Abstract
Background and Aims
Nitrogen (N) availability in the forest soil is extremely low and N economy has a special importance in woody plants that are able to cope with seasonal periods of growth and development over many years. Here we report on the analysis of amino acid pools and expression of key genes in the perennial species Populus trichocarpa during autumn senescence.
Methods
Amino acid pools were measured throughout senescence. Expression analysis of arginine synthesis genes and cationic amino acid transporter (CAT) genes during senescence was performed. Heterologous expression in yeast mutants was performed to study Pt-CAT11 function in detail.
Key Results
Analysis of amino acid pools showed an increase of glutamine in leaves and an accumulation of arginine in stems during senescence. Expression of arginine biosynthesis genes suggests that arginine was preferentially synthesized from glutamine in perennial tissues. Pt-CAT11 expression increased in senescing leaves and functional characterization demonstrated that Pt-CAT11 transports glutamine.
Conclusions
The present study established a relationship between glutamine synthesized in leaves and arginine synthesized in stems during senescence, arginine being accumulated as an N storage compound in perennial tissues such as stems. In this context, Pt-CAT11 may have a key role in N remobilization during senescence in poplar, by facilitating glutamine loading into phloem vessels.
Keywords: Nitrogen metabolism, senescence, glutamine, arginine, cationic amino acid transporters, storage protein, Populus trichocarpa
INTRODUCTION
Seasonal nitrogen (N) cycling is an adaptation of plants to winter cold seasonal climates in which nutrients (mostly N) are often considered to be the major growth-limiting factor (Cooke and Weih, 2005). Nitrogen translocation from senescing leaves to over-winter storage sites is a common feature of temperate deciduous trees (Ryan and Bormann, 1982). Poplar is extremely efficient at N conservation since >80 % of the whole-tree nitrogen content is conserved during dormancy (Pregitzer et al., 1990). During autumnal leaf senescence, there is a functional shift in leaf metabolism from resource assimilation to resource remobilization and export. N-rich amino acids and other mobile nutrients are transported via the phloem from senescing leaves to perennial tissues where they are used to synthesize proteins (Sauter et al., 1989; Hörtensteiner and Feller, 2002). Proteins represent the major fraction of the stored N, and vegetative storage proteins (VSPs) represent the major form of reduced N storage in vegetative tissues of both annual and perennial plants (Staswick, 1994; Stepien et al., 1994). The bark storage protein (BSP) family comprises the major VSPs in Populus. During autumn, BSPs accumulate in the bark parenchyma and xylem cells of the main stem, branches and roots of the tree (Sauter et al., 1989).
Amino acids are the currency of N exchange between source and sink tissues in plants (Bush, 1999). Glutamine is the predominant translocated form for organic N in poplar (Dickson, 1979; Sauter and van Cleve, 1992) and is preferentially transported through the stem to developing leaves via a xylem to phloem transfer facilitated by ray cells (Dickson et al., 1985). Nevertheless, the amino acid composition of xylem sap exhibits seasonal variations. During the wintering phase, arginine is the major amino acid in bark and xylem of poplar, whereas at the time of budding and growing, glutamine and glutamate become dominant (Sagisaka, 1974). These variations in amino acid pools could be associated with variations in expression of amino acid transporter genes not only in storage tissues but also in sieve elements which allow amino acid distribution in the whole plant.
In plants, the majority of genes encoding putative amino acid transporters can be classified into two major groups: the amino acid transporter family (ATF) and the amino acid polyamine choline (APC) superfamily (Wipf et al., 2002). Most of the amino acid transporters from plants that have been characterized functionally belong to the ATF superfamily, with the amino acid permease (AAP) family being the best studied sub-family (Boorer et al., 1995; Fischer et al., 1995, 2002; Boorer and Fischer, 1997; Okumoto et al., 2002, 2004). In plants, APC amino acid transporters are poorly understood and have been described only in Arabidopsis thaliana. APC transporters of the l-type amino acid transporter (LAT) sub-family (five members) have not been characterized and only a few members of the cationic amino acid transporters (CAT) family have been studied (Frommer et al., 1995; Su et al., 2004; Hammes et al., 2006). They contain between 11 and 14 putative transmembrane (TM) domains and they are high-affinity basic amino acid transporters. They are located in the plasma membrane or in the vacuolar membrane (Frommer et al., 1995; Su et al., 2004; Hammes et al., 2006). It has been demonstrated that At-CAT1 is expressed in leaves, flowers and developing siliques, and transcripts were specifically localized in major veins of leaves and roots (Frommer et al., 1995). It has been suggested that At-CAT1 might play multiple roles in phloem physiology, from phloem loading to providing amino acids for developing embryos. Moreover, At-CAT1 is likely to be a proton-driven high-affinity transporter that transports mainly cationic amino acids (Frommer et al., 1995). At-CAT2 is probably localized to the tonoplast and may be the long-sought vacuolar amino acid transporter (Su et al., 2004). At-CAT5 functions as a high-affinity, basic amino acid transporter at the plasma membrane. Expression profiles suggest that At-CAT5 may function in the re-uptake of leaked amino acids at the leaf margin (Su et al., 2004). At-CAT8 is expressed in young and rapidly dividing tissues such as young leaves and root apical meristem. At-CAT8 is also localized to the plasma membrane (Su et al., 2004). At-CAT6 has a high affinity for cationic amino acids and is also likely to be energized by protons (Hammes et al., 2006). At-CAT6 transports large, neutral and cationic amino acids in preference to other amino acids and plays a role in supplying amino acids to sink tissues of plants and nematode-induced feeding structures.
As exemplified above, N storage and cycling have traditionally been investigated at the molecular physiology and ecophysiology scales. Taking advantage of the annotated Populus trichocarpa (Nisqually 1) genome (Tuskan et al., 2006), we present here the analysis of amino acid pools in different organs of poplar during autumn and winter, combined with the expression analysis of genes encoding enzymes of arginine biosynthesis and genes encoding CAT members. Finally, we also characterize Pt-CAT11 by heterologous expression in yeast and show that it preferentially transports glutamine.
MATERIALS AND METHODS
Plant material
Leaves from 1- and 2-year-old stems were sampled from free-growing Populus trichocarpa trees at the University of Nancy campus. About 20 leaves and four stems were sampled at 14 h for every time point, frozen in liquid nitrogen and stored at −80 °C. Leaves were sampled on 27 October, 23 November, 5 December and 12 December. This latter point corresponds to a period just before leaf fall. Stems were also sampled on 8 January and 2 February. These two dates correspond to the wintering phase.
Semi-quantitative RT–PCR
Total RNA extraction was performed with the RNeasy Plant Mini kit (Qiagen, Darmstadt, Germany) from approx. 100 mg of frozen tissues of poplar. To remove contaminating genomic DNA, the samples were treated with DNase I (Qiagen), as recommended by the manufacturer. To obtain cDNA, 500 ng of total RNA were annealed to oligo(dT) primers (Promega, Madison, WI, USA) and reverse transcribed using reverse transcriptase (Eppendorf, Hamburg, Germany) at 42 °C for 90 min. Each reaction was set up in three biological replicates. For each Pt-CAT, the PCR program was as follows: 94 °C for 3 min and 35 cycles of 94 °C for 30 s, 58 °C for 45 s and 72 °C for 1 min. The whole set of Pt-CAT genes (12 genes) was tested by reverse transcription–PCR (RT–PCR) in every experiment performed, but only Pt-CAT genes detected and well expressed are retained in figures for greater clarity. To study the expression of genes involved in the pathway of arginine biosynthesis, cDNA corresponding to argininosuccinate lyase (AL), argininosuccinate synthase (AS), ornithine transcarbamoylase (OTC) and carbamoyl-phosphate synthase (CPS) were also amplified using the same PCR program as described above. The numbers of genes coding for AL, AS, OTC and CPS were one, two, one and two, respectively. When two genes were coding for an enzyme, primers were designed for the gene with the highest expressed sequence tag (EST) numbers in poplar databases. Control PCRs were sequenced to ensure that only one gene was amplified.
A cDNA fragment corresponding to the constitutively expressed ubiquitin gene was amplified simultaneously (28 cycles) and used as a control. Cysteine protease (CP) was amplified (28 cycles) and used as control of the senescence state of leaves. The sequences of the gene-specific oligonucleotides, designed in the non-conserved regions of the genes and used for RT–PCR, are listed in Table 1. The ethidium bromide-stained agarose gels were imaged on a Bio-Rad GelDoc 2000 transilluminator, and quantitative data were determined using Quantity One software (Bio-Rad, Hercules, CA, USA). Signal intensities were normalized to the constitutively expressed poplar ubiquitin gene.
Table 1.
Primers used for RT–PCR analysis
| Name | Sequence |
|---|---|
| CAT1 f | ACCATTTATGCCATATGATGTCCG |
| CAT1 r | GGTTCAACTTGTGATGACACAAC |
| CAT2 f | TTCCTCTGCATTCGCTGCATAT |
| CAT2 r | TAGTGACATCTGGGCTACCTGTA |
| CAT3 f | GTCCTCTTCGTTTTACAACG |
| CAT3 r | TTTCTCCAGAGCTCCGATAA |
| CAT4 f | TTTGCATAGGAGAAGGTGCAGCAT |
| CAT4 r | GACAAAGCAACGCCTATACCT |
| CAT5 f | ACAGCACTGAATACTGCTGTA |
| CAT5 r | GCTAGCTTCAAGAGGTTTGTT |
| CAT6 f | TACATGTGTGTTATCGGACGGTC |
| CAT6 r | TTACACTTTGAAAGAATTAATATGGTCCTCGC |
| CAT7 f | CTGTCTTTGCCATAGCACAAAG |
| CAT7 r | CTGGCCTTTAGTGTGGTCATG |
| CAT8 f | GCCTCTATTGCTACTGCTTTTATC |
| CAT8 r | TCCAAGTGATCCAACCATTAAGCT |
| CAT9 f | CAGCTTTCAATGAGCTTACTGCTT |
| CAT9 r | ACAAGACTTCCAATGATGCCT |
| CAT10 f | ACAGCTTCAATTGCACTCTTTACC |
| CAT10 r | TCATAGCAGCTGAATATCTAGC |
| CAT11 f | TCATCAAGAAGGTGGAGACCAAGA |
| CAT11 r | GGCAGCACAAACAAAAACAGAT |
| CAT12 f | TGATCATCAAGAAGAAGGGCTG |
| CAT12 r | CACAACACCAACAAGAACAGCA |
| Ubq f | GCACCTCTGGCAGACTACAA |
| Ubq r | TAACAGCCGCTCCAAACAGT |
| CP f | AGTCACTGAGAAAGGCTGTGG |
| CP r | CCAAATGGATTGTTCTTGCTC |
| AS f | AGCGGAAATACTTATTGGGGACGT |
| AS r | ACAAGTTCCTGTCCCTGCTATA |
| AL f | GTTCCTGGTTACACACATTTGCAA |
| AL r | ACAGGTTCCTTGTCTTCCTGCAAA |
| OTC f | ATGGCCTGAACTATAACCATCC |
| OTC r | CTCGATCTTGCTGATTCCAGC |
| CPS f | CGGTGTCCTAACCACAGAAGAATT |
| CPS r | CCTCAGGATGGTATTGTAGAGA |
Amino acid extraction and analysis
Amino acids were extracted twice from 10–20 mg of freeze-dried plant tissues with 300 µL of 70 % (v/v) cold ethanol. The samples were dried under N2 using a Reacti-Therm Heating Module (Pierce, Rockford, IL, USA) and resuspended in 400 µL of 0·1 n HCl. Extracts and standards were loaded onto a Dowex 50WX-8 cation ion exchange column (Sigma-Aldrich, St Louis, MO, USA). After two successive washing steps with sterile water, amino acids were eluted with 4·5 n ammonia. Aliquots of purified samples were then transferred to microvials, dried in a Reacti-Therm Heating Module (Pierce) and derivatized according to Javelle et al. (2003). Gas chromatography and mass spectrometry (GC-MS) analysis was performed as described previously (Javelle et al., 2003).
Statistical analysis
The effects of the senescence state on tissue amino acid concentrations, soluble protein concentrations and gene expression were tested with a one-way analysis of variance (ANOVA) using the SYSTAT statistical package (SYSTAT Inc., Evanston, IL, USA). The Tukey test was used for all pairwise comparisons of the mean responses to the different treatment groups.
Protein extraction and analysis
Small pieces (about 50–100 mg) of stems were ground with a mortar and pestle cooled in liquid nitrogen in 2 mL of 50 mm Tris–HCl pH 8·0, 1 mm PMSF (phenylmethylsulfonyl fluoride) and 50 mm mercaptoethanol. Samples were then mixed by vortexing, and held at 4 °C for 30 min. Samples were centrifuged at 13 000 rpm for 15 min and the supernatants collected. Proteins were precipitated with acetone at −20 °C for 2 h. Aliquots of 100 µL were centrifuged at 13 000 rpm for 15 min and proteins were resuspended with 50 µL of 0·2 % SDS. Protein concentration was determined by the bicinchoninic acid (BCA) colorimetric assay kit (Interchim, Montluçon, France; Brown et al., 1989). The BCA procedure followed the manufacturer's recommendations, with bovine serum albumin as a standard and absorbance measured at 562 nm. Protein concentrations were determined for duplicate sub-samples for each replicate.
DNA constructs
The predicted coding sequence corresponding to Pt-CAT11 (1767 bp) was amplified by PCR using cDNA generated for RT–PCR studies (see above) and the following primers: Pt-CAT11fow (5′-CCCGAATTCATGAGGAGGAGGAGGGGATGT-3′) and Pt-CAT11rev (5′-CCCCTCGAGTCATGAACCATTCCGGGAAGG-3). The amplification product was cloned into the EcoRI/XhoI sites of the yeast expression vector pYES2 and sequenced to confirm that no modifications occurred.
Yeast transformation
The yeast strains 22Δ8AA (MATα, ura3-1, gap1-1, put4-1, uga4-1, can1::HisG, lyp1/alp1::HisG, hip1::HisG, dip5::HisG, ura3-1) (Fischer et al., 2002) and JA248 (MATα ura3Δ gap1Δ gnp1Δ agp1Δ) (Velasco et al., 2004) were transformed with pYES2 harbouring the cDNA sequence of Pt-CAT11. Yeast transformants were selected on synthetic dextrose minimal medium. Yeast strain 22Δ8AA complementation tests were performed on N-free medium supplemented with 20 g L−1 Gal and either 1, 3 or 6 mm l-proline, l-citrulline, l-aspartate or l-glutamate as sole N source, whereas yeast strain JA248 complementation tests were performed on N-free medium supplemented with 20 g L−1 Gal and either 0·5, 1, 2 or 5 mm l-glutamine as sole N source.
Transport measurements
For Saccharomyces cerevisiae uptake studies, yeast cells were grown to logarithmic phase. Cells were harvested at an OD600 of 0·5, washed twice in water, and resuspended in buffer A (0·6 m sorbitol, 50 mm potassium phosphate, at the desired pH) to a final OD600 of 5. Prior to the uptake measurements, the cells (100 µL) were supplemented with 5 µL of 1 m galactose and incubated for 5 min at 30 °C. To start the reaction, 100 µL of this cell suspension was added to 100 µL of the same buffer containing at least 18·5 kBq of [3H]glutamine, and unlabelled glutamine to the concentrations used in the experiments. Sample aliquots of 50 µL were removed after 30, 60 and 120 s, transferred to 4 mL of ice-cold buffer A, filtered on glass fibre filters and washed twice with 4 mL of buffer A. The uptake of tritium was determined by liquid scintillation spectrometry.
Phylogenetic analyses
CAT sequences were retrieved by text and Blast searches from the P. trichocarpa whole genome database (version 1·1) at the US Department of Energy Joint Genome Institute (JGI) (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). The curated poplar amino acid sequences were used to search against five other genomes from photosynthetic organisms using BLASTP or TBLASTN. The genomes are available at the following websites, for A. thaliana (http://www.arabidopsis.org/), Oryza sativa (http://rice.plantbiology.msu.edu/), Vitis vinifera (http://www.genoscope.cns.fr/spip/Vitis-vinifera-whole-genome.html) and Sorghum bicolor (http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html). Amino acid sequences were aligned by CLUSTALW and imported into the Molecular Evolutionary Genetics Analysis (MEGA) package version 4·1 (Tamura et al., 2007). Phylogenetic analyses were conducted using the neighbor–joining (NJ) method implemented in MEGA, with the pairwise deletion option for handling alignment gaps, and with the Poisson correction model for distance computation. Bootstrap tests were conducted using 1000 replicates. Branch lengths are proportional to phylogenetic distances. All protein sequences and corresponding accession numbers can be found in the databases mentioned above and as Supplementary Data (available online).
RESULTS
Glutamine–arginine relationships during senescence
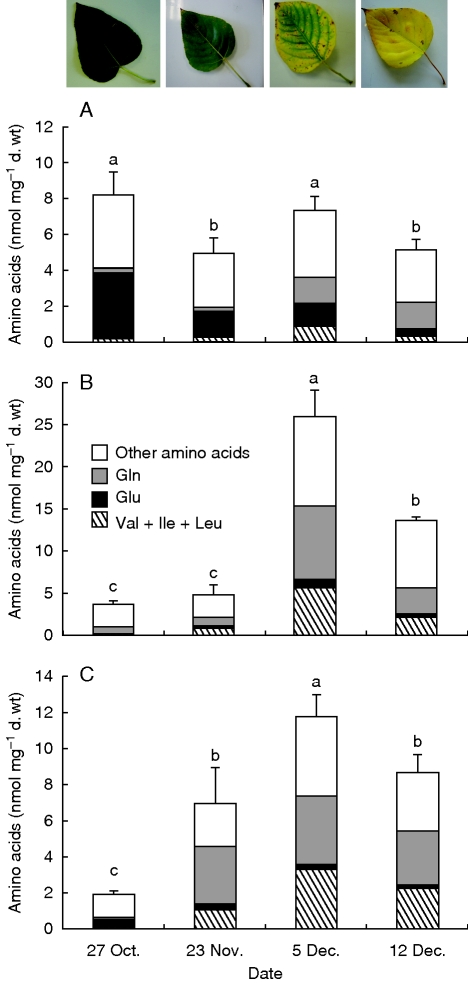
Amino acid concentrations were investigated in the lamina (Fig. 1A), central vein (Fig. 1B) and petiole (Fig. 1C) of poplar leaves. In the lamina, the total amino acid concentration did not change much during senescence, varying between 5 and 8 nmol mg−1 d. wt. Amino acid profiling indicated that the glutamate (and aspartate; not shown) concentration decreased whereas that of glutamine (and asparagine; not shown) increased during leaf senescence (Fig. 1A). In the central vein and petiole, the total amino acid concentration increased by approx. 6-fold from 27 October to 5 December, thereafter decreasing at the latest sampling date (Fig. 1B, C). Glutamine was the predominant amino acid before leaf fall, representing 23 and 34 % of the total amino acid pool in the central vein and petiole, respectively, followed by leucine, isoleucine and valine.
Fig. 1.
Quantification of amino acids by gas chromatography–mass spectrometry in (A) laminae, (B) central veins (C) and petioles of poplar during senescence. Values are expressed as the mean ± s.e. of three replicate experiments. For a given tissue, amino acid concentration values with the same letter are not significantly different, according to ANOVA at P < 0·05.
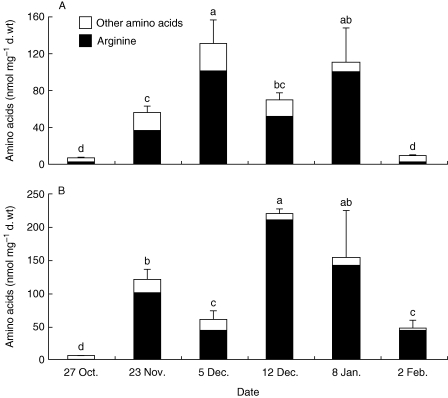
Amino acid pools were investigated in 1-year-old (Fig. 2A) and 2-year-old (Fig. 2B) stems. In October, total amino acid concentrations were <15 nmol mg−1 d. wt and arginine was almost undetectable in 2-year-old stems. During autumnal senescence, total amino acid pools increased by 20- and 37-fold in 1- and 2-year-old stems, respectively, when measured at their maximal level. Noticeably, arginine rapidly became the predominant amino acid accumulated in stems, accounting for 91 and 92 % in 1- and 2-year-old stems, respectively, on 8 January. Arginine accumulation was slightly delayed in 2-year-old stems, peaking on 5 December in 1-year-old stems and on 12 December in 2-year-old stems (Fig. 2).
Fig. 2.
Quantification of amino acids by gas chromatography–mass spectrometry in (A) 1-year-old and (B) 2-year-old stems of poplar during senescence. Values are expressed as the mean ± s.e. of three replicate experiments. For a given tissue, amino acid concentration values with the same letter are not significantly different, according to ANOVA at P < 0·05.
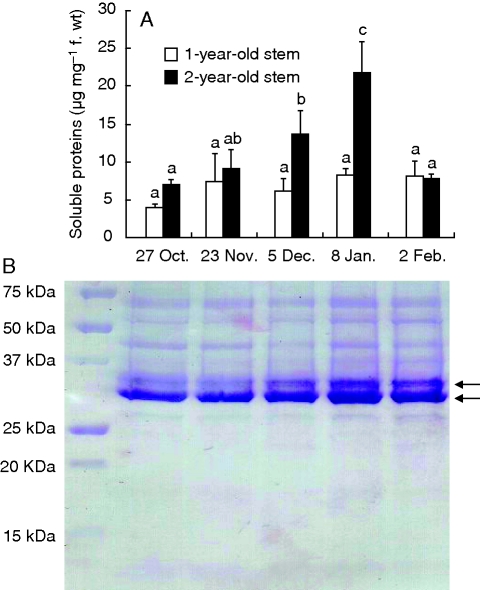
The amount of total soluble protein was investigated in 1- and 2-year-old stems (Fig. 3). During autumnal senescence, soluble protein content of 2-year-old stems increased by >3-fold between 27 October and 8 January. In contrast, there were no statistically significant changes in soluble protein content of 1-year-old stems during senescence (Fig. 3A). Soluble proteins from 2-year-old stems were analysed by SDS–PAGE (Fig. 3B). Analysis revealed the presence of two major proteins with relative molecular masses of between 30 and 37 kDa. Interestingly, the content of these two proteins increased during autumn and winter. These accumulating proteins correspond to the well-characterized BSPs of poplar.
Fig. 3.
Quantification of soluble proteins in 1- and 2-year-old stems of poplar during autumnal senescence and winter. Protein concentration (A) was determined for duplicate sub-samples from each replicate. Values are expressed as the mean ± s.e. of three replicate experiments. For a given tissue, protein concentration values with the same letter are not significantly different, according to ANOVA at P < 0·05. Soluble proteins from 2-year-old (B) stems of poplar were separated by SDS–PAGE. The presence of two major proteins with a relative molecular mass of between 30 and 37 kDa is indicated by arrows.
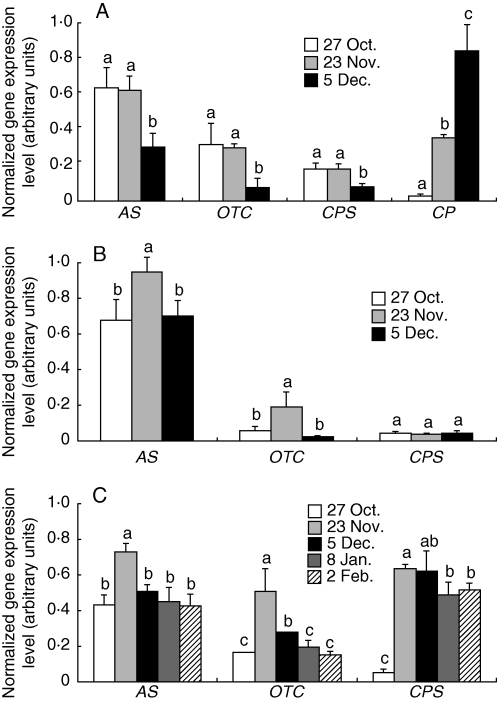
The metabolic route to arginine synthesis in plants involves two distinct processes: synthesis of ornithine from glutamate and synthesis of arginine from the ornithine intermediate (Slocum, 2005). Considering the striking accumulation of arginine during senescence, some of the genes involved in its biosynthesis were investigated: CPS, OTC, AS and AL genes. In order to investigate their expression during senescence in poplar, total RNAs were extracted from laminae, petioles and 2-year-old stems sampled at different times during autumn and winter. AL transcripts were undetectable under conditions used in these experiments (not shown). In laminae, AS, CPS and OTC transcripts decreased at the end of the senescence period (Fig. 4A). The senescence state of leaves was confirmed by the parallel amplification of a CP transcript (Bhalerao et al., 2003; Andersson et al., 2004), which was highly expressed on 5 December. In petioles, CPS and OTC transcripts were less abundant than AS transcripts during autumnal senescence (Fig. 4B). Moreover, AS and OTC were maximal on 23 November. In 2-year-old stems, as observed in laminae and petioles, OTC was weakly expressed and, as observed for AS, its expression decreased after 23 November (Fig. 4C). In contrast, CPS expression increased by 11-fold during leaf senescence and was maximal on 5 December (Fig. 4C).
Fig. 4.
Expression of arginine biosynthetic genes in various poplar tissues during or after senescence. (A) Expression of AS, OTC, CPS and CP genes in laminae. Cysteine protease (CP) was used as a marker of the senescence state (Bhalerao et al., 2003). (B) Expression of AS, OTC and CPS genes in petioles. (C) Expression of AS, OTC and CPS in 2-year-old stems. Total RNAs were extracted at different time periods during autumn and winter and 300 ng of total RNAs were reverse transcribed into cDNA. The ubiquitin (Ubq) gene was amplified and used as internal control. Analyses were performed by RT–PCR in triplicate. Signal intensities were normalized to the constitutively expressed poplar ubiquitin gene. Values are expressed as the mean ± s.e. of three replicate experiments. For a given gene, values with the same letter are not significantly different, according to ANOVA at P < 0·05.
Pt-CAT11 is a glutamine transporter upregulated during senescence
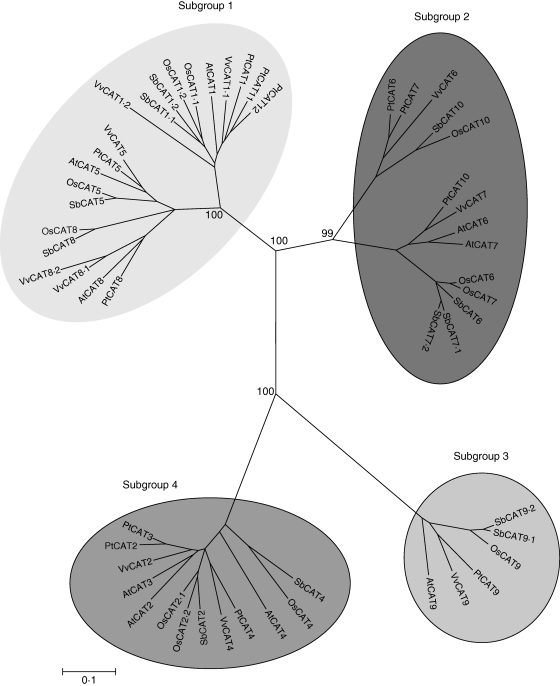
The JGI P. trichocarpa gene search mode revealed the existence of 12 CAT gene models. As described for the Arabidopsis CAT family, plant CAT members can be phylogenetically grouped into four small sub-groups (Fig. 5). Sub-group 1 contains the members CAT1, CAT5, CAT8, CAT11 and CAT12, whereas sub-group 2 includes the members CAT6, CAT7 and CAT10. Interestingly sub-group 3 only includes CAT9 whereas sub-group 4 contains CAT2, CAT3 and CAT4. Analysis of the assembled genome revealed relatively recent whole-genome duplication shared among all modern taxa in Salicaceae. A second, older duplication appears to be shared with the Arabidopsis lineage (Tuskan et al., 2006). These duplicated genes originated through very recent small-scale gene duplications and one relatively recent large-scale gene duplication event (Sterck et al., 2005). A detailed analysis of duplication events for the Pt-CAT members revealed that poplar CAT6, CAT7 and CAT9 derived from a common ancestor through an ancient and a recent duplication event, and that poplar CAT2 and CAT3 derived from a common ancestor through a recent duplication event. The same analysis also revealed that poplar CAT11 and CAT12 derived from a common ancestor through a recent duplication event.
Fig. 5.
An unrooted, neighbor–joining (NJ)-based tree of the cationic amino acid transporter (CAT) family. The analysis was performed as described in the Materials and Methods. Branch lengths (drawn in the horizontal dimension only) are proportional to phylogenetic distances. Corresponding gene loci or protein IDs are given in the Supplementary Data (available online).
In Arabidopsis, members of the CAT family have been characterized as high affinity basic amino acid transporters. For instance, At-CAT1 and At-CAT5 mediate high-affinity transport of arginine, lysine and histidine (Frommer et al., 1995; Su et al., 2004). To complement previous expression studies, we extracted GENEVESTIGATOR (Zimmermann et al., 2004; www.genevestigator.ethz.ch) data for the Arabidopsis CAT gene family, which indicated that At-CAT2 and At-CAT5 were mostly upregulated during leaf senescence.
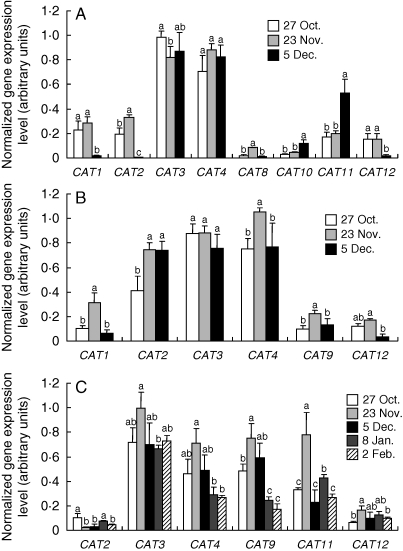
To investigate the potential role of Pt-CAT members during senescence, transcript levels were estimated in laminae, petioles and 2-year-old stems sampled at different times during autumn and winter. Pt-CAT3 and Pt-CAT4 transcripts remained high throughout the season and were not affected by leaf senescence (Fig. 6A). Similarly, Pt-CAT8 was weakly expressed and poorly affected by senescence in these experiments (Fig. 6A). Pt-CAT1, Pt-CAT2 and Pt-CAT12 were expressed in leaves in October and November but not expressed at the end of the senescing period (Fig. 6A). Conversely, Pt-CAT10 and Pt-CAT11 showed increased expression levels in senescing leaves in December compared with leaves collected in October (Fig. 6A). These expression patterns could be related to amino acid concentration and more particularly to glutamine. Indeed, the amino acid concentration of laminae displayed the same variations during autumn (Fig. 2A), and regression analysis between glutamine concentration and Pt-CAT11 expresssion in laminae revealed a good correlation (R2 = 0·994). All other poplar CAT members were also analysed but were not detected in these samples.
Fig. 6.
Expression of CAT transporter genes in various poplar tissues during or after senescence. (A) Expression of poplar CAT1, CAT2, CAT3, CAT4, CAT8, CAT10, CAT11 and CAT12 genes in laminae. (B) Expression of poplar CAT1, CAT2, CAT3, CAT4, CAT9 and CAT12 genes in petioles. (C) Expression of poplar CAT2, CAT3, CAT4, CAT6, CAT11 and CAT12 genes in in 2-year-old stems. Total RNAs were extracted at different time periods during autumn and winter and 300 ng of total RNAs were reverse transcribed into cDNA. The ubiquitin (Ubq) gene was amplified and used as internal control. Analyses were performed by RT–PCR in triplicate. Signal intensities were normalized to the constitutively expressed poplar ubiquitin gene. Values are expressed as the mean ± s.e. of three replicate experiments. For a given gene, values with the same letter are not significantly different, according to ANOVA at P < 0·05.
As observed in laminae, Pt-CAT3 and Pt-CAT4 were strongly expressed in petioles but transcript levels of these genes were poorly affected by leaf senescence (Fig. 6B). Pt-CAT1, Pt-CAT9 and Pt-CAT12 showed a similar expression pattern with more transcripts detected on 23 November (Fig. 6B). Pt-CAT2 showed increased expression levels in petioles in December compared with petioles collected in October (Fig. 6B).
Interestingly, although not expressed in laminae and only poorly expressed in petioles, Pt-CAT9 was highly expressed in 2-year-old stems, just before leaf fall. In contrast, Pt-CAT2 was very weakly expressed in 2-year-old stems in autumn and in winter (Fig. 6C). Pt-CAT12 presented the same expression pattern in 2-year-old stems, in laminae and in petioles (Fig. 6C). As observed in laminae and petioles, Pt-CAT3 transcript levels were high and barely affected by senescence in 2-year-old stems (Fig. 6C). Interestingly, Pt-CAT4 and Pt-CAT9 transcript levels were high during autumn senescence and decreased in 2-year-old stems after leaf fall (Fig. 6C). More surprising, Pt-CAT11 expression was subjected to quite high variations (Fig. 6C) which could be related to amino acid concentration. Indeed, the amino acid concentration of 2-year-old stems displayed the same variations as Pt-CAT11 expression during autumn and winter (Fig. 2B). Regression analysis between total amino acid concentration and Pt-CAT11 expresssion did not reveal a correlation between these two parameters (R2 = 0·295). Regression analysis between glutamine concentration and Pt-CAT11 expression, however, reveals a better correlation (R2 = 0·730). As observed in laminae, Pt-CAT11 expression in stems seems to be related to glutamine concentration. All other poplar CAT genes were also analysed but were not detected in these experiments.
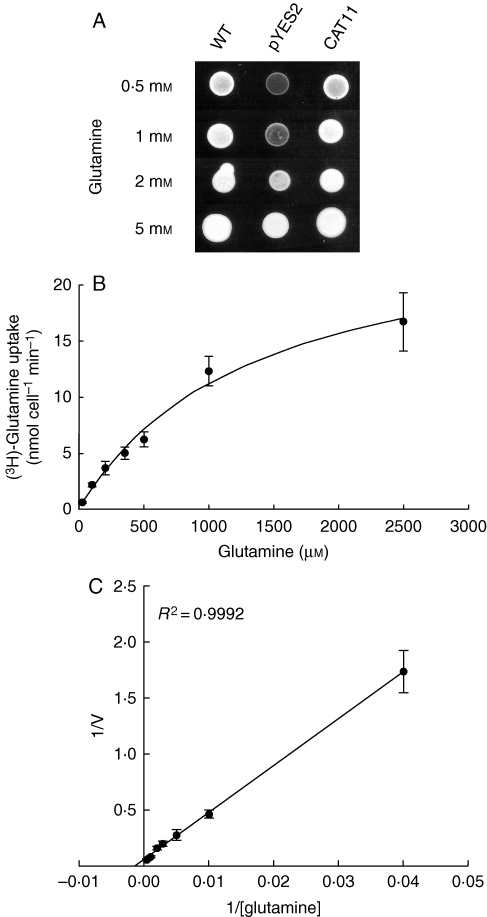
In order to determine the function of Pt-CAT11, yeast complementation experiments were performed with the yeast mutants 22Δ8AA and JA248. The 22Δ8AA strain is unable to use arginine, aspartate, citrulline, γ-aminobutyric acid (GABA), glutamate and proline efficiently as sole N sources (Fischer et al., 2002) and the JA248 strain is unable to use glutamine efficiently as sole N source (Velasco et al., 2004). As controls, strains 22Δ8AA and JA248 were transformed with the expression vector pYES2. Transformation with the yeast expression vector pYES2 bearing the Pt-CAT11 coding sequence under the control of the GAL1 promoter conferred the ability of JA248 to grow in the presence of 0·5 mm glutamine (Fig. 7A). The transport of glutamine by Pt-CAT11 was further confirmed by uptake experiments, which demonstrated that Pt-CAT11-mediated [3H]glutamine uptake was concentration dependent and showed saturable kinetics with an apparent Km value of 690 µm (Fig. 7B, C). Transformation with the yeast expression vector pYES2 bearing the Pt-CAT11 coding sequence under the control of the GAL1 promoter conferred the ability of 22Δ8AA to grow when supplied 3 and 6 mm proline, GABA or citrulline as the sole N source but not when supplied aspartate or glutamate (not shown). Yeast transformed with Pt-CAT11 showed no growth on medium containing 1 mm arginine as sole N source (data not shown).
Fig. 7.
Functional characterization of poplar CAT11 by heterologus expression in a yeast mutant strain. (A) Yeast strain JA248 was transformed with the yeast expression vector pYES2 or pYES2 harbouring the coding sequence of Pt-CAT11. Growth was assayed on N-free medium containing 20 g L−1 Gal and either 0·5, 1, 2 or 5 mm l-glutamine as sole N source. Pictures were taken after 2 d of growth at 30 °C and are representative of three replicates. (B) Concentration-dependent kinetics of [3H]glutamine uptake by yeast strain JA248 expressing Pt-CAT11. The Michaelis–Menten constant for glutamine is 690 µm. Values are expressed as the mean ± s.e. of three replicate experiments. (C) Lineweaver–Burk representation of [3H]glutamine uptake by yeast strain JA248 expressing Pt-CAT11. Values are expressed as the mean ± s.e. of three replicate experiments.
DISCUSSION
Glutamine is the key metabolite to transfer N from senescing leaf to perennial tissues
Nitrogen economy has a special importance in woody plants that are able to cope with seasonal periods of growth and development over many years. As N availability in the forest soil is extremely low, efficient mechanisms are required for the assimilation, storage, mobilization and recycling of inorganic and organic forms of N. Seasonal N cycling is an adaptation of plants to winter cold seasonal climates in which nutrients (mostly N) are often considered to be the major growth-limiting factor (Cooke and Weih, 2005). In the N metabolism of conifers, the cyclic interconversion of arginine and the amides glutamine and asparagine plays a central role, and its regulation is critical to maintain the N economy of these long-living plants (Canovas et al., 2007).
At the beginning of autumn, the major amino acids found in laminae were glutamate and glutamine. During senescence the glutamate concentration decreased whereas that of glutamine increased. The same variations were observed for aspartate and asparagine but to a smaller extent (Fig. 1A). It has been demonstrated that N content decreased in autumn leaves of aspen and about 80 % of total leaf N was withdrawn during autumn senescence (Keskitalo et al., 2005). Amino-N pools may contribute only slightly to this decrease, while other N-containing compounds (chlorophyll for instance) may be of more importance in this process.
On the other hand, qualitative changes in amino acid concentration also revealed this N remobilization process. In senescing leaves, a large amount of ammonium is produced as a result of protein hydrolysis (Hörteinsteiner and Feller, 2002). Ammonium is assimilated into the glutamine amide group, and the specific expression of the glutamine synthetase gene NtGLN1;3 was observed in senescing leaves of Nicotiana tabacum (Brugiere et al., 2000). Moreover, it has been demonstrated recently that expression of several ammonium transporter genes in poplar (PtAMT1;5, PtAMT1;6 and PtAMT3;1) increased with leaf maturation, suggesting that they are specifically recruited to ensure ammonium assimilation during the process of leaf senescence (Couturier et al., 2007). Whereas glutamate decreased with ageing, the glutamine pool increased, suggesting that glutamine biosynthesis had exhausted the glutamate pool.
In the central vein and petiole, amino acid concentrations increased during senescence until 5 December, before leaf fall (Fig. 1B, C). Glutamine was the major amino acid found in the central vein and petiole, whereas glutamate represented <3 % and 6 %, respectively in these tissues. The increase in glutamine concentration in the central vein and petiole suggests an export from leaves to perennial organs during autumn (Fig. 1). It can be also noted that leucine, isoleucine and valine concentrations increased strongly in the central vein and petiole during senescence, which was not observed in the lamina. Interestingly, pools of leucine and isoleucine also increased with ageing in Arabidopsis leaves (Diaz et al., 2005). It has been suggested that isoleucine and leucine biosynthesis exhausted the aspartate pool. The same processes could occur in poplar leaves during senescence.
During autumn leaf senescence, there is a functional shift in leaf metabolism from resource allocation to resource remobilization and export. Rubisco breakdown during autumn leaf senescence in poplar (Brendley and Pell, 1998) accounts for a notable proportion of the N exported from leaves (Titus and Kang, 1982; Millard and Thompson, 1989). N-rich amino acids are transported via the phloem from senescing leaves to perennial tissues, where they are used to synthesize proteins (Sauter et al., 1989). During autumn, BSPs accumulate in perennating tissues such as bark, wood and roots (Sauter and van Cleve, 1990; Langheinrich and Tischner, 1991). Interestingly, arginine and soluble protein contents increased during autumn and were higher after leaf fall (Figs 2 and 3). This was mostly evidenced for arginine, which increased from undetectable levels in October to >200 nmol mg−1 d. wt on 12 December in 2-year-old stems (Fig. 2). Arginine concentration decreased after leaf fall. Furthermore, as observed in several Populus species (Langheinrich and Tischner, 1991), two major polypeptides accumulated in 1- and 2-year-old stems during autumn and winter (Fig. 3B). Previous studies have demonstrated that during the wintering phase, arginine was the major amino acid in both bark and xylem (Sagisaka, 1974). It can be noted that storage proteins are particularly rich in arginine and in amide-containing amino acids (Müntz, 1998). Arginine accumulation in poplar stems during autumn and winter could therefore be considered as a temporary N storage form that could be used thereafter for storage protein synthesis.
Arginine is preferentially synthesized in perennial tissues
The fact that arginine was not detected in laminae, central veins and petioles during senescence suggests that arginine synthesis may occur in perennial tissues such as stems. The metabolic route to arginine synthesis in plants involves two distinct processes: synthesis of ornithine from glutamate and synthesis of arginine from the ornithine intermediates (Slocum, 2005). The second process requires the carbamoyl-phosphate intermediate, which is generated from glutamine via CPS which also contributes to nucleotide metabolism. The CPS protein is made up of a small and a large subunit (Slocum, 2005). Three others enzymes are involved in arginine synthesis: OTC, AS and AL. A detailed expression analysis of AS and OTC revealed that these genes were expressed in leaves and petioles during autumn (Fig. 4A, B). However none of these gene expression patterns followed the CP marker gene. AS transcript levels were preferentially higher in autumn than in winter (Fig. 4). OTC transcripts did not show variations during senescence in leaves, petioles and stems (Fig. 4). In contrast, the CPS gene expression level strongly increased in stems during senescence (Fig. 4C), whereas it was weakly detected in leaves and petioles (Fig. 4A, B). It also clearly matched the expression of CP in leaves. Interestingly, stem glutamine concentration increased from November to January (data not shown) and carbamoyl-phosphate is generated from glutamine via CPS. In stems, glutamine could be used for carbamoyl-phosphate synthesis and consequently for arginine synthesis. In leaves, glutamine could be preferentially used as a transport component from senescing leaves to perennial poplar tissues. Nevertheless, it cannot be ruled out that arginine could also be synthesized in leaf but to a smaller extent. Indeed, in Arabidopsis senescing leaves, it has been demonstrated that arginine content increased and represented around 1 % of total amino acid content (Diaz et al., 2005).
Pt-CAT11 is a candidate for glutamine transfer during the senescing process
In senescing leaves, production of glutamine increases and glutamine is further loaded into central veins and petioles to reach perennial tissues where it may be used for arginine synthesis. We therefore looked at the genetic potential for loading glutamine into the phloem, and more specifically we looked at the expression levels of AAP and CAT amino acid transporters. The AAP members were either not expressed in senescing tissues or even not expressed in leaves at all. We therefore did not focus much attention on this family.
Expression data for the Arabidopsis CAT gene family indicated that At-CAT2 and At-CAT5 were upregulated during leaf senescence. At-CAT2 is probably located in the tonoplast and may be the long-sought vacuolar amino acid transporter (Su et al., 2004). At-CAT5 functions as a high-affinity, basic amino acid transporter in the plasma membrane and At-CAT5 may function in reuptake of leaking amino acids at the leaf margin (Su et al., 2004). In contrast to their Arabidopsis orthologues (Fig. 5), Pt-CAT5 transcripts were not detected and Pt-CAT2 was only expressed at the beginning of senescence (Fig. 6A). However, Pt-CAT2 was strongly upregulated in the petiole during senescence and very weakly expressed in stems (Fig. 6B, C). Nevertheless, expression of the orthologous genes of amino acid transporters may not be similar because the pool of amino acids available for phloem transport is differentially regulated in different species (Delrot et al., 2001). A detailed analysis of each amino acid transporter gene must be made before conclusions can be drawn about the role of the different orthologues. Pt-CAT3 was highly expressed in laminae, petioles and stems and was only slightly affected by senescence in leaves (Fig. 6). Pt-CAT4 was also highly expressed in laminae, petioles and stems but, in contrast to Pt-CAT3, it was downregulated in stems during winter (Fig. 6). Pt-CAT9 transcripts were weakly detected in petioles and strongly in stems (Fig. 6B, C). Pt-CAT9 seems to be preferentially expressed in organs containing sieve elements. In poplar senescing leaves, Pt-CAT10 and Pt-CAT11 were upregulated during senescence (Fig. 6A). Pt-CAT11 was not expressed in petioles but was expressed in laminae and stems (Fig. 6). Pt-CAT11 expression was upregulated in senescing leaves (Fig. 6A) and subject to quite high variations in stems (Fig. 6C). Interestingly, regression analyses have shown that in the lamina and stem, Pt-CAT11 expression and glutamine concentration are related and displayed variations of the same order (Figs 1A and 2B).
Functional analysis demonstrated that Pt-CAT11 restored growth of the yeast mutant JA248 on low glutamine medium (Fig. 7A). Additionally, Pt-CAT11 allowed growth of the yeast mutant 22Δ8AA on medium containing neutral amino acids (proline, citrulline and GABA) but not medium containing acid (aspartate and glutamate) amino acids or arginine. Determination of kinetic parameters for [3H]glutamine uptake by Pt-CAT11 in yeast revealed that it can transport glutamine efficiently, with an apparent Km value of 690 µm (Fig. 7B, C).
Most importantly, recent analysis of expression data showed that Pt-CAT11 was highly and preferentially expressed in phloem tissues (Courtois-Moreau et al., 2009). Taken together, these data suggest that the major function of Pt-CAT11 is related to the transport of amino acids, and notably glutamine, from senescing leaves to sink tissues such as stems, thus facilitating N remobilization during senescence in poplar.
Conclusions
The analysis of amino acid pools in different organs showed that N remobilization from leaves to perennial organs occurs in poplar during autumn senescence. N-rich amino acids, such as glutamine, are transported via the phloem from senescing leaves to perennial organs, such as stems, where they are used to synthesize storage proteins. The glutamine pools in late autumn correlate with increased Pt-CAT11 expression, which may function as a glutamine transporter for amino acid transfer between source and sink tissues during senescence processes in poplar. Arginine was being accumulated, probably as an N storage compound, and would be preferentially synthesized in stems, as indicated by the strong arginine accumulation in stems during autumn and at the beginning of winter and the large increases in CPS transcript levels during autumn. Whether arginine would be further metabolized to provide N for protein biosynthesis remains to be demonstrated.
The elucidation of amino acid concentrations and profiles together with the characterization of a new amino acid transporter (Pt-CAT11) may present a comprehensive foundation for future studies on amino acid transport and metabolism during autumn N remobilization in perennial plants.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
J.C. was supported by a PhD fellowship from the ‘Ministère délégué à l'Enseignement supérieur et à la Recherche’. Aude Migeon is gratefully acknowledged for help in sampling leaves, petioles and stems of poplar and assistance with statistical analysis. We thank Dr Nicolas Rouhier (University Nancy, France) for help with protein extraction.
LITERATURE CITED
- Andersson A, Keskitalo J, Sjodin A, et al. A transcriptional timetable of autumn senescence. Genome Biology. 2004;5:R24. doi: 10.1186/gb-2004-5-4-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, et al. Gene expression in autumn leaves. Plant Physiology. 2003;131:430–442. doi: 10.1104/pp.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorer KJ, Fischer WN. Specificity and stoichiometry of the Arabidopsis H+/amino acid transporter AAP5. Journal of Biological Chemistry. 1997;29:13040–13046. doi: 10.1074/jbc.272.20.13040. [DOI] [PubMed] [Google Scholar]
- Boorer KJ, Frommer WB, Bush DR, Loo DDF, Wright EM. Kinetics and specificity of a H+/amino acid transporter from Arabidopsis thaliana. Journal of Biological Chemistry. 1995;29:2213–2220. doi: 10.1074/jbc.271.4.2213. [DOI] [PubMed] [Google Scholar]
- Brendley BW, Pell EJ. Ozone-induced changes in biosynthesis of Rubisco and associated compensation to stress in foliage of hybrid poplar. Tree Physiology. 1998;18:81–90. doi: 10.1093/treephys/18.2.81. [DOI] [PubMed] [Google Scholar]
- Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: elimination of interfering substances. Analytical Biochemistry. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Brugiere N, Dubois F, Masclaux C, Sangwan RS, Hirel B. Immunolocalization of glutamine synthetase in senescing tobacco (Nicotiana tabacum L.) leaves suggests that ammonia assimilation is progressively shifted to the mesophyll cytosol. Planta. 2000;211:519–527. doi: 10.1007/s004250000309. [DOI] [PubMed] [Google Scholar]
- Bush DR. Sugar transporters in plant biology. Current Opinion in Plant Biology. 1999;2:187–191. doi: 10.1016/S1369-5266(99)80034-X. [DOI] [PubMed] [Google Scholar]
- Canovas FM, Avila C, Canton FR, Canas RA, de la Torre F. Ammonium assimilation and amino acid metabolism in conifers. Journal of Experimental Botany. 2007;58:2307–2318. doi: 10.1093/jxb/erm051. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Weih M. Nitrogen storage and seasonal nitrogen cycling in Populus: bridging molecular physiology and ecophysiology. New Phytologist. 2005;167:19–30. doi: 10.1111/j.1469-8137.2005.01451.x. [DOI] [PubMed] [Google Scholar]
- Courtois-Moreau CL, Pesquet E, Sjödin A, et al. A unique program for cell death in xylem fibers of Populus stem. Plant Journal. 2009;58:260–274. doi: 10.1111/j.1365-313X.2008.03777.x. [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. The expanded family of ammonium transporters in the perennial poplar plant. New Phytologist. 2007;174:137–150. doi: 10.1111/j.1469-8137.2007.01992.x. [DOI] [PubMed] [Google Scholar]
- Delrot S, Rochat C, Tegeder M, Frommer WB. Amino acid transport. In: Lea P, Gaudry JFM, editors. Plant nitrogen. Paris, France: INRA-Springer; 2001. pp. 215–235. [Google Scholar]
- Diaz C, Purdy S, Christ A, Morot-Gaudry JF, Wingler A, Masclaux-Daubresse C. Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. A metabolic profiling approach. Plant Physiology. 2005;138:898–908. doi: 10.1104/pp.105.060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RE. Xylem translocation of amino acids from roots to shoots in cottonwood plants. Canadian Journal of Forest Research. 1979;9:374–378. [Google Scholar]
- Dickson RE, Vogeman TC, Larson PR. Glutamine transfer from xylem to phloem and translocation to developing leaves of Populus deltoides. Plant Physiology. 1985;77:412–417. doi: 10.1104/pp.77.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. Journal of Biological Chemistry. 1995;29:16315–16320. doi: 10.1074/jbc.270.27.16315. [DOI] [PubMed] [Google Scholar]
- Fischer WN, Loo DD, Koch W, et al. Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. The Plant Journal. 2002;29:717–731. doi: 10.1046/j.1365-313x.2002.01248.x. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O. Seed and vascular expression of a high affinity transporter for cationic amino acids in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1995;29:12036–12040. doi: 10.1073/pnas.92.26.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes UZ, Nielsen E, Honaas LA, Taylor CG, Schachtman DP. AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. The Plant Journal. 2006;48:414–426. doi: 10.1111/j.1365-313X.2006.02880.x. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U. Nitrogen metabolism and remobilization during senescence. Journal of Experimental Botany. 2002;53:927–937. doi: 10.1093/jexbot/53.370.927. [DOI] [PubMed] [Google Scholar]
- Javelle A, Morel M, Rodriguez-Pastrana BR, et al. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Molecular Microbiology. 2003;47:411–430. doi: 10.1046/j.1365-2958.2003.03303.x. [DOI] [PubMed] [Google Scholar]
- Keskitalo J, Bergquist G, Gardestrom P, Jansson S. A cellular timetable of autumn senescence. Plant Physiology. 2005;139:1635–1648. doi: 10.1104/pp.105.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langheinrich U, Tischner R. Vegetative storage proteins in poplar. Induction and characterization of a 32- and 36-kilodalton polypeptide. Plant Physiology. 1991;97:1017–1025. doi: 10.1104/pp.97.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P, Thomson CM. The effect of the autumn senescence of leaves on the internal cycling of nitrogen for the spring growth of apple trees. Journal of Experimental Botany. 1989;40:1285–1289. [Google Scholar]
- Müntz K. Deposition of storage protein. Plant Molecular Biology. 1998;38:77–99. [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, et al. High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. Journal of Biological Chemistry. 2002;277:45338–45346. doi: 10.1074/jbc.M207730200. [DOI] [PubMed] [Google Scholar]
- Okumoto S, Koch W, Tegeder M, et al. Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. Journal of Experimental Botany. 2004;55:2155–2168. doi: 10.1093/jxb/erh233. [DOI] [PubMed] [Google Scholar]
- Pregitzer KS, Dickmann DI, Hendrick R, Nguyen PV. Whole-tree carbon and nitrogen partitioning in young hybrid poplars. Tree Physiology. 1990;7:79–93. doi: 10.1093/treephys/7.1-2-3-4.79. [DOI] [PubMed] [Google Scholar]
- Ryan DF, Bormann FH. Nutrient resorption in northern hardwood forests. Bioscience. 1982;32:29–32. [Google Scholar]
- Sagisaka S. Effect of low temperature on amino acid metabolism in wintering poplar: arginine–glutamine relationships. Plant Physiology. 1974;53:319–322. doi: 10.1104/pp.53.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter JJ, van Cleve B, Wellenkamp S. Ultrastructural and biochemical studies results on the localization and distribution of storage proteins in a poplar tree and in twigs of other tree species. Holzforschung. 1989;43:1–6. [Google Scholar]
- Sauter JJ, van Cleve B. Biochemical, immunochemical, and ultrastructural studies of protein storage in poplar (Populus × canadensis ‘robusta’) wood. Planta. 1990;183:92–100. doi: 10.1007/BF00197572. [DOI] [PubMed] [Google Scholar]
- Sauter JJ, van Cleve B. Seasonal variation of amino acids in the xylem sap of Populus × canadensis and its relation to protein body remobilisation. Trees. 1992;7:26–32. [Google Scholar]
- Slocum RD. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiology and Biochemistry. 2005;43:729–745. doi: 10.1016/j.plaphy.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Staswick PE. Storage proteins of vegetative plant tissues. Annual Review of Plant Physiology and Plant Molecular Biology. 1994;45:303–322. [Google Scholar]
- Stepien V, Sauter JJ, Martin F. Vegetative storage proteins in woody plants. Plant Physiology and Biochemistry. 1994;32:185–192. [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouze P, van de Peer Y. EST data suggest that poplar is an ancient polyploid. New Phytologist. 2005;167:165–170. doi: 10.1111/j.1469-8137.2005.01378.x. [DOI] [PubMed] [Google Scholar]
- Su YH, Frommer WB, Ludewig U. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiology. 2004;136:3104–3113. doi: 10.1104/pp.104.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Titus JS, Kang S-M. Nitrogen metabolism, translocation, and recycling in apple trees. Horticultural Reviews. 1982;4:204–246. [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Velasco I, Tenreiro S, Calderon IL, André B. Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryotic Cell. 2004;3:1492–1503. doi: 10.1128/EC.3.6.1492-1503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB. Conservation of amino acid transporters in fungi, plants and animals. Trends in Biochemical Sciences. 2002;27:139–147. doi: 10.1016/s0968-0004(01)02054-0. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.