Abstract
Background and Aims
Aluminium (Al) resistance in common bean is known to be due to exudation of citrate from the root after a lag phase, indicating the induction of gene transcription and protein synthesis. The aims of this study were to identify Al-induced differentially expressed genes and to analyse the expression of candidate genes conferring Al resistance in bean.
Methods
The suppression subtractive hybridization (SSH) method was used to identify differentially expressed genes in an Al-resistant bean genotype (‘Quimbaya’) during the induction period. Using quantitative real-time PCR the expression patterns of selected genes were compared between an Al-resistant and an Al-sensitive genotype (‘VAX 1’) treated with Al for up to 24 h.
Key Results
Short-term Al treatment resulted in up-regulation of stress-induced genes and down-regulation of genes involved in metabolism. However, the expressions of genes encoding enzymes involved in citrate metabolism were not significantly affected by Al. Al treatment dramatically increased the expression of common bean expressed sequence tags belonging to the citrate transporter gene family MATE (multidrug and toxin extrusion family protein) in both the Al-resistant and -sensitive genotype in close agreement with Al-induced citrate exudation.
Conclusions
The expression of a citrate transporter MATE gene is crucial for citrate exudation in common bean. However, although the expression of the citrate transporter is a prerequisite for citrate exudation, genotypic Al resistance in common bean particularly depends on the capacity to sustain the synthesis of citrate for maintaining the cytosolic citrate pool that enables exudation.
Keywords: Aluminium resistance, aluminum, citrate exudation, common bean, MATE, Phaseolus vulgaris, transcriptomic analysis, differential gene expression
INTRODUCTION
Common bean (Phaseolus vulgaris) is produced in the tropics on small-scale farms where unfavourable edaphic factors limit the yield potential. Among others, soil acidity which affects about 40 % of the world arable land (Von Uexküll and Mutert, 1995) accounts for 30–40 % yield reduction in Africa and Latin America (CIAT, 1992). The crop yield on acid soils is mainly limited by aluminium (Al) toxicity. In addition, other soil acidity-related stresses, such as proton toxicity, Mn toxicity, and nutrient deficiencies particularly of P, Mg, Ca and Mo are also important constraints (Marschner, 1995). Al toxicity causes inhibition of root growth by injuring primarily the root apex of the growing plant (Ryan et al., 1993; Sivaguru and Horst, 1998; Rangel et al., 2007).
Common bean is generally less adapted to acid soil environments and improving Al resistance of common bean to reduce the dependence of small-scale farmers on lime and nutrient inputs is a major challenge (Rao, 2001). However, efforts to develop adapted genotypes indicate that there are genotypic differences in Al resistance in the bean germplasm (Rao, 2001; Rangel et al., 2005; Manrique et al., 2006). Comparing two contrasting bean cultivars ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive), Rangel et al. (2010) found that Al resistance in common bean is attributed to the release of citrate by the root apex. Organic acid anions such as citrate, malate and oxalate detoxify Al through forming a non-phytotoxic organic acid–Al complex. Ma et al. (2001) described two patterns of organic acid secretion: pattern I plants release organic anions immediately after the onset of Al treatment while in pattern II plants, organic anion release starts after a lag phase of several hours. This suggests that in pattern I the organic anion release mechanism is constitutively expressed, whereas in pattern II plants the induction of the resistance mechanism involves gene expression, and new protein synthesis. Common bean proved to be a typical pattern II plant (Rangel et al., 2007, 2010). The delay in citrate exudation was not due to the limitation of internal citrate reserve but to the absence of citrate permeases. The role of organic anion permeases in Al resistance was recently reviewed by Delhaize et al. (2007). Aluminium resistance genes of several plant species have been identified and found to encode membrane proteins which mediate the exudation of organic acid anions from the root. These proteins belong to two families, ALMT (Al-activated malate transporter) and MATE (multidrug and toxic compound extrusion). The ALMT facilitates malate efflux in plant species that depend on malate exudation as the Al resistance mechanism (Sasaki et al., 2004; Hoekenga et al., 2006; Ligaba et al., 2006). On the other hand, the MATE proteins are citrate transporters which play a decisive role in Al-induced citrate exudation (Furukawa et al., 2007; Magalhaes et al., 2007; Ryan et al., 2009).
On the basis of the results published by Rangel et al. (2010), it was hypothesized that the expression of a citrate transporter and the enhanced synthesis of citrate are crucial for sustained Al resistance in common bean. Thus, the objectives of this work were: (a) to study the transcriptional changes occurring between the onset of Al treatment and the beginning of citrate release in the Al-resistant common bean genotype ‘Quimbaya’ using suppression subtractive hybridization (SSH); and (b) to analyse the expression of selected candidate genes which may have significant roles in Al resistance using quantitative real-time PCR (qRT-PCR).
