AtNRT1·2 is constitutively expressed only in the root epidermis and participates in the constitutive low-affinity system (Huang et al., 1999). Once taken up by root cells, nitrate must be transported across several cell membranes and distributed in various tissues. AtNRT1·5, located on the plasma membrane of root pericycle cells close to the xylem, is involved in long-distance transport of nitrate from the root to the shoot (Lin et al., 2008). The AtNRT1·4 gene is only expressed in the leaf petiole, and in the mutant the level of nitrate content in the petiole is half that in the wild-type (Chiu et al., 2004). The AtNRT1·6 gene, expressed in the vascular tissue of the silique and funiculus, is thought to deliver nitrate from maternal tissue to the developing embryo (Almagro et al., 2008). A striking property of the NRT1 family is that certain members belonging to the group II (reviewed in Tsay et al., 2007) are able, in heterologous systems, to transport not only nitrate but also di- or tripeptides, while OPT (oligo-peptide transporter) proteins transport tetra/pentapeptides.
The high-affinity transport system (HATS), acting when the external nitrate concentration is low, relies on the activity of the so-called NRT2 family (reviewed in Williams and Miller, 2001). Although the functional characterization of almost all higher plant NRT2 transporters remains to be done, it is now well documented that AtNRT2·1, in interaction with an NAR2 protein (Orsel et al., 2006), is a major component of the HATS in Arabidopsis, as shown by the fact that a mutant disrupted for the AtNRT2·1 gene has lost up to 75 % of the high-affinity NO3− uptake activity and showed a lower leaf nitrate content (Filleur et al., 2001). As a consequence, growth of these mutants is severely impaired at low NO3− concentration (Orsel et al., 2004, 2006). A lower nitrate content was also found in a clce mutant, affected in a protein belonging to the Arabidopsis AtCLC (ChLoride Channel) family. De Angeli et al. (2006) have demonstrated that another member of the family, the vacuolar AtCLCa protein, behaves as a NO3−/H+ exchanger allowing the accumulation of nitrate within the vacuole. Insertion mutants within the AtCLCa gene exhibit normal development but show a reduced capacity to store nitrate but not other anions. This phenotype was also recently found when the expression of AtNRT2·7 was affected. This AtNRT2 gene is expressed in aerial organs and also highly induced in dried seeds. In two allelic atnrt2·7 mutants, less nitrate is accumulated in the seed. In contrast, seeds from plants over-expressing the AtNRT2·7 coding region accumulate more nitrate and as a consequence are less dormant than the corresponding wild-type seeds (Chopin et al., 2007).
Electrophysiological studies together with the pH-dependent equilibrium between the uncharged NH3 and charged NH4+ forms suggest that the ion is predominant under all physiological conditions and is the dominant species for controlled membrane transport (Ludewig et al., 2007). With functional complementation of a yeast mutant defective in methylammonium uptake and recent efforts in sequencing the genome of model species, 6 genes belonging to the same family of ammonium transporters were found in Arabidopsis (Gazzarini et al., 1999), 10 in rice (Sonoda et al., 2003) – a species adapted to ammonium nutrition (Yoshida, 1981) – and 14 in poplar (Couturier et al., 2007). In order to analyse the function of each of the AMT (ammonium transporter) genes separately in planta, physiological and ammonium influx studies were carried out on single, double, triple and quadruple mutants, obtained by T-DNA insertion or by RNAi approach, or by complementing the quadruple mutant by single genes (Yuan et al., 2007). An additive contribution of each protein to ammonium transport was shown, AMT1·1 and AMT1·3 conferring a similar capacity of 30–35 % while AMT1·2 conferred a lower capacity of 18–25 %. A second saturable transport system with a low Km of 4·5 mm and a very low capacity is thought to be coded by the AMT1·5 gene. A complex picture is now emerging from these studies. There is a spatial organization of AMT1 proteins (Fig. 1), the transporters possessing the highest ammonium affinities being located in outer root cells or root hairs where they can take up ammonium from the soil solution (AMT1·1, AMT1·3, AMT1·5). The lower affinity of AMT1·1, and its location in the endodermis along the root hair zone, suggests a function in the retrieval of ammonium that is released from the cortex or that enters the root via the apoplastic route. The electrochemical gradient between the vacuole and cytosol would drive NH3 import to and NH4+ export out of the vacuole. Indeed, tonoplast intrinsic proteins from the TIP family were shown to play a role in NH3 transport into the vacuole (Loque et al., 2005). Vacuolar loading with NH4+ should require an electrogenic ammonium transporter, which is not yet identified.
Thus far, putative plant amino acid transporters have been identified as members of at least five gene families. In Arabidopsis these comprise at least 67 genes (reviewed in Rentsch et al., 2007). Substrate specificities as well as gene expression patterns or subcellular localization of the protein may give a good indication of the function of each protein. Forward and reverse genetic approaches were used to identify transporters involved in root amino acid uptake (Hirner et al., 2006; Svennerstam et al., 2007). Both studies led to the conclusion that LHT1 (lysine/histidine transporter), belonging to the ATF (amino acid transport) family, is crucial for root uptake of acidic and neutral amino acids. The AAP1 (amino acid permease 1) protein was also shown to transport uncharged amino acids but only when they are supplied at high concentrations in the external medium (Lee et al., 2007). Uptake of cationic amino acids such as l-lysine or l-arginine is mediated by AAP5 within the concentration range relevant for field conditions (Svennerstam et al., 2008). The precise localization of these transporter mRNAs within different cell types in the root led Näsholm et al. (2009) to propose a hypothetic mode of root amino acid uptake in non-mycorrhizal plants.
NITROGEN ASSIMILATION: CROSS-TALK WITH CARBON METABOLISM
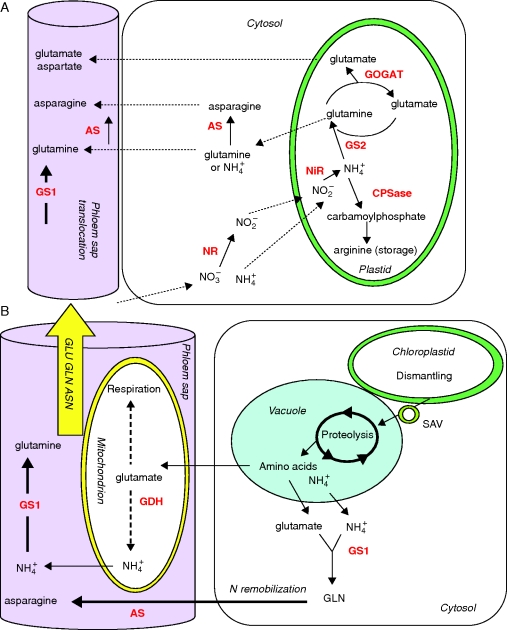
The nitrogen sources taken up by higher plants are nitrate or ammonium as inorganic nitrogen sources and amino acids under particular conditions of soil composition. Nitrogen assimilation requires the reduction of nitrate to ammonium, followed by ammonium assimilation into amino acids (Fig. 2A).
Fig. 2.
Schematic presentation of key enzymes involved in nitrogen management in (A) young and (B) senescing leaves. (A) Nitrate reductase (NR) and asparagine synthetase (AS) are localized in the cytosol, and nitrite reductase (NiR), glutamine synthetase 2 isoenzyme (GS2), glutamate synthase (GOGAT) and carbamoylphosphate synthetase (CPSase) within the plastids of mesophyll cells. Glutamine synthetase isoenzyme 1 (GS1) and AS are located in the cytosol of companion cells. (B) Senescence-associated events include chloroplast degradation and translocation of plastid proteins to the central vacuole via senescence-associated vacuole (SAV) trafficking. Amino acid recycling occurred in mitochondria and cytosol of mesophyll cells and companion cells. Glutamate dehydrogenase (GDH), GS1 and AS are the major enzymes involved in the synthesis of glutamine, glutamate and asparagine in the phloem.
Nitrate reduction takes place in both roots and shoots but is spatially separated between the cytoplasm where the reduction takes place and plastids/chloroplasts where nitrite reduction occurs. Nitrate reduction into nitrite is catalysed in the cytosol by the enzyme nitrate reductase (NR) (Meyer and Stitt, 2001). This enzyme is a homodimer, each monomer being associated with three prosthetic groups: flavin adenine dinucleotide (FAD), a haem and a molybdenum cofactor (MoCo). Characterization of mutants resistant to chlorate, which can be reduced into toxic chlorite by NR, identified two classes of genes, the Nia genes encoding the NR apoenzyme and the Cnx genes encoding the MoCo cofactor. Since 1993, considerable work has been done to characterize the NR in different species (reviewed by Meyer and Stitt, 2001). Although the NR enzyme is thought to be localized in the cytosol, an association with the plasma membrane (PM-NR) has been found in corn roots and barley (Ward et al., 1989).
After nitrate reduction, nitrite is translocated to the chloroplast where it is reduced to ammonium by the second enzyme of the pathway, the nitrite reductase (NiR). The Nii genes encoding the NiR enzyme have been cloned from various species, the number of genes varying from one to two copies (Meyer and Stitt, 2001).
Ammonium, originating from nitrate reduction, and also from photorespiration or amino acid recycling, is mainly assimilated in the plastid/chloroplast by the so-called GS/GOGAT cycle (Lea and Miflin, 1974; Lea and Forde, 1994). The glutamine synthetase (GS) fixes ammonium on a glutamate molecule to form glutamine. This glutamine reacts subsequently with 2-oxoglutarate to form two molecules of glutamate, this step being catalysed by the glutamine 2-oxoglutarate amino transferase (or glutamate synthase, GOGAT). The decameric structure of maize GS was described by Unno et al. (2006). Two classes of nuclear genes code for GS: the GLN2 and GLN1 genes. GLN2, present as a single nuclear gene in all the species studied so far, codes for the chloroplastic GS2, thought to be involved in the primary assimilation of ammonium coming from nitrate reduction in both C3 and C4 plants and in the re-assimilation of ammonium produced from photorespiration in C3 plants. The magnitude of the ammonium flux through the photorespiration pathway in the leaves of C3 plants was indeed estimated to exceed that produced from nitrate reduction by five- to ten-fold (Keys et al., 1978).
Conversely, the GLN1 gene family codes for cytosolic GS1 isoforms, present in different organs such as roots or stems and thought to be involved in ammonium recycling during particular developmental steps such as leaf senescence and in glutamine synthesis for transport into the phloem sap (reviewed by Bernard and Habash, 2009). Two different forms of glutamate synthase are present in plants: Fd-GOGAT and NADH-GOGAT use ferredoxin and NADH as the electron donors, respectively (Vanoni et al., 2005). Fd-GOGAT is predominantly localized in leaf chloroplasts whereas NADH-GOGAT is primarily located in plastids of non-photosynthetic tissues, such as roots, etiolated leaf tissues and companion cells. The structural, mechanistic and regulatory properties of GOGAT enzymes and their role in amino acid metabolism have been reviewed by Suzuki and Knaff (2005).
In addition to the GS/GOGAT cycle, three enzymes probably participate in ammonium assimilation. Cytosolic asparagine synthetase (AS) catalyses the ATP-dependent transfer of the amido group of glutamine to a molecule of aspartate to generate glutamate and asparagine (Fig. 2A; Lam et al., 2003). Masclaux-Daubresse et al. (2006) provided evidence that AS can also use ammonia as a substrate. Indeed, providing 15NH4+ to leaf discs in the presence of azaserine, the authors blocked glutamate biosynthesis but not asparagine biosynthesis, thus showing that asparagine production is not amino transferase dependent. In Arabidopsis, three genes encode AS (ASN1, ASN2 and ASN3). Asparagine has a higher N/C ratio than glutamine and can be used as a long-range transport and storage compound, especially in legumes (Rochat and Boutin 1991; Lam et al., 2003). AS could in certain situations compensate for the reduced GS-dependent ammonium assimilatory activity.
Carbamoylphosphate synthase (CPSase) forms carbamoylphosphate, a precursor of citrulline and arginine, within plastids using bicarbonate, ATP and ammonium or the amide group of glutamine. In Arabidopsis, a single copy each of carA and of carB encode the small subunit and the large subunit of CPSase, respectively, to form a single heterodimeric enzyme (Potel et al., 2009).
Finally, the mitochondrial NADH-glutamate dehydrogenase could alternatively incorporate ammonium into glutamate in response to high levels of ammonium under stress (Skopelitis et al., 2006). However, the major catalytic activity for GDH in plant cells has been reported to be glutamate deamination (Masclaux-Daubresse et al., 2006; Purnell and Botella, 2007). GDH activity was shown to be essential for plant survival in dark conditions (Miyashita and Good, 2008).
NR, NiR and GOGAT require reducing power as either NADH or ferredoxin (Fdx) according to the enzyme. Glutamine synthetase and asparagine synthetase need ATP. In addition, carbon skeletons and especially keto-acids are essential to form organic nitrogen as amino acids. The availability of carbon skeletons for ammonium condensation and the supply of ATP, Fdx and NADH as products of photosynthesis, respiration and photorespiration pathways are thus essential for nitrogen assimilation.
NITROGEN REMOBILIZATION: A KEY FACTOR FOR NITROGEN USE EFFICIENCY
Nitrogen remobilization and yield dilemma
Leaf proteins and in particular photosynthetic proteins of plastids are extensively degraded during senescence, providing an enormous source of nitrogen that plants can tap to supplement the nutrition of growing organs such as new leaves and seeds. Nitrogen remobilization has been studied in several plant species through the ‘apparent remobilization’ method, which is the determination of the amount of total nitrogen present in the different plant organs at different times of development and through 15N long-term labelling, which allows the determination of fluxes (Gallais et al., 2006). In Arabidopsis and oilseed rape, it has been shown that nitrogen can be remobilized from senescing leaves to expanding leaves at the vegetative stage as well as from senescing leaves to seeds at the reproductive stage (Malagoli et al., 2005; Diaz et al., 2008; Lemaître et al., 2008). In Arabidopsis, for which sequential (at vegetative stage) and monocarpic (after flowering) senescence phases can be distinguished easily, it was shown that the N remobilization rate was correlated with leaf-senescence severity at the vegetative stage only (Diaz et al., 2008). Experiments of 15N tracing at the reproductive stage showed that the rate of nitrogen remobilization from the rosettes to the flowering organs and to the seeds was similar in early- and late-senescing lines (Diaz et al., 2008). At the reproductive stage, NRE is mainly related to harvest index (C. Masclaux-Daubresse, unpubl. res.).
Studies using 15N tracing in cereals, oilseed rape and legumes showed that the onset of grain filling was a critical phase because N uptake and N2 fixation declined during plant maturation and seed filling (Salon et al., 2001). Nitrogen fluxes and 15N remobilization experiments performed by Cliquet et al. (1990) showed that leaf blades, stalks, cob, husk and shank serve successively as N-sinks and N-sources. Nitrogen uptake and assimilation during the grain filling period is generally insufficient for the high demand of the seeds, and the progressive and numerous remobilization steps, occurring successively in the different plant organs, are needed to route nitrogen to the seeds. The contribution of leaf N remobilization to rice, wheat or maize grain N content is cultivar dependent, varying from 50 to 90 % (Masclaux et al., 2001). N remobilization is also environment dependent and favoured under limiting nitrate supplies (Lemaître et al., 2008). Although 15N remobilization is a step-by-step mechanism that involves the different plant organs, evidence shows that grain nitrogen content is correlated with flag leaf senescence in maize, wheat and barley. Flag leaf senescence seems then to have a special role in nitrogen availability for grain filling. The onset and the speed of flag leaf senescence are likely to be essential for NRE (Martin et al., 2005; Uauy et al., 2006). Leaf senescence is not only essential for nitrogen mobilization. Evidence has shown that leaf senescence is also important for yield. Delaying leaf senescence results in a prolongation of photosynthesis that increases grain yield and carbon filling into seeds. Breeding plants have then to cope with the dilemma that delayed senescence could lead to higher yields but also to a decrease in NRE and to a decrease in grain protein content. On the other hand, increasing nitrogen remobilization has the advantage of re-using nitrogen from the vegetative parts and of lowering nitrogen loss in the dry remains.
Nitrogen sources for N remobilization
Chloroplasts are the main source of nutrients used during senescence. Rubisco accounts for 50 % of the total soluble protein content in the leaves of C3 plants and 20 % in C4 plants (Mae et al., 1983; Sage et al., 1987). Together with other photosynthesis-related proteins, Rubisco is a major source of nitrogen for remobilization. Over-investment in Rubisco is thus important for N-source management at the whole-plant level.
Although chloroplasts show the first symptoms of deterioration during senescence, whereas other organelles are degraded later, the mechanisms responsible for chloroplast degradation are largely unknown. Chloroplast dismantling does not mean chaotic decay. Controlled and coordinated degradation is needed to prevent cell damage due to the highly photodynamic nature of some of the breakdown products and to maintain export capacity and remobilization. The initial steps of chlorophyll and chloroplast protein degradation are likely to take place within the plastid itself (review by Martinez et al., 2008). Later steps may take place within the central vacuole.
Degradation of chloroplast proteins within the organelle is supported by the fact that chloroplasts contain a rather high number of proteases, some of them being upregulated during senescence. Chloroplastic DegP and FstH proteases seem to be responsible for D1 protein degradation during senescence. The FstH6 protease may be implicated in LHCII degradation (for a review see Martinez et al., 2008). The exact processes leading to Rubisco degradation remain unclear. It was shown that plastids isolated from senescing leaves can degrade photosynthetic proteins to some extent, particularly in the light (Feller et al., 2008). Proteolysis might be initiated by the deleterious effects of reactive oxygen species (ROS) over-produced within the organelle during senescence (Zimmermann and Zentgraf, 2005). Toxic effects of ROS are usually balanced by a battery of anti-oxidative enzymes. During leaf senescence, this equilibrium is altered and ROS over-production may occur. The degradation of stromal proteins such as Rubisco and chloroplastic glutamine synthetase (GS2) generated via the action of ROS was shown to release 37- and 16-kDa and 39- and 31-kDa degradation products, respectively (Ishida et al., 1997; Palatnik et al., 1999).
The senescence-induced aspartic protease CND41 encoded by the nuclear genome has been localized specifically to the chloroplast and an in vitro analysis confirmed that CND41 showed a Rubisco proteolytic activity at physiological pH (Kato et al., 2005). However, data suggest that CND41 could not act on Rubisco unless its structure was denatured. Therefore, active Rubisco in the chloroplast would be resistant to CND41-catalysed degradation until leaf senescence was under way. CND41 homologues identified in Arabidopsis accumulate with ageing. Their role in the regulation of Rubisco turnover and senescence in Arabidopsis remains to be studied (Kato et al., 2005; Diaz et al., 2008).
It seems likely that degradation of chloroplast protein starts within the plastid but it is far less clear whether chloroplastic proteases drive protein degradation to the end. The central vacuole that remains intact and for which compartmentation is maintained during senescence might be the end point of chloroplast protein degradation. The increase of vacuolar protease mRNA levels in senescing leaves is in good agreement with this scenario (Martinez et al., 2008). Several possible routes for internalization of chloroplast components by the central vacuole are proposed, such as autophagosome and senescence-associated vesicle (SAV) trafficking.
Autophagy is a well-known process in yeast and animals but it has only been recently established in plants. Macro-autophagy involves formation of autophagosomes, which are double membrane-bound structures enclosing macromolecules and organelle residues. Transcriptome analysis has shown that most of the autophagy genes are upregulated during senescence and in response to nitrogen limitation (Thompson and Vierstra, 2005; Wingler et al., 2009). Chiba et al. (2003) demonstrated that Rubisco is released from the chloroplast into Rubisco-containing bodies (RCBs) in naturally senescent leaves. Concanamycin A is an inhibitor of vacuolar H+-ATPase activity that blocks autophagosome degradation. When leaves of transgenic Arabidopsis plants expressing stromal-targeted fluorescent proteins were incubated with concanamycin A, spherical bodies exhibiting protein-specific fluorescence were observed within the vacuolar lumen. These bodies – corresponding to RCBs – were, however, not observed in the concanamycin A-treated leaves of the atg5 T-DNA insertion mutant impaired for autophagy. In addition, stromal-targeted DsRed proteins and GFP-ATG8 fusion proteins were observed together in autophagic bodies within the vacuole. The authors concluded that Rubisco and stromal proteins can be mobilized to the vacuole through an ATG gene-dependent autophagic process without prior chloroplast destruction (Ishida and Yoshimoto, 2008). The role of autophagy in recycling cell proteins is now accepted, although the premature leaf senescence and accelerated chloroplast degradation observed in autophagy knockout lines is less well understood. As shown by yeast and animal studies, autophagy might have a dual role preventing or triggering cell death depending on its fine-tuning. By removing cell waste, autophagy could be essential for cell longevity. In contrast, excessive autophagy activity could lead to cell death.
SAVs differ from autophagosomes in that they occur only in chloroplast-containing cells whereas autophagosomes have been observed in leaf and root cells (Otegui et al., 2005; Xiong et al., 2005). As with RCBs, evidence has shown that SAVs contain chloroplast proteins such as Rubisco and GS2 (Martinez et al., 2008). Because of their high protease activity, SAVs are a likely site for chloroplast protein degradation. The senescence-associated cysteine-protease encoded by the SAG12 (senescence-associated gene) gene has been detected in SAVs (Otegui et al., 2005). SAG12 may then participate in the intense proteolytic activity contained in this type of vesicle. Because SAG12 is the only SAG whose expression is uniquely controlled by natural senescence, a specific role of SAV in the natural senescence process has been proposed.
Regulation of proteolysis during senescence is then likely to integrate the regulation of chloroplastic and vacuolar proteases and the regulation of various trafficking pathways. Desclos et al. (2009) showed that during the nitrogen remobilization phase occurring in oilseed rape leaf senescence, the downregulation of a protease inhibitor named BnD22 parallels the increase of numerous proteases, including SAG12. The protease activation state might thus be tightly controlled during leaf development in relation with N remobilization (Etienne et al., 2007; Desclos et al., 2009).
Plant nitrogen uptake and metabolism during senescence
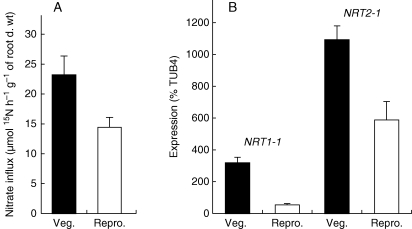
Depending on the species, nitrogen uptake could be negatively regulated or even in some cases totally inhibited during seed production. For example, in oilseed rape, the transition between the vegetative and reproductive phases is characterized by a drastic decrease of HATS and HATS + LATS activities (Malagoli et al., 2004). This change is correlated with a strong repression of gene expression, the BnNRT2 mRNA being undetectable at the flowering stage (Beuve et al., 2004). In contrast, in maize, post-silking nitrogen uptake contributes 30–70 % of the total N in the seeds, according to the environment and the genotype (Coque and Gallais, 2007). In Arabidopsis, when plants are grown under non-limiting N supply (10 mm nitrate), nitrate influx (HATS + LATS) is lower in the reproductive than in the vegetative stage but still operates during seed maturation (Fig. 3). This residual influx is correlated with a decreased yet significant expression of both NRT2·1 and NRT1·1 genes (Fig. 3B; J. Dechorgnat et al., unpubl. res.). However, due to this decrease in N uptake activities, another nitrogen source such as remobilization is necessary to cope with the strong N demand from seed filling.
Fig. 3.
Nitrate uptake (HATS + LATS) is lower during seed maturation than in the vegetative stage but still operates. (A) Root nitrate influx in plants at the vegetative (Veg.) and reproductive (Repro.) stage. Nitrate influx was measured by supplying 6 mm 15NO3− for 5 min to plants grown with 10 mm nitrate. (B) Expression of NRT1-1 (left) and NRT2-1 (right) genes, at the vegetative (Veg.) and reproductive (Repro.) stage. Expression of nitrate transporter genes was measured using quantitative PCR and expressed as a percentage of the tubulin 4 gene, used as a control.
There is evidence that plants share common N remobilization mechanisms whether they are monocotyledonous, dicotyledonous, C3 or C4 photosynthesis types. Grain N accumulation usually appears to be regulated by the N source. In wheat, the kinetics of Rubisco content and grain N accumulation suggest that during grain filling N translocation from the vegetative organs is mainly limited by the availability of the substrate in the source organs (Bertheloot et al., 2008, and references therein). To investigate whether NRE is controlled by source availability or by the transfer processes located in the source leaves and the phloem pathway efficiency, functional genomic and mutant approaches have been used.
Genes encoding enzymes involved in nitrogen metabolism and specifically induced during N remobilization have been identified (Masclaux et al., 2001; Buchanan-Wollaston et al., 2003; Guo et al., 2004). These enzymes are a major focus of plant physiologists, with special focus on cytosolic glutamine synthetase (GS1), glutamate dehydrogenase (GDH) and AS (reviewed by Masclaux-Daubresse et al., 2008). Nitrogen assimilation and recycling in young leaves mainly takes place within the chloroplast where nitrite reduction occurs and ammonium is assimilated by the GS/GOGAT cycle involving chloroplastic GS2 and Fd-GOGAT (Fig. 2A). Chloroplast breakdown during senescence involves de facto NiR, GS2 and GOGAT proteolysis. In senescing leaves, nitrogen recycling and re-assimilation needs then to be catalysed by enzymes other than chloroplastic ones. The metabolic model proposes that glutamine is mainly synthesized in senescing leaves by newly expressed GS1 isoforms (Fig. 2B). Using the amino acid pool released via the proteolysis of chloroplast proteins, a series of transamination reactions would lead to an increase in the glutamate pool that could serve immediately as a substrate for GDH, deaminating activity thus providing 2-oxoglutarate and ammonia. Ammonia released this way is in turn re-assimilated by GS1 to produce glutamine for export.
The importance of GS1 in nitrogen management, growth rate, yield and grain filling has been emphasized by functional genomics and quantitative trait loci (QTL) approaches mainly performed on rice and maize (Hirel et al., 2001; Obara et al., 2004; for a review see Bernard and Habash, 2009). Correlation between GS activity and the amount of N remobilized from shoot to the grain was demonstrated in wheat using cultivars exhibiting contrasted NUE (Kichey et al., 2007) and using a quantitative genetic approach (Habash et al., 2007). However, the role of the GS1 enzyme is complex – numerous isoforms encoded by a multigenic family exist. In rice, three genes have been identified, while maize and Arabidopsis contain five GLN1 genes coding for GS1 (Bernard and Habash, 2009). These genes are not regulated in a similar manner and GS1 isoforms are located in different plant tissues and do not have the same kinetic properties (Ishiyama et al., 2004). It is thus clear that not all GS1 isoforms participate equally in N remobilization.
In Arabidopsis, transcriptomic data show that all GLN1 genes except GLN1·5 are induced by leaf senescence (Guo et al., 2004). GLN1·1 was induced more than five-fold. Functional genomics are in progress to determine the extent of the participation of each of the four senescence-induced GLN1 genes in N remobilization. Our recent results on gln1·2 mutants show that GLN1·2 is expressed in companion cells and that the mutant plants accumulate amides in their old leaves. However, 15N labelling experiments did not show significant differences between mutant and wild-type for N-remobilization (J. Lothier et al., INRA, Versailles, France, unpubl. res.).
Among the five GS1-encoding genes (Gln1-1 to Gln1-5) in maize, only Gln1-4 is upregulated during senescence (Martin et al., 2005, 2006). Gln1-3 and Gln1-4 knockout mutants have been isolated (Martin et al., 2006). The gln1-3, gln1-4 and gln1-3.gln1-4 double mutants showed a sharp reduction of kernel yield whereas nitrogen concentration in the kernels was increased. The gln1-3 and gln1-4.gln1-3 mutants accumulated large amount of amino acids and ammonia in the source leaf located below the ear and dedicated to grain feeding. Amino acid accumulation in the blade was mainly due to an increase in glutamate and asparagine levels as a consequence of a dysfunction in N export that reduced the total amino acid concentration and especially glutamine amounts in the phloem sap of mutants. Interestingly, the Gln1-4 locus co-localized with a maize QTL for thousand-kernel weight, and the Gln1-3 locus co-localized with two QTLs for thousand-kernel weight and yield. More recently, 15N tracing experiments showed that the Gln1-2 and Gln1-4 loci co-localized with a QTL for N remobilization (Hirel et al., 2001; Coque et al., 2008). The role of Gln1-2 remains to be determined.
In rice, mutants lacking OsGS1·1 were severely impaired in growth rate and grain filling. Total free amino acid concentration was reduced in leaf blades of this mutant due to low glutamine levels. The OsGS1·1 gene product, which is located in companion cells and parenchyma cells of leaf tissues, is likely to be responsible for the generation of glutamine for remobilization via the phloem (Obara et al., 2001, 2004).
Efforts to study nitrogen management during leaf senescence have mainly focused on GS1 and to a lesser extent on GDH. However, much data support the idea that GS1 and GDH are not the only limiting factors in N remobilization (for a review see Masclaux et al., 2001). Transcriptomic studies of leaf senescence have shown that several aminotransferase and AS genes are also induced during senescence. In sunflower, expression of two AS genes (HAS1 and HAS1·1) detected only during leaf senescence, when asparagine amounts increased, suggest a role of these enzymes in N remobilization (Herrera-Rodriguez et al., 2006).
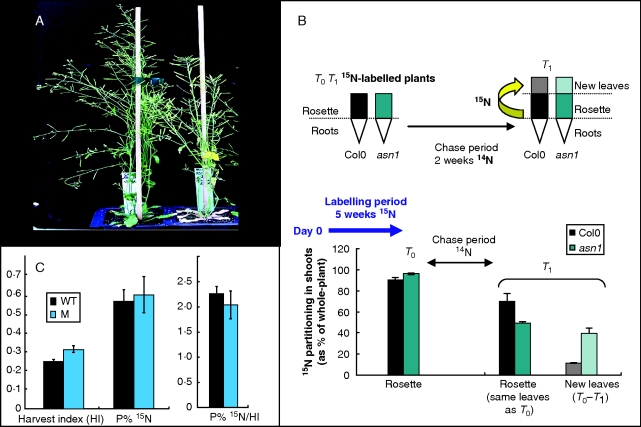
In Arabidopsis, among the three asparagine synthetase genes ASN1, ASN2 and ASN3, only one is over-expressed during leaf senescence (Lam et al., 2003; Buchanan-Wollaston et al., 2005). A study of the asn1 mutant in our laboratory revealed early senescing phenotypes (Fig. 4A). Although Lam et al. (2003) reported that protein and amino acid concentrations were increased in the seeds of ASN1 over-expressor lines, no significant difference between the wild-type and asn1 mutant could be found for seed protein concentration. The role of ASN1 in nitrogen remobilization from leaf to leaf and from rosette to seeds was investigated using 15N tracing as described by Diaz et al. (2008). Results presented in Fig. 4B show that nitrogen remobilization from old leaves to younger ones was higher in the asn1 mutant than in the wild-type. Such a finding is in good agreement with the relationship between severity of leaf-senescence and NRE at the vegetative stage described by Diaz et al. (2008) and suggests that N-remobilization from leaf to leaf does not depend only on ASN1. By contrast, we found that 15N-remobilization from rosette to seeds was slightly impaired in the asn1 mutant (Fig. 4C). Although the partitioning of 15N in seeds (P%15N) did not differ between the asn1 mutant and the wild-type, harvest index (HI; partitioning of dry matter in seeds) was significantly higher for the mutant than wild-type. As a result, the ratio (P%15N/HI) appeared to be slightly lower in the mutant than in the wild-type, indicating that ASN1 might have a role in harvest index and in N-remobilization from vegetative tissues to the seeds. This preliminary result shows that the role of asparagine synthetase is certainly complex and further investigations taking into account the contribution of the other asparagine synthetases, ASN2 and ASN3, are needed to understand the role of AS in nitrogen recycling and mobilization at the whole-plant level.
Fig. 4.
Asparagine synthetase AS1 might play a role in nitrogen recycling and mobilization. (A) Phenotypes of asn1 mutant (Gabi 829B05) and Col0 wild-type grown in greenhouse with 10 mm nitrate. (B) Hydroponically grown asn1 mutant (Gabi 829B05) and Col0 were labelled using 15NO3−-containing nutritive solution for 4 weeks (from sowing to the end of the pulse period T0). At T0, plants were transferred to a 15NO3−-free solution and chase period was performed over 2 weeks (T1 = T0 + 2 weeks). At T1, partitioning of 15N (15N%, as percentage of whole plant) in roots, rosette (already emerged at T0) and new leaves (emerged between T0 and T1) was monitored in ans1 mutant and Col0. Nitrogen remobilization from rosette to the new leaves occurring during the chase period (T0 to T1) was higher in asn1 mutant than in wild-type, as shown by the greater decrease of 15N% in rosette and the higher 15N% in new leaves of mutant. (C) Nitrogen remobilization from the vegetative tissues to the seeds was monitored according to Diaz et al. (2008). 15N labelling was performed once at the vegetative stage. At the end of the plant cycle, the dry weight of seeds and dry remains were recorded and used to calculate harvest index (HI, seed d. wt as a percentage of the whole-plant d. wt). The amount of 15N remobilized from the rosette to the seeds was estimated through the calculation of 15N partitioning in seeds (P%15N; 15N in seeds as a percentage of total 15N in the whole plant). Comparing P%15N/HI ratios facilitates comparison between plants with different HI. The mutant (M) displayed significantly higher HI than wild-type (WT) and might be impaired for N-remobilization to the seeds as shown by its slightly lower P%15N/HI ratio. Such preliminary finding needs confirmation using a second mutant allele.
Phloem, a key factor for amino acid translocation from sources to sinks
Prior to phloem loading the central vacuole of mesophyll cells might be a site for transient storage of amino acids released from protein degradation. In tobacco, the total amount of amino acids exported from leaf blades increased five-fold during leaf ageing (Masclaux-Daubresse et al., 2006). Asparagine is the major translocated amino acid in pea. In cereals, tomato and tobacco, glutamine is the preferentially exported N-compound. In Arabidopsis, phloem sap mainly contains asparagine, glutamate and glutamine (J. Lothier, INRA, Versailles, France, unpubl. res.). During senescence, both asparagine and glutamine concentrations increase in the phloem sap and both amino acids are likely to play a key role in rendering nitrogen available for remobilization from the senescing leaf. The Arabidopsis genome encodes 67 (putative) amino acid transporters belonging to 11 gene families (reviewed by Rentsch et al., 2007). The nature of the amino acid transporter involved in phloem loading during senescence is, however, poorly understood (van der Graaff et al., 2006; for a review see Masclaux-Daubresse et al., 2008).
N-storage and remobilization are essential for plant survival
N-storage and remobilization potential are important for both annual and perennial plants. For annual plants, as mentioned above, nitrogen remobilization is important for seed production and seed nitrogen content. Nitrogen content in the seeds further determines germination efficiency and survival of young seedlings.
Nitrogen remobilization is also important for perennial plant survival. Trees, which grow in low nitrogen environments most of the time, have two phases of nitrogen remobilization. Nitrogen is remobilized from the senescing leaves in autumn to be stored in trunks during winter. N is remobilized a second time from trunks to developing organs in spring before root N uptake becomes the main process to meet tree N needs. As trees age, the internal cycling of N becomes more and more important in the whole-tree N-budget. Both nitrogen withdrawal from senescing leaves and root N uptake contribute to the build-up of N storage pools and to the efficient nitrogen management that are essential for plant survival over years (Millard et al., 2006, 2007).
Forage grasses are subject to frequent defoliation by herbivores or mechanical harvesting. Recovery of grasses after defoliation is related to the availability of carbon and nitrogen reserves in the remaining tissues (Volenec et al., 1996). Decreasing mineral N supply before defoliation was shown to decrease the availability of N reserves in leaves and as a result the absolute amount of N subsequently remobilized to roots.
Interestingly, it was shown that N remobilization and senescence can be induced prematurely by environmental factors, such as pathogen attack or heavy metals. Evidence corroborates that the N remobilization process is enhanced by biotic and abiotic stresses through the induction of the GLN1, GDH and ASN genes (Pérez-Garcia et al., 1998a, b; Olea et al., 2004). Chaffei et al. (2004) showed that cadmium toxicity induced senescence-like symptoms and the expression of the N remobilization enzymes GS1 and GDH in tomato leaves. The induction of AS in roots might facilitate amino acid recycling and storage of asparagine in this organ. The co-ordinated leaf N remobilization and root N storage is certainly essential for plant recovery. Pageau et al. (2006) showed that GS1 and GDH genes in tobacco leaves responded to ethylene, jasmonic acid and several other plant defence elicitors as well as to virus and bacterial infections. N mobilization promoted by infection could be considered on the one hand as part of a slash-and-burn strategy that should deprive the pathogen of nutrients by exporting nutrients away from the developing infection site, and on the other hand as a strategy to save nutrients in healthy organs involved in recovery.
REGULATION OF NITROGEN UPTAKE, ASSIMILATION AND REMOBILIZATION BY NITRATE AND CARBON AVAILABILITIES
N uptake by the roots and further N assimilation are integrated in the plant to match the nutrient demand of the whole organism. External stimuli or stresses as well as nutritional status of the plant modulate the expression and/or the activity of transport systems and enzymes by various regulatory mechanisms.
The first mechanism operates at the transcriptional level and includes the induction by the substrates and the repression exerted by endogenous N assimilates. This results in an upregulation when N is low and a downregulation when N is high. Accordingly, several NRT2 and AMT1 transporters as well as Nia and Nii genes were found to be transcriptionally repressed by N metabolites such as amino acids like glutamine (Tsay et al., 2007; Meyer and Stitt, 2001). On the other hand, in response to N deprivation, expression of many ammonium and high-affinity nitrate transporters is induced or repressed (reviewed by Tsay et al., 2007). In response to N deprivation, expression of GLN and GDH genes is also up- or downregulated. In tobacco, it was shown that ammonium and amino acids regulate GLN and GDH transcript levels (Masclaux-Daubresse et al., 2005). Glutamate feeding over 5 h increased GLN1 mRNA while both glutamate and proline feeding decreased GLN2 mRNA. Ammonium, proline, glutamine and glutamate increased GDH transcripts. Effects of N-metabolites on GLN and GDH transcript levels proved to be sensitive to calcium blockers and the K252a protein-kinase inhibitor. Evidence showed that ammonium itself regulates GLN genes at the transcriptional level. In soybean, co-operation between three distinct promoter regions is necessary for ammonium-stimulated expression of the GS15 gene. The interaction among these regions may be facilitated by an HMG A (high-mobility group A)-like protein that binds to the proximal and distal promoter regions of the soybean GS15 gene (Reisdorf-Cren et al., 2002, and references therein). Global transcriptome studies after nitrate induction (Scheible et al., 2004) confirmed regulation of N uptake and assimilation by nitrate at the expression level and showed a large action spectrum of nitrate as a regulator of gene expression, co-ordinating for example C and N metabolism. Using NR mutants, it was shown that much of this regulation is exerted by nitrate itself (Wang et al., 2004).
The stimulation of N uptake and N assimilation by photosynthesis (for a review see Lillo, 2008) ensures that N uptake is correlated with C status. For example, nitrate uptake and reduction are co-ordinately regulated by a circadian control. This control has often been attributed to the regulatory action on gene expression of sugars produced by photosynthesis and transported downward to the roots. This has been shown for the ammonium and nitrate transporters, NR and NiR. The regulation of nitrate uptake and transporters seems to be independent of the known sugar regulation pathways, such as hexokinase signalling (Lillo, 2008). Wirth et al. (2007) showed that upregulation of nitrate transporters (AtNRT2·1 and AtNRT1·1) was related to the concentration of glucose 6-phosphate. In contrast, the diurnal regulation of Nia transcripts is governed not only by sugars but also by light regulation via phytochrome (Lillo, 2008). In addition, it was observed that Nia expression is controlled by signals from photosynthetic electron flow, which adds a new facet to the intracellular cross-talk between chloroplasts and the nucleus (Lillo, 2008).
HY5 and its homologue HYH, two transcription factors from the bZIP family, are essential for phytochrome-dependent light-activated expression of NR (Lillo, 2008). ChIPchip analyses showed a binding site for HY5 in the Nia2 promoter (Lillo, 2008). The NRT1·1 promoter also has three binding sites for HY5, although HY5 seems to have a negative effect on transcription in this case (Lillo, 2008). Castaings et al. (2009) identified NLP7 as a regulator of Nia expression in Arabidopsis. Arabidopsis nlp7 mutants are defective in the nitrate induction of Nia genes and NRT2·1 and NRT2·2. Interestingly, mutants in the CIPK8 gene, which encode a protein kinase (Hu et al., 2009), are also unable to fully induce expression of several genes by nitrate, such as the Nia genes, NRT2·1, NRT1·1 and several others. It is tempting to speculate that CIPK8 might be involved in the same regulation pathway as NLP7. NLP7 belongs to a gene family with nine different members, but the functions of the other NLP proteins are still unknown.
In Arabidopsis, compelling evidence shows that SnRK1s (Snf1-related protein kinases) are central integrators of transcription networks in plant stress and energy signalling that are inactivated by sugars (Baena-Gonzalez et al., 2007). SnRK1 proteins were shown to specifically regulate the ASN1 gene, encoding the dark-induced asparagine synthetase also known as DIN6. ASN1/DIN6 is induced by darkness, hypoxia, DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] and other stress within 3–6 h. The protein kinase inhibitor K252a abolishes such induction and the two ubiquitously expressed members of the SnRK1 group, Kin10 and Kin11, specifically activate a DIN6promoter::LUC fusion. Mutation of the G-box (CACGTG, G1) proximal to the TATA box abolished most of the activation of DIN6promoter::LUC by KIN10, hypoxia, darkness and DCMU.
Rapid post-translational regulation such as protein modification is the second mechanism that controls nitrogen uptake and assimilation. Post-transcriptional regulation of nitrate transporters by phosphorylation has recently been described for the nitrate transporter NRT1·1. When phosphorylated, AtNRT1·1 functions as a high-affinity transporter whereas it is active in the low affinity range when dephosphorylated (for a review see Tsay et al., 2007). Tsay and co-workers obtained significant insight into the role of the CIPK23 kinase in the specific phosphorylation of the AtNRT1·1 protein in response to nitrate levels, demonstrating AtNRT1·1-mediated sensing in the primary nitrate response (Ho et al., 2009).
The best studied post-translational regulation in N metabolism is the regulation of NR in higher plants. NR is inactivated by a two-step process that involves the phosphorylation of ser543, as shown in spinach, followed by the binding of an inhibitory 14-3-3 protein kinase. In addition, both CDPK (calcium-dependent protein kinases) and SnRK1 protein kinases are able to phosphorylate NR at least in vitro (reviewed by Lillo, 2008). When a modified form of NR, no longer susceptible to post-translational dark inactivation, was over-expressed, the resulting protein did not decline in the second part of the photoperiod. The inactive phosphorylated form is re-activated by dephosphorylation, probably by PP2A. Moreover, evidence showed that there is a correlation between the phosphorylation state or the activation state of NR and the rate at which NR protein decreases.
Plastidic glutamine synthetase from Medicago truncatula is also regulated through phosphorylation and 14-3-3 interactions. The GS2 phosphorylation site ser97, critical for the interaction with 14-3-3 and subsequent proteolysis, was identified by directed mutagenesis. Cytosolic glutamine synthetases from M. truncatula are also regulated by phosphorylation but by calcium-independent kinases (Lima et al., 2006, and references therein). Phosphorylation occurs at more than one residue and increases affinity for the substrate glutamate.
In addition to phosphorylation, several chloroplastic enzymes of nitrogen assimilation such as NIR, GS2 and Fd-GOGAT are also redox-regulated through the thioredoxin system (for reviews see Lemaire et al., 2007; Lillo, 2008).
TRANSGENIC EFFORTS TO MANIPULATE NUE
With the aim of improving NUE, many critical candidate genes have been manipulated, over-expressing them or using knockout mutations, in order to test their effects on biomass and plant nitrogen status. Several good reviews have been written on this subject that provide more detail than mentioned in this section (Andrews et al., 2004; Good et al., 2004; Pathak et al., 2009).
Until now, probably because of strong post-transcriptional controls (see above), manipulating nitrate uptake through the over-expression of HATS-like NRT2·1 led to increased nitrate influx under some conditions but did not change the phenotypic NUE or nitrate utilization (Fraisier et al., 2000). NR has long been considered to be the rate-limiting step in nitrate assimilation. The utility of transgenic over-expression of NR/NiR for major improvements of NUE, however, remains uncertain. Nicotiana tabaccum plants constitutively expressing NR from N. plumbaginifolia showed delay in NR-activity loss under drought, which allowed them to present a more rapid recovery after short-term water deficit (Ferrario-Mery et al., 1998). Then, under field conditions of fluctuating water availability, constitutive NR expression may confer some physiological advantage. Over-expressing NR or NiR in Arabidopsis, potato or tobacco reduced nitrate levels in plant tissues but did not increase biomass yield, tuber numbers or seed yields. Over-expression of Nia or Nii genes in plants increased mRNA levels and often affected N uptake without modifying yield or plant growth regardless of the nitrogen source available. This is believed to be due in part to the complex post-transcriptional regulation of NR (reviewed by Pathak et al., 2009).
Over-expression of cytosolic glutamine synthetase GS2 genes was performed in N. tabaccum and Oryza sativa using the Rubisco small subunit promoter and the CaMV 35S promoter, respectively (Hoshida et al., 2000; Migge et al., 2000). In N. tabaccum, over-expression enhanced growth rate and in O. sativa it increased photorespiration and drought tolerance. Attempts to over-express GS1 genes are more numerous and have used different promoter combinations, including CaMV 35S, RolD and small Rubisco subunit (rbcS). Effects on plant biomass and grain yield were also more successful. For example, over-expression of the Phaseolus vulgaris GS1 gene under the control of the rbcS promoter in wheat resulted in significantly higher root and grain yield with higher N content in grain in some lines (Habash et al., 2001). Over-expression of the Pisum sativum GS1 gene under the control of the CaMV 35S promoter in N. tabaccum resulted in biomass and leaf protein increases (Oliveira et al., 2002). More recently, over-expressing a GS1 isoenzyme of maize in maize under the control of the CsVMV promoter increased kernel number and kernel size, thus increasing yield by 30 % [patent AU2007306040 (A1); http://v3.espacenet.com/publicationDetails/biblio?CC=AU&NR=2007306040A1&KC=A1&FT=D&date=20080417&DB=EPODOC&locale=en_EP]. In summary, several studies have demonstrated a direct correlation between an enhanced GS activity in transgenic plants and biomass or yield (Good et al., 2004, and references therein). Although over-expression of GOGAT genes has been rare, Yamaya et al. (2002) reported a spectacular effect of the over-expression of NADH-GOGAT under the control of its own promoter in rice. Transgenic plants showed an increase (up to 80 %) in grain weight. In conclusion, studies show that over-expression of GS or GOGAT genes can improve biomass and grain yields depending on which gene allele and which promoters are used. This indicates that further characterization is required to demonstrate the beneficial effects of such strategies for crops and in field conditions.
Attempts to over-express AS were carried out in tobacco and Arabidopsis (for a review see Good et al., 2004). Interestingly, over-expression of ASN1 in Arabidopsis enhanced soluble seed protein content and total protein and increased fitness of plants grown under nitrogen-limiting conditions (Lam et al., 2003).
Alanine is a major amino acid for nitrogen storage under anaerobic stress such as flooding. Over-expression of barley alanine amino transferase under the control of root promoters in canola and rice had interesting effects, considerably increasing plant biomass, seed yield, NUE and shoot nitrogen concentration when plants were grown at low nitrate supply (Good et al., 2007; Shrawat et al., 2008). These results are of particular interest, showing that it is possible to improve NUE by manipulating downstream steps in N-remobilization.
In addition to manipulating enzymes involved in nitrogen assimilation of amino acid metabolism, the generation of plants modified for the expression of transcription factors has also been attempted. For example, ectopic expression of the maize Dof1 transcription factor, which regulates the expression of genes involved in organic acid metabolism, led in Arabidopsis to the accumulation of amino acids and to an increase of growth under N-limiting conditions. These effects suggest that NUE could also be improved by manipulating carbon metabolism pathways. PII-like, NLP7 and TOR (target of rapamycin) proteins, which are potentially linked to C and N sensing in plants, are other candidates for further engineering as shown by the increased plant growth, yield and stress resistance acquired by TOR-overexpressing plants (Ferrario-Mery et al., 2006; Deprost et al., 2007; Castaings et al., 2009).
Hibberd et al. (2008) suggested that engineering C4 rice or C4 wheat would be a good way to improve NUE. The Rubisco protein is known to be used as a storage protein in C3 herbaceous plants and trees (Millard et al., 2006). In elevated atmospheric CO2, Rubisco carboxylase activity is increased and Rubisco protein content is decreased. The selective loss of Rubisco enzyme under elevated CO2 thus benefits NUE without necessarily significantly changing the leaf C assimilation rate. The lower Rubisco levels in C4 plants explains why NUE is higher in C4 crops than in C3.
NATURAL VARIATION OF NITROGEN UPTAKE AND NITROGEN REMOBILIZATION IN ARABIDOPSIS
More than 300 accessions of Arabidopsis, originating from various locations worldwide, are available in stock centres. Probably due to a selective adaptation to original edaphic and climatic environments, they show natural variation of their development and they constitute large genetic and phenotypic resources (McKhann et al., 2004). Several recent papers have presented the first evidence that natural variation exists for nitrogen metabolism, including nitrogen uptake and nitrogen remobilization. The first clue was provided by the analysis of root plasticity. Studies in Arabidopsis have shown the stimulation of root growth by a localized source of nitrate (Robinson, 1994; Forde and Lorenzo, 2001). Walch-Liu and Forde (2008) assayed the extent of root stimulation using a small collection of six accessions. A large range of responses was obtained from one extreme, which was not significantly affected, to the other extreme that showed a 90 % increase of primary root growth. This observed variation of root adaptation to nitrogen availability should have consequences for nitrogen uptake in plants.
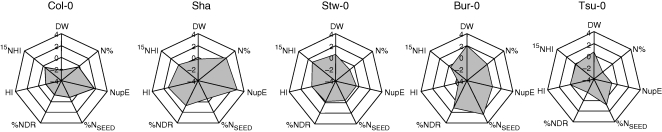
Our recent investigation of natural variation in nitrate uptake and nitrogen remobilization in Arabidopsis gives the second clue (Fig. 5; C. Masclaux-Daubresse and F. Chardon, unpubl. res.). A core-collection of 18 accessions of Arabidopsis was grown under limiting and ample nitrogen nutritive conditions. The aim of this study was to collect data allowing us to monitor the natural variation of N uptake at the vegetative stage and the N-remobilization to the seeds at the reproductive stage depending of nitrogen availability, and also to measure traits related to NUE such as biomass of rosettes at the vegetative stage, seed yield, harvest index and nitrogen concentrations in the different plant material collected (at vegetative and reproductive stages). Using all the data collected at low and high nitrogen supply, groups of accessions can be clustered, assembling plants that have similar responses depending on nitrate availability. Surprisingly, for most of the traits measured or computed the variation was higher at high nitrate supply when N uptake and N remobilization are not forced by nitrate limitation. Figure 5 shows schematically the large variation observed between the classes of accessions. The differences between the lowest and highest performing accessions were three-fold for N uptake and six-fold for N remobilization. We also noted interesting correlations between how plants manage N uptake and N metabolism and their biomass. Considering the relative NUE at the vegetative stage as the ratio DW/N% (optimal NUE: plants with large biomass and low nitrogen concentration), we found that the estimated NUE in the core-collection was not different at limiting and ample nitrogen supply, suggesting that NUE at the vegetative stage was determined solely genetically (C. Masclaux-Daubresse and F. Chardon, unpubl. res.).
Fig. 5.
Nitrogen absorption and nitrogen remobilization profiling of five Arabidopsis accessions. A core collection of 18 accessions of Arabidopsis grown with 10 mm nitrate was used to measure traits related to biomass, N uptake, N remobilization and NUE (C. Masclaux-Daubresse and F. Chardon, unpubl. res.). Five accessions representative of the main classes found are presented. Traits measured at the vegetative stage (40 d after sowing) are rosette dry weight (DW), nitrogen concentration in the rosette (N%) and N-uptake efficiency (NupE) as the quantity of 15N absorbed at 40 d after sowing divided by plant biomass. Traits measured at the end of the plant cycle are N remobilization efficiency (15NHI) expressed as the partitioning of 15N in seeds compared with whole plant (seeds + dry remains; see Diaz et al., 2008), harvest index (HI) expressed as the partitioning of biomass (dry matter) in seeds (DM of seeds/DM of the whole plant), N concentration in dry remains (%NDR) and N concentration in the seeds (%NSEED). Individual values ranged between [−4; + 4] and centred around the mean value of the core collection (value 0). The small sample of accessions highlights the variation of performances. Plants, such as Col0 or Sha, have a relatively good N uptake. The highest N remobilization score was found in Stw-0 while plants with high N percentage and high biomass were Bur-0 and Tsu-0.
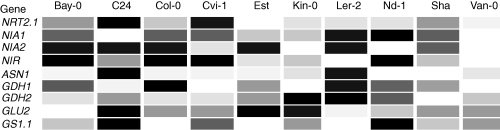
The last clue is provided by the Arabidopsis eFP-Browser database, which combines microarray analyses (Winter et al., 2007). Some experiments included in this database used several accessions (Lempe et al., 2005). It is possible to select specifically the pattern of genes involved in primary N metabolism. Although plants were cultivated in the same conditions (4-day-old seedlings grown in soil in the glasshouse), the signal intensities of some N genes indeed varied among these accessions (Fig. 6), revealing the existence of genetic variation for the transcription level of N genes. Whether such variation is correlated with N-dependence and NUE remains to be determined.
Fig. 6.
Heat map illustrating the natural variation in expression of genes involved in nitrogen metabolism. Signal levels of N genes in ten accessions were obtained from the Arabidopsis eFP-Browser. For each gene, high expression is depicted as dark shading, and low expression is depicted as light shading.
Experiments presented above provide ideas about the various traits that can be measured to explore NUE (N-gene transcription levels, N uptake efficiency, NRE, nitrogen content, enzyme activities, biomass) and encourage computing data according to a systems biology approach, in order to reveal the functioning of the different modules that constitute nitrogen metabolism adaptation in plants. A better characterization of edaphic environments and the metabolism of different Arabidopsis accessions would consequently allow a better understanding of how these modules are regulated according to the nitrogen availability in soil.
Natural variation also exists in crops. Approaches currently performed by breeders to improve varieties for many agronomical traits include QTL mapping and marker-assisted selection. However, it is worth noting that the experiments performed on Arabidopsis presented above have been carried out on wild plants, meaning that the plants studied have not been modified for agronomic criteria and that no adaptive selection has decreased differences between them. Using Arabidopsis rather than highly selected plants such as crops for such approaches should then be more informative. Natural variation of N uptake and N remobilization identified in the model plant Arabidopsis is a source of knowledge that can be useful to transfer to crops.
QUANTITATIVE GENETIC AND GENETIC BASES OF NITROGEN UPTAKE, ASSIMILATION AND REMOBILIZATION
QTLs for NUE and other agronomic traits have been mapped in numerous plant species, including maize, rice and Arabidopsis (Hirel et al., 2001; Obara et al., 2001; Loudet et al., 2003). QTL mapping for nitrogen-related enzyme activities such as nitrate reductase or glutamine synthetase are rarer. Even more rare is the mapping of N-remobilization or N-influx QTLs because of the difficulty in performing 15N tracing on large populations.
The first report of mapping QTL for N remobilization was in barley using the N-balance method, which requires monitoring the difference in flag leaf N-content between anthesis and maturity [ΔN (mg) per leaf] (Mickelson et al., 2003). QTLs explaining the variation of this trait were mapped on chromosomes 6 and 7. Mickelson et al. (2003) also mapped two QTLs for grain protein concentration on chromosomes 2 and 6. There was no co-localization between the QTLs for ΔN per leaf and grain protein content. However, the most prominent QTL for grain protein content on barley chromosome 6 appeared to be a potential homologue of the grain protein QTL from durum wheat mapped by Joppa et al. (1997). Recently, a wheat QTL was cloned through positional cloning and fine mapping (Uauy et al., 2006). The locus encodes an NAC transcription factor, NAM-B1, which accelerates leaf senescence and increases nutrient filling in developing grains. The ancestral wild wheat allele is functional whereas modern wheat varieties carry a non-functional NAM-B1 allele. The role of NAM-B1 was confirmed by reducing the RNA levels of multiple NAM homologues by RNA interference, resulting in delayed senescence (by more than 3 weeks) and reduced wheat grain protein, zinc and iron contents (by more than 30 %). The effect of the chromosome Gpc-B1 region including the NAM-B1 gene was studied further by introgressing the Gcp-B1 locus in hexaploid near-isogenic lines. As a result of Gcp-B1 introgression, significantly lowered straw N concentration at maturity and higher nitrogen harvest index (NHI) were measured, suggesting that the functional Gcp-B1 allele improves N remobilization and diminishes the amount of nitrogen lost in residual dry remains (Brevis and Dubcovsky, University of California, Davis, CA, unpubl. res.). Comparing NAM-B1 knockdown RNAi and control lines showed similar results (Waters et al., 2009).
In the same barley population used by Mickelson et al. (2003) to determine grain protein and leaf N storage and remobilization, QTLs for leaf amino-, carboxy- and endo-peptidase activities were determined (Yang et al., 2004). QTL co-localization strongly suggested that the major endo- or amino-peptidases were not involved in leaf N remobilization or in the control of grain protein content. By contrast, QTL co-localization suggested that vacuolar carboxy-peptidase isoenzymes are involved in leaf N remobilization.
More recently, N-uptake and N-remobilization QTLs have been mapped in maize (Coque and Gallais, 2007; Coque et al., 2008). The authors monitored an impressive number of traits for NUE, leaf senescence, enzyme activities, yield, biomass, N uptake and N remobilization, which, together with the former data from Hirel et al. (2001) and additional gene positional cloning, provide a huge amount of information about the coincidence of agronomical, physiological, biochemical and genomic traits. The study by Coque et al. (2008) is also the first and to date the only study to have used the 15N tracing method to map N-remobilization QTLs. QTL clustering showed an antagonism between N remobilization and N uptake at several loci. Positive coincidences between N uptake, root system architecture and leaf greenness were also found in eight clusters, while N-remobilization QTLs mainly coincided with leaf senescence QTLs. Co-localization with N-related genes showed that the two NR loci (chromosomes 1 and 4, see Hirel et al., 2001) coincided with QTLs affecting kernel number at high N input and kernel weight at both low and high N. Grain yield-related traits coincided with the three GS1 loci corresponding to Gln1-2, Gln1-3 and Gln1-4. The GS2 locus (chromosome 10) coincided with a leaf senescence QTL (Coque et al., 2008).
QTLs for NUE and N-enzyme activities were also recently explored in wheat (Habash et al., 2007; Laperche et al., 2007; Fontaine et al., 2009). Interestingly, Habash et al. (2007) showed that QTLs for GS activity were invariably co-localized with those for grain N, and that increased activity was associated with higher grain N. This finding was confirmed by Fontaine et al. (2009) on another population. Unlike in maize, there was no correlation between GS activity and yield components in wheat.
CONCLUSIONS
Improving global plant productivity and product quality together with taking care of environmental quality and human wellbeing are the main challenges for the immediate future (Vitousek et al., 2009). Such a goal depends on agricultural development and policy and can be achieved by providing the right nutrient source at the right rate, the right time and the right place. To improve sustainable agricultural production, it is also necessary to grow crops that can remove the nutrient applied to soil efficiently, and therefore require less fertilizer. Such global ‘resource use efficiency’ necessitates having a global view of plant physiology, plant uptake capacity, plant metabolism and plant response to restrictions, as well as a view of soil physical and chemical properties. This review gives an overview of the different metabolic and physiological clues that agronomical research has provided. The enzymes and regulatory processes that can be manipulated to control NUE are presented. The last results obtained from natural variation and QTL studies show the complexity of NUE and open new perspectives. With regard to the complexity of the challenge we have to face and with regard to the numerous approaches available, the integration of data coming from transcriptomic studies, functional genomics, quantitative genetics, ecophysiology and soil science into explanatory models of whole-plant behaviour in the environment has to be encouraged.
ACKNOWLEDGEMENT
We are grateful to Dr Heather I. McKhann (INRA, Paris, France) for carefully reading the manuscript.
LITERATURE CITED
- Almagro A, Lin S, Tsay Y. Characterization of the Arabidopsis nitrate transporter NRT1·6 reveals a role of nitrate in early embryo development. The Plant Cell. 2008;20:3289–3299. doi: 10.1105/tpc.107.056788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M, Lea PJ, Raven JA, Lindsay K. Can genetic manipulation of plant nitrogen assimilation enzymes result in increased crop yield and greater N-use efficiency? An assessment. Annals of Applied Biology. 2004;145:25–40. [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytologist. 2009;182:608–620. doi: 10.1111/j.1469-8137.2009.02823.x. [DOI] [PubMed] [Google Scholar]
- Bertheloot J, Martre P, Andrieu B. Dynamics of light and nitrogen distribution during grain filling within wheat canopy. Plant Physiology. 2008;148:1707–1720. doi: 10.1104/pp.108.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuve N, Rispail N, Laine P, Cliquet J-B, Ourry A, Le Deunff E. Putative role of gamma-aminobutyric acid (GABA) as a longdistance signal in up-regulation of nitrate uptake in Brassica napus L. Plant Cell Environment. 2004;27:1035–1046. [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, et al. The molecular analysis of leaf senescence – a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark / starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. The Plant Journal. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- Chaffei C, Pageau K, Suzuki A, Gouia H, Ghorbel MH, Masclaux-Daubresse C. Cadmium toxicity induced changes in nitrogen management in Lycopersicon esculentum leading to a metabolic safeguard through an amino acid storage strategy. Plant and Cell Physiology. 2004;45:1681–1693. doi: 10.1093/pcp/pch192. [DOI] [PubMed] [Google Scholar]
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T. Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant and Cell Physiology. 2003;44:914–921. doi: 10.1093/pcp/pcg118. [DOI] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF. Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant and Cell Physiology. 2004;45:1139–1148. doi: 10.1093/pcp/pch143. [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, et al. The Arabidopsis ATNRT2·7 nitrate transporter controls nitrate content in seeds. The Plant Cell. 2007;19:1590–1602. doi: 10.1105/tpc.107.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliquet J-B, Deléens E, Mariotti A. C and N mobilization from stalk to leaves during kernel filling by 13C and 15N tracing in Zea mays L. Plant Physiology. 1990;94:1547–1553. doi: 10.1104/pp.94.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque M, Gallais A. Genetic variation for nitrogen remobilization and postsilking nitrogen uptake in maize recombinant inbred lines: heritabilities and correlations among traits. Crop Science. 2007;47:1787–1796. [Google Scholar]
- Coque M, Martin A, Veyrierias JB, Hirel B, Gallais A. Genetic variation of N-remobilization and postsilking N-uptake in a set of maize recombinant inbred lines. IIi. QTL detection and coincidences. Theoretical and Applied Genetics. 2008;117:729–747. doi: 10.1007/s00122-008-0815-2. [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. The expanded family of ammonium transporters in the perennial poplar plant. New Phytologist. 2007;174:137–150. doi: 10.1111/j.1469-8137.2007.01992.x. [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F, Filleur S, Caboche M. Nitrate transport: a key step in nitrate assimilation. Current Opinion in Plant Biology. 1998;1:235–239. doi: 10.1016/s1369-5266(98)80110-6. [DOI] [PubMed] [Google Scholar]
- De Angeli A, Monachello D, Ephritikhine G, et al. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos M, Etienne P, Coquet L, et al. A combined 15N tracing/proteomics study in Brassica napus reveals the chronology of proteomics events associated with N remobilisation during leaf senescence induced by nitrate limitation or starvation. Proteomics. 2009;9:3580–3608. doi: 10.1002/pmic.200800984. [DOI] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ C, et al. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiology. 2008;147:1437–1449. doi: 10.1104/pp.108.119040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne P, Desclos M, Le Gou L, et al. N-protein mobilisation associated with the leaf senescence process in oilseed rape is concomitant with the disappearance of trypsin inhibitor activity. Functional Plant Biology. 2007;34:895–906. doi: 10.1071/FP07088. [DOI] [PubMed] [Google Scholar]
- Feller U, Anders I, Mae T. Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. Journal of Experimental Botany. 2008;59:1615–1624. doi: 10.1093/jxb/erm242. [DOI] [PubMed] [Google Scholar]
- Ferrario-Mery S, Valadier M, Foyer C. Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiology. 1998;117:293–302. doi: 10.1104/pp.117.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario-Mery S, Besin E, Pichon O, Meyer C, Hodges M. The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Letters. 2006;580:2015–2020. doi: 10.1016/j.febslet.2006.02.075. [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe M, Cerezo M, et al. An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Letters. 2001;489:220–224. doi: 10.1016/s0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- Fontaine JX, Ravel C, Pageau K, et al. A quantitative genetic study for elucidating the contribution of glutamine synthetase, glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theoretic and Applied Genetics. 2009;119:645–662. doi: 10.1007/s00122-009-1076-4. [DOI] [PubMed] [Google Scholar]
- Forde B, Lorenzo H. The nutritional control of root development. Plant and Soil. 2001;232:51–68. [Google Scholar]
- Fraisier V, Gojon A, Tillard P, Daniel-Vedele F. Constitutive expression of a putative high affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for a post transcriptional regulation by a reduced nitrogen source. The Plant Journal. 2000;23:489–496. doi: 10.1046/j.1365-313x.2000.00813.x. [DOI] [PubMed] [Google Scholar]
- Gallais A, Coque M, Quillere I, Prioul JL, Hirel B. Modelling postsilking nitrogen fluxes in maize (Zea mays) using 15N labelling field experiments. The New Phytologist. 2006;172:696–707. doi: 10.1111/j.1469-8137.2006.01890.x. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wiren N. Three functional transporters for constitutive, diurnally regulated, and starvation induced uptake of ammonium into Arabidopsis roots. The Plant Cell. 1999;11:937–947. doi: 10.1105/tpc.11.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends in Plant Science. 2004;9:597–605. doi: 10.1016/j.tplants.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Good AG, Johnson SJ, De Pauw M, et al. Engineering nitrogen use efficiency with alanine aminotransferase. Canadian Journal of Botany. 2007;85:252–262. [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell and Environment. 2004;27:521–549. [Google Scholar]
- Habash D, Massiah A, Rong H, Wallsgrove R, Leigh R. The role of cytosolic glutamine synthetase in wheat. Annals of Applied Biology. 2001;138:83–89. [Google Scholar]
- Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA. The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theoretical and Applied Genetics. 2007;114:403–419. doi: 10.1007/s00122-006-0429-5. [DOI] [PubMed] [Google Scholar]
- Herrera-Rodriguez MB, Maldonado JM, Perez-Vicente R. Role of asparagine and asparagine synthetase genes in sunflower (Helianthus annuus) germination and natural senescence. Journal of Plant Physiology. 2006;163:1061–1070. doi: 10.1016/j.jplph.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice – rationale and feasibility. Current Opinion in Plant Biology. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hirel B, Bertin P, Quillere I, et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiology. 2001;125:1258–1270. doi: 10.1104/pp.125.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany. 2007;58:2369–2387. doi: 10.1093/jxb/erm097. [DOI] [PubMed] [Google Scholar]
- Hirner A, Ladwig F, Stransky H, et al. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. The Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C, Lin S, Hu H, Tsay Y. CHL1 functions as a nitrate sensor in plants. Cell. 2009;18:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Hodge A, Robinson D, Fitter A. Are microorganisms more effective than plants at competing for nitrogen? Trends in Plant Science. 2000;5:304–308. doi: 10.1016/s1360-1385(00)01656-3. [DOI] [PubMed] [Google Scholar]
- Hoshida H, Tanaka Y, Hibino T, et al. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Molecular Biology. 2000;43:103–111. doi: 10.1023/a:1006408712416. [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. The Plant Journal. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. The Plant Cell. 1999;11:1381–1392. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K. Chloroplasts are partially mobilized to the vacuole by autophagy. Autophagy. 2008;4:961–962. doi: 10.4161/auto.6804. [DOI] [PubMed] [Google Scholar]
- Ishida H, Nishimori Y, Shimizu S, Makino A, Mae T. The large subunit of ribulose-1,5-biphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant and Cell Physiology. 1997;38:471–479. doi: 10.1093/oxfordjournals.pcp.a029191. [DOI] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H. Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. The Journal of Biological Chemistry. 2004;279:16598–16605. doi: 10.1074/jbc.M313710200. [DOI] [PubMed] [Google Scholar]
- Joppa LR, Du C, Hart GE, Hareland GA. Mapping gene(s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Science. 1997;37:1586–1589. [Google Scholar]
- Kato Y, Yamamoto Y, Murakami S, Sato F. Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta. 2005;222:643–651. doi: 10.1007/s00425-005-0011-4. [DOI] [PubMed] [Google Scholar]
- Keys A, Bird I, Cornelius M, Lea P, Wallsgrove R, Miflin B. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- Kichey T, Hirel B, Heumez E, Dubois F, Le Gouis J. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crop Research. 2007;102:22–32. [Google Scholar]
- Lam HM, Wong P, Chan HK, et al. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiology. 2003;132:926–935. doi: 10.1104/pp.103.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A, Brancourt-Hulmel M, Heumez E, et al. Using genotype × nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theoretical and Applied Genetics. 2007;115:399–415. doi: 10.1007/s00122-007-0575-4. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Forde BG. The use of mutants and transgenic plants to study amino acid metabolism. Plant, Cell and Environment. 1994;17:541–556. [Google Scholar]
- Lea P, Miflin B. Alternative route for nitrogen assimilation in higher plants. Nature. 1974;251:614–616. doi: 10.1038/251614a0. [DOI] [PubMed] [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M. AAP1 transports uncharged amino acids into roots of Arabidopsis. The Plant Journal. 2007;50:305–319. doi: 10.1111/j.1365-313X.2007.03045.x. [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Michelet L, Zaffagnini M, Massot V, Issakidis-Bourguet E. Thioredoxins in chloroplasts. Current Genetic. 2007;51:343–365. doi: 10.1007/s00294-007-0128-z. [DOI] [PubMed] [Google Scholar]
- Lemaître T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C. Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant and Cell Physiology. 2008;49:1056–1065. doi: 10.1093/pcp/pcn081. [DOI] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genetics. 2005;1 doi: 10.1371/journal.pgen.0010006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C. Signalling cascades integrating light-enhanced nitrate metabolism. Biochemical Journal. 2008;415:11–19. doi: 10.1042/BJ20081115. [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. Journal of Experimental Botany. 2006;57:2751–2761. doi: 10.1093/jxb/erl036. [DOI] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, et al. Mutation of the Arabidopsis NRT1·5 nitrate transporter causes defective root-to-shoot nitrate transport. The Plant Cell. 2008;20:2514–2528. doi: 10.1105/tpc.108.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell. 1999;11:865–874. doi: 10.1105/tpc.11.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loque D, Ludewig U, Yuan L, von Wiren N. Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiology. 2005;137:671–680. doi: 10.1104/pp.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiology. 2003;131:345–358. doi: 10.1104/pp.102.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig U, Neuhauser B, Dynowski M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Letters. 2007;581:2301–2308. doi: 10.1016/j.febslet.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Maathuis F. Physiological functions of mineral nutrients. Current Opinion in Plant Biology. 2009;12:250–258. doi: 10.1016/j.pbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Mae T, Makino A, Ohira K. Changes in the amounts of ribulose biphosphate carboxylase synthesized and degraded during the life span of rice leaf (Oryza sativa L.) Plant and Cell Physiology. 1983;24:1079–1086. [Google Scholar]
- Malagoli P, Laine P, Le Deunff E, Rossato L, Ney B, Ourry A. Modeling nitrogen uptake in oilseed rape cv Capitol during a growth cycle using influx kinetics of root nitrate transport systems and field experimental data. Plant Physiology. 2004;134:388–400. doi: 10.1104/pp.103.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagoli P, Laine P, Rossato L, Ourry A. Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. Annals of Botany. 2005;95:853–861. doi: 10.1093/aob/mci091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Belastegui-Macadam X, Quillere I, et al. Nitrogen management and senescence in two maize hybrids differing in the persistence of leaf greenness: agronomic, physiological and molecular aspects. New Phytologist. 2005;167:483–492. doi: 10.1111/j.1469-8137.2005.01430.x. [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. The Plant Cell. 2006;18:3252–3274. doi: 10.1105/tpc.106.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DE, Costa ML, Guiamet JJ. Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biology (Stuttgart) 2008;10(Suppl. 1):15–22. doi: 10.1111/j.1438-8677.2008.00089.x. [DOI] [PubMed] [Google Scholar]
- Masclaux C, Quilleré I, Gallais A, Hirel B. The challenge of remobilisation in plant nitrogen economy. A survey of physio-agronomic and molecular approaches. Annals of Applied Biology. 2001;138:68–81. [Google Scholar]
- Masclaux-Daubresse C, Carrayol E, Valadier M-H. The two nitrogen mobilisation- and senescence-associated GS1 and GDH genes are controlled by C and N metabolites. Planta. 2005;221:580–588. doi: 10.1007/s00425-004-1468-2. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, et al. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiology. 2006;140:444–456. doi: 10.1104/pp.105.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M. Leaf nitrogen remobilisation for plant development and grain filling. Plant Biology. 2008;10(Suppl. 1):23–36. doi: 10.1111/j.1438-8677.2008.00097.x. [DOI] [PubMed] [Google Scholar]
- McKhann IH, Camilleri C, Bérard A, et al. Nested core collections maximizing genetic diversity in Arabidopsis thaliana. The Plant Journal. 2004;38:193–202. doi: 10.1111/j.1365-313X.2004.02034.x. [DOI] [PubMed] [Google Scholar]
- Meyer C, Stitt M. Nitrate reductase and signalling. In: Lea PJ, Morot-Gaudry J-F, editors. Plant nitrogen. New York: Springer; 2001. pp. 37–59. [Google Scholar]
- Mickelson S, See D, Meyer FD, et al. Mapping of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. Journal of Experimental Botany. 2003;54:801–812. doi: 10.1093/jxb/erg084. [DOI] [PubMed] [Google Scholar]
- Migge A, Carrayol E, Hirel B, Becker T. Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta. 2000;210:252–260. doi: 10.1007/PL00008132. [DOI] [PubMed] [Google Scholar]
- Millard P, Wendler R, Grassi G, Grelet GA, Tagliavini M. Translocation of nitrogen in the xylem of field-grown cherry and poplar trees during remobilization. Tree Physiology. 2006;26:527–536. doi: 10.1093/treephys/26.4.527. [DOI] [PubMed] [Google Scholar]
- Millard P, Sommerkorn M, Grelet GA. Environmental change and carbon limitation in trees: a biochemical, ecophysiological and ecosystem apraisal. The New Phytologist. 2007;175:11–28. doi: 10.1111/j.1469-8137.2007.02079.x. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. Journal of Experimental Botany. 2008;3:667–680. doi: 10.1093/jxb/erm340. [DOI] [PubMed] [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Obara M, Kajiura M, Fukuta Y, et al. Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.) Journal of Experimental Botany. 2001;52:1209–1217. [PubMed] [Google Scholar]
- Obara M, Sato T, Sasaki S, et al. Identification and characterization of a QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice. Theoretical and Applied Genetics. 2004;110:1–11. doi: 10.1007/s00122-004-1828-0. [DOI] [PubMed] [Google Scholar]
- Olea F, Pérez-Garcia A, Canton FR, et al. Up-regulation and localization of asparagine synthetase in tomato leaves infected by the bacterial pathogen Pseudomonas syringae. Plant Cell and Physiology. 2004;45:770–780. doi: 10.1093/pcp/pch092. [DOI] [PubMed] [Google Scholar]
- Oliveira I, Brears T, Knight T, Clark A, Coruzzi G. Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiology. 2002;129:1170–1180. doi: 10.1104/pp.020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F. Disruption of the nitrate transporter genes AtNRT2·1 and AtNRT2·2 restricts growth at low external nitrate concentration. Planta. 2004;219:714–721. doi: 10.1007/s00425-004-1266-x. [DOI] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, et al. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiology. 2006;142:1304–1317. doi: 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Noh YS, Martinez DE, et al. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. The Plant Journal. 2005;41:831–844. doi: 10.1111/j.1365-313X.2005.02346.x. [DOI] [PubMed] [Google Scholar]
- Pageau K, Reisdorf-Cren M, Morot-Gaudry J-F, Masclaux-Daubresse C. The two senescence-related markers GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilisation are differentially regulated during pathogen attack, by stress hormones and reactive oxygen species in Nicotiana tabacum L. Journal of Experimental Botany. 2006;57:547–557. doi: 10.1093/jxb/erj035. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Carillo N, Valle EM. The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiology. 1999;121:471–478. doi: 10.1104/pp.121.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RR, Ahmad A, Lochab S, Raghuram N. Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Current Science. 2009;94:1394–1403. [Google Scholar]
- Pérez-Garcia A, De Vicente A, Canton FR, et al. Light-dependent changes of tomato glutamine synthetase in response to Pseudomonas syringae infection or phosphinothricin treatment. Physiologia Plantarum. 1998a;102:377–384. [Google Scholar]
- Pérez-Garcia A, Pereira S, Pissara J, et al. Cytosolic localization in tomato mesophyll cells of a novel glutamine synthetase induced in response to bacterial infection of phosphinothricin treatment. Planta. 1998b;206:426–434. [Google Scholar]
- Potel F, Valadier MH, Ferrario-Mery S, et al. Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana – roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS Journal. 2009;276:4061–4076. doi: 10.1111/j.1742-4658.2009.07114.x. [DOI] [PubMed] [Google Scholar]
- Purnell M, Botella J. Tobacco isozyme 1 of NAD(H)-dependent glutamate dehydrogenase catabolizes glutamate in vivo. Plant Physiology. 2007;143:530–539. doi: 10.1104/pp.106.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorf-Cren M, Carrayol E, Terce-Laforgue T, Hirel B. A novel HMG A-like protein binds differentially to the AT-rich regions located in the far distal and proximal parts of a soybean glutamine synthetase gene (GS15) promoter. Plant Cell and Physiology. 2002;43:1006–1016. doi: 10.1093/pcp/pcf123. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Letters. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Robinson D. The responses of plants to non-uniform supplies of nutrients. New Phytologist. 1994;127:635–674. doi: 10.1111/j.1469-8137.1994.tb02969.x. [DOI] [PubMed] [Google Scholar]
- Rochat C, Boutin J-P. Metabolism of phloem-borne amino acids in maternal tissues of fruit of nodulated or nitrate-fed pea plants (Pisum sativum L.) Journal of Experimental Botany. 1991;42:207–214. [Google Scholar]
- Sage RF, Pearcy RW, Seeman JR. The nitrogen use efficiency in C3 and C4 plants. Plant Physiology. 1987;85:355–359. doi: 10.1104/pp.85.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salon C, Munier-Jolain N, Duc G, et al. Grain legume seed filling in relation to nitrogen acquisition: A review and prospects with particular reference to pea. Agronomie. 2001;21:539–552. [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat AK, Carroll RT, DePauw M, Taylor GJ, Good AG. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnology Journal. 2008;6:722–732. doi: 10.1111/j.1467-7652.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Skopelitis D, Paranychianakis N, Paschalidis K, Pliakonis E, Delis I, Yakoumakis D, Kouvarakis A, Papadakis A, Stephanou E, Roubelakis-Anfelakis K. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell. 2006;18:2767–2781. doi: 10.1105/tpc.105.038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, Yamaya T, Yamaguchi J. Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell and Physiology. 2003;44:1396–1402. doi: 10.1093/pcp/pcg169. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Knaff DB. Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynthesis Research. 2005;83:191–217. doi: 10.1007/s11120-004-3478-0. [DOI] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Bellini C, Nasholm T. Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiology. 2007;143:1853–1860. doi: 10.1104/pp.106.092205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Nasholm T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytologist. 2008;180:620–630. doi: 10.1111/j.1469-8137.2008.02589.x. [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD. Autophagic recycling: lessons from yeast help define the process in plants. Current Opinion in Plant Biology. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell. 1993;72:705–713. doi: 10.1016/0092-8674(93)90399-b. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Letters. 2007;581:2290–2300. doi: 10.1016/j.febslet.2007.04.047. [DOI] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno H, Uchida T, Sugawara H, et al. Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. Journal of Biological Chemistry. 2006;281:29287–29296. doi: 10.1074/jbc.M601497200. [DOI] [PubMed] [Google Scholar]
- Vanoni M, Dossena L, van den Heuvel R, Curti B. Structure–function studies on the complex iron-sulfur flavoprotein glutamate synthase: the key enzyme of ammonia assimilation. Photosynthesis Research. 2005;83:219–238. doi: 10.1007/s11120-004-2438-z. [DOI] [PubMed] [Google Scholar]
- Vitousek PM, Naylor R, Crews T, et al. Nutrient imbalances in agricultural development. Science. 2009;19:1519–1520. doi: 10.1126/science.1170261. [DOI] [PubMed] [Google Scholar]
- Volenec J, Ourry A, Joern B. A role for nitrogen reserves in forage regrowth and stress tolerance. Physiologia Plantarum. 1996;97:185–193. [Google Scholar]
- Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1·1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. The Plant Journal. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proceedings of the National Academy of Sciences USA. 1998;95:15134–15139. doi: 10.1073/pnas.95.25.15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiology. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M, Grimes H, Huffaker R. Latent nitrate reductase activity is associated with the plasma membrane of corn roots. Planta. 1989;177:470–475. [PubMed] [Google Scholar]
- Waters B, Uauy C, Dubcovsky J, Grusak M. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. Journal of Experimental Botany. 2009;60:4263–4274. doi: 10.1093/jxb/erp257. [DOI] [PubMed] [Google Scholar]
- Williams L, Miller A. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:659–688. doi: 10.1146/annurev.arplant.52.1.659. [DOI] [PubMed] [Google Scholar]
- Wingler A, Masclaux-Daubresse C, Fischer AM. Sugars, senescence, and ageing in plants and heterotrophic organisms. Journal of Experimental Botany. 2009;60:1063–1066. doi: 10.1093/jxb/erp067. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000718. e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth J, Chopin F, Santoni V, et al. Regulation of root nitrate uptake at the NRT2·1 protein level in Arabidopsis thaliana. Journal of Biological Chemistry. 2007;282:23541–23552. doi: 10.1074/jbc.M700901200. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. The Plant Journal. 2005;42:535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- Yamaya T, Obara M, Nakajima H, Saski S, Hayakawa T, Sato T. Genetic manipulation and quantitative trait loci mapping for nitrogen recycling in rice. Journal of Experimental Botany. 2002;53:917–925. doi: 10.1093/jexbot/53.370.917. [DOI] [PubMed] [Google Scholar]
- Yang L, Mickelson S, See D, Blake TK, Fischer AM. Genetic analysis of the function of major leaf proteases in barley (Hordeum vulgare L.) nitrogen remobilization. Journal of Experimental Botany. 2004;55:2607–2616. doi: 10.1093/jxb/erh267. [DOI] [PubMed] [Google Scholar]
- Yuan L, Loque D, Ye F, Frommer WB, von Wiren N. Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiology. 2007;143:732–744. doi: 10.1104/pp.106.093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. Fundamentals of rice crop science. IRRI, Los IRRI, Los Banos, Philippines. 1981;22:1067–1074. [Google Scholar]
- Zimmermann P, Zentgraf U. The correlation between oxidative stress and leaf senescence during plant development. Cellular and Molecular Biology Letters. 2005;10:515–534. [PubMed] [Google Scholar]