MATERIALS AND METHODS
Plant material and growth condition
Two common bean (Phaseolus vulgaris L.) genotypes with known differential Al resistance (Rangel et al., 2005, 2010) were used in this study. The seeds of Al-resistant genotypes ‘Quimbaya’ and Al-sensitive genotype ‘VAX 1’ were germinated in filter papers sandwiched between sponges soaked with tap water. After 4 d, uniform seedlings were transferred to 18-L pots with a continuously aerated nutrient solution (containing 5 mm CaCl2, 1 mm KCl and 8 µm H3BO3) in a controlled climate chamber, with a 16/8 h light/dark regime, 27/25 °C day/night temperatures, 70 % relative air humidity and a photon flux density of 230 µmol m−2 s−1 (photosynthetic active radiation) at the plant canopy. The pH of the nutrient solution was gradually lowered to 4·5 within 2 d. Then the plants were treated with or without 20 µm AlCl3 for various durations of time up to 24 h. Root growth was measured at 4, 8 and 24 h of Al treatment.
Determination of citrate exudation and citrate contents of root apices
Citrate exudatation from root tips and citrate contents of root tips were determined as described by Rangel et al. (2010). Briefly, plants were pre-treated with or without 20 µm Al for 3, 7 and 23 h at pH 4·5 as described above. To collect root exudates from intact root apices, 12 pre-treated plants were bundled in filter paper soaked with nutrient solution. Approximately 1 cm of the main root apex of each plant was immersed into 18 mL of a constantly aerated collection solution containing 5 mm CaCl2, 8 µm H3BO3 and 0 or 40 µm AlCl3, pH 4·5, in 20-mL poly prep filtration columns (BioRad Laboratories, Richmond, CA, USA). The Al concentration in the collection medium was doubled in order to compensate for the small volume and thus low total Al supply. After 2 h of incubation, the collection solution containing the root exudates was immediately frozen at −20 °C for later citrate determination. At the end of incubation period, the root tips (1 cm) were excised with a razor blade, rinsed with double deionized water, transferred to Eppendorf reaction vials and fixed immediately in liquid nitrogen to measure the citrate content in the root tissue. The citrate concentrations in the root exudates as well as in the root tissue extracts were measured by isocratic high-pressure liquid chromatography (Kroma System 3000; Kontron Instruments, Munich, Germany).
RNA isolation and construction of the SSH library
For construction of the SSH library, only the Al-resistant genotype ‘Quimbaya’ was used. The plants were treated with or without 20 µm AlCl3 for 4 h as described above. At the end of the treatment time, roots were rinsed with distilled water and ten root tips (1 cm long) per plant were harvested and shock-frozen in liquid nitrogen. Root tips of 15 plants per treatment were bulked and ground to powder in liquid nitrogen. Total RNA was isolated using the NucleoSpin RNA plant kit (MACHEREY-NAGEL GmbH and Co., KG, Düren, Germany) following the manufacturer's protocol. Tester and driver cDNAs were prepared from the total RNA of each treatment (control and 4 h Al treatment) using the Super SMART PCR cDNA synthesis kit (Clontech Laboratories, Inc., Mountain View, CA, USA). SSH was performed using PCR-Select cDNA subtraction kit (Clontech Laboratories, Inc.). Forward and reverse subtraction libraries were constructed using cDNA samples of control (no Al) versus 4 h Al treatment. The subtracted cDNAs were subjected to two rounds of PCR to normalize and enrich cDNA populations. The PCR products were sub-cloned into the pCR2·1-TOPO Vector (Invitrogen, Carlsbad, CA, USA) by T-A cloning. The vector was used to transform Escherichia coli TOP10 (Invitrogen) competent cells. Transformed clones were grown on LB plates containing X-gal and ampicillin for blue/white screening. Positive clones were checked for the presence of gene inserts after plasmid isolation and EcoR1 digestion. A total of 144 clones containing putative differentially expressed genes were further analysed and the gene inserts were sequenced (ITT, Bielefeld, Germany).
Sequence homology search
In order to identify putative gene functions of the differentially expressed genes, the cDNA sequences were compared with the GenBank database using the online Basic Local Alignment Search Tool (BLAST) program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Candidate-gene selection and primer design
Candidate genes were selected either from the authors' SSH library or from a public database based on previous results on the physiological characterization of the mechanisms of Al resistance in common bean (Rangel et al., 2005, 2007, 2010). The sequences of candidate genes were initially obtained from the arabidopsis database (TAIR). Then, similar sequences of known genes of legumes and expressed sequence tags (ESTs) of common bean (P. vulgaris) were searched and gathered for sequence alignment. Finally, primers were designed in such a way that they anneal to part of the sequence which is well conserved among the legume species. Primers were designed using Primer3 software (Rozen and Skaletsky, 2000). The list of candidate genes and their respective primer pairs are shown in Table 1.
Table 1.
List of genes and specific primer pairs used for quantitative gene expression analysis
| Candidate gene | Primer pairs (5′ → 3′)* | Amplicon size (bp) | GenBank accession no. |
|---|---|---|---|
| PFK (phosphofructokinase) | (+) ACCCTTGCAAGTCGAGATGT | 171 | FE688067 |
| (−) CTGCACACTCTCGGAAACAA | |||
| PEPC (phosphoenolpyruvate carboxylase) | (+) TGGCTCCTTCCAAAGTGAGT | 100 | AF288382 |
| (−) TCCTCCCCGTGTAAATTCTG | |||
| CS (citrate synthase) | (+) CGACGGATATTCAAGGATGG | 142 | FE693132 |
| (−)CGTGATCACTGTGGATGGAA | |||
| Aco (aconitase) | (+) TCCAGTGTGTTGCCTGACAT | 116 | CV536664 |
| (−) GACTTGGGGTCCCATGAGTA | |||
| ICDH (isocitrate dehydrogenase) | (+) GCTCATTTTGCCTTTCCTG | 165 | SSH library |
| (−) TTCACACGAGCTTCATCTGG | |||
| MDH (malate dehydrogenase) | (+) CCAACTGCAAGGTTCTGGTT | 124 | FE677903 |
| (−) GCCCTGTTGTGATCCAATCT | |||
| STOP1 (transcription factor) | (+) GCTCTAACTGCCGATGAGAA | 159 | SSH library |
| (−) TCTCTCCAGCTCCTCCTGAA | |||
| MATE-a (citrate transporter family) | (+) GCTGGATGCAGTTTCAAGAGAG | 138 | CV535133 |
| (−) ACTCCAGCAGCTGCAAGTTC | |||
| MATE-b (citrate transporter family) | (+) TGCTGTTCAAGCCATTCTAGC | 124 | CV534527 |
| (−) TCCAACAGCAAGAGAGAGTCC | |||
| MATE-c (citrate transporter family) | (+) GTGACACTGGCTGCATCATT | 91 | FD792891 |
| (−) GAGAAACTGCCAACCAAACC | |||
| ALS3 (Al-sensitive3) | (+) ACAAGCTTGGCCTCCAGATA | 106 | CV532021 |
| (−) GCGTTGTCCTGGTTGAAGAT | |||
| ALMT1 (Al-activated malate transporter) | (+) TTCGCCCCATCTGGGCTGGT | 118 | CV543751 |
| (−) TCCGGGGTTTCACTGCCATCA | |||
| VDAC (voltage dependent anion channel) | (+) TGCCTCGTTGACTCTGAATG | 146 | SSH library |
| (−) CCGAGGTACCAAGGATGTGA | |||
| ACC-oxidase† | (+) GAAGATGGCGCAAGAAGAAG | 105 | SSH library |
| (−) TGGAGCAAAGGTTCAAGGAG | |||
| β-Tubulin | (+) CCGTTGTGGAGCCTTACAAT | 117 | CV530631 |
| (−) GCTTGAGGGTCCTGAAACAA |
* (+) and (−) indicate forward and reverse primers, respectively.
† ACC-oxidase = aminocyclopropane-1-carboxylic acid oxidase.
The citrate transporter gene MATE is a member of a large gene family. Several ESTs of P. vulgaris which have similarity with known MATE genes were gathered and aligned to assess their homology. Based on the alignment result (see Fig. S1 in Supplementary data, available online), they were grouped into three classes (MATE-a, MATE-b and MATE-c) and appropriate primers were designed as described above. MATE-a and MATE-c have nucleotide-sequence similarities of 81 % and 75 %, respectively, with the arabidopsis MATE gene (locus: AT1G51340). Likewise, MATE-b has 72 % similarity with the arabidopsis FRD3 (ferric reductase defective 3) gene (locus: AT3G08040).
Quantitative RT-PCR
Two common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) were grown and treated with or without 20 µm AlCl3 for 1, 2, 3, 4, 8 and 24 h as described above. At the end of each treatment time, the roots were rinsed with distilled water and ten root tips (1 cm) per plant were harvested and shock-frozen in liquid nitrogen. Root tips of 15 plants per treatment were bulked and ground to powder in liquid nitrogen. Total RNA was isolated from the root tips as described above. First-strand cDNA was synthesized using RevertAid H-Minus first strand cDNA synthesis kit (Fermentas, www.fermentas.com). Random hexamer primers were used for this purpose. The reaction was stopped by heating at 70 °C for 10 min followed by 20 min incubation at 37 °C after addition of 10 U RNase-H (EPICENTRE Biotechnologies, www.epibio.com). Quantitative RT-PCR was undertaken using the Applied Biosystems StepOne Plus thermo-cycler (www.appliedbiosystems.com). The SYBR Green detection system was used with self-prepared SYBR Green master mix including a passive reference dye, ROX. The constituents of the qRT-PCR reaction mix were 1× hot-start PCR buffer, 3·6 mm MgCl2, 0·5 µM ROX, 0·1× SYBR Green-I, 200 µm each dNTP (dATP, dTTP dCTP dGTP), 252 nm each forward and reverse primer, 0·75 U hot-start Taq DNA polymerase, 2 ng μL−1 cDNA template and ultra-pure DNase/RNase-free distilled water in a final volume of 25 µL. The qRT-PCR cycling stages consist of initial denaturation at 95 °C (3 min), followed by 45 cycles of 95 °C (15 s), 60 °C (30 s), 72 °C (30 s), and a final melting curve stage of 95 °C (15 s), 60 °C (1 min) and 95 °C (15 s). The fluorescence signal was recorded during the strand elongation step at 72 °C and the melting curve stage at every 0·3 °C temperature ramp. Samples for qRT-PCR were run in three biological replicates and two technical replicates. Relative gene expression was calculated using the comparative ΔΔCT method according to Livak and Schmittgen (2001). For the normalization of gene expression, three housekeeping genes, namely 18S rRNA, actin and β-tubulin, were tested; β-tubulin was found to be most stable. Accordingly, β-tubulin was used as an internal standard and the control plants of the Al-resistant genotype ‘Quimbaya’ were used as reference sample. The PCR efficiencies of the β-tubulin and the target genes were comparable and thus relative gene expression was calculated without efficiency correction.
Statistical analysis
A completely randomized experimental design with three to six replicates was used for each experiment. The general linear model procedure of the statistical program SAS 9·1 (SAS Institute, Cary, NC, USA) was used for analysis of variance as well as multiple mean comparison using the Tukey test. For qRT-PCR data, relative gene expression and standard deviation were calculated using StepOne Plus software (Applied Biosystems). Expression levels were considered to be significantly different only if the fold change is at least three times higher/lower than the control and if the standard deviations are not overlapping.
RESULTS
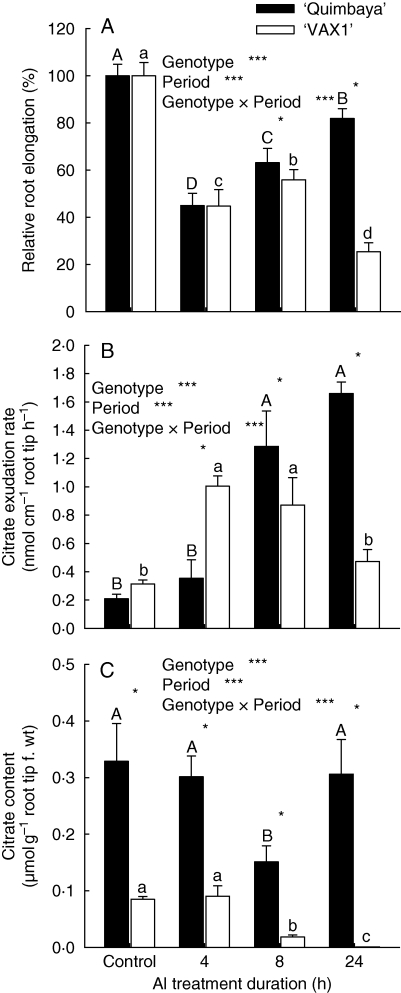
Root growth of both genotypes ‘Quimbaya’ and ‘VAX 1’ was reduced by >50 % within 4 h of Al treatment (Fig. 1A). However, ‘Quimbaya’ gradually recovered from the initial Al stress and recovered to nearly full growth within 24 h. In contrast, ‘VAX 1’ showed a transient recovery after 8 h, but later the growth was again severely inhibited. Citrate exudation from root tips was determined after 4, 8 and 24 h Al treatment (Fig. 1B). Citrate exudation was induced by Al treatment after a lag phase of 4 h in genotype ‘VAX 1’, but in genotype ‘Quimbaya’ the lag phase lasted >4 h. After 8 h the exudation rate remained constant in genotype VAX-1, but in genotype ‘Quimbaya’ the citrate exudation-rate steeply increased up to 24 h of Al treatment. The Al-induced exudation of citrate might be related to the citrate contents of the root-tip tissue. Therefore, the citrate contents of the 1-cm root tips were determined after the collection of root exudates in order to study the effect of Al-treatment duration on the dynamics of the citrate content in the root tissue (Fig. 1C). Genotype ‘Quimbaya’ had constitutively higher citrate contents than ‘VAX 1’. After 8 h Al treatment the citrate content of both genotypes were significantly reduced compared with their respective controls. This was related to the enhanced citrate exudation (Fig. 1B). Striking differences between the two genotypes were observed in the ability to restore the tissue citrate content. While the Al-resistant genotype ‘Quimbaya’ was able to restore its tissue citrate level, in the Al-sensitive genotype ‘VAX 1’ the citrate pool was depleted after 24 h. A close relationship existed between the dynamics of citrate exudation after the lag phase (Fig. 1B) and the recovery of root growth after 8–24 h Al treatment (Fig. 1A).
Fig. 1.
(A) Root growth, (B) citrate exudation rate and (C) citrate content of 1-cm root tips of two common bean genotypes, ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive), grown in simplified nutrient solution treated with or without 20 µm Al, pH 4·5, for up to 24 h. For the determination of citrate exudation and root-tissue citrate-content, plants were pre-cultured in nutrient solution with or without 20 µm Al, pH 4·5 for 3, 7, or 23 h. Root exudates were collected for a period of 2 h in simplified collection solution with or without 40 µm Al, pH 4·5. Citrate content of the root tissue was determined at the end of collection period. Bars are means + standard deviation of four to six replicates. For the analysis of variance, three asterisks denote significance at P < 0·001. Means with the same letter are not significantly different between exudation periods for ‘Quimbaya’ (upper case) and VAX-1 (lower case); asterisks over data points show significant differences between genotypes within each treatment time (Tukey test, P < 0·05). Control plants were not treated with Al.
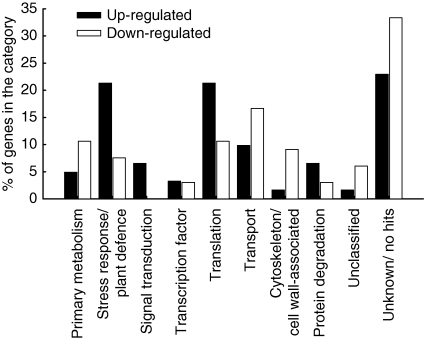
Differential gene expression in Al-resistant genotype ‘Quimbaya’, in response to Al stress was assessed by using SSH. Through forward and reverse subtraction of cDNA samples (4 h Al treatment vs. control) 127 Al-induced differentially expressed transcripts, 61 up-regulated and 66 down-regulated genes were obtained (see Tables S1 and S2 in Supplementary data, available online). These genes were grouped into several functional categories as shown in Fig. 2. Al treatment affected the transcription of genes involved in a wide range of functions. Transcripts which were up-regulated due to Al stress include genes involved in stress response, plant defence, signal transduction and translation. In contrast, genes involved in primary metabolism, transport processes, cytoskeleton and cell wall organization were largely down-regulated. This indicates that Al toxicity induces manifold changes in the plant root, ranging from perception of stress signals, gene transcription and translation to downstream physiological processes. The up-regulated stress-responsive genes include those encoding peroxidases, heat shock proteins and dehydrins. Peroxidases may contribute to the plant Al resistance by detoxifying reactive oxygen species which are produced as a result of oxidative stress induced by Al. Down-regulated genes such as auxin influx-carrier and cytoskeleton-associated proteins confirm the physiological characterization of the mechanism of Al toxicity. However, this study was restricted to the genes which could be involved in citrate exudation and metabolism which was the focus of the physiological studies (Fig. 1). These include genes encoding isocitrate dehydrogenase, an anion-selective channel, a mitochondrial ATP synthase and cytochrome P450 monooxygenase. Their possible contribution to Al resistance is briefly summarized in Table 2.
Fig. 2.
Functional categories of genes differentially expressed (SSH) by 4 h Al treatment in root tips of Al-resistant bean genotype ‘Quimbaya’. Up-regulated genes are those whose transcripts were more abundant in Al-treated than in the control sample and vice versa for down-regulated genes.
Table 2.
Differentially expressed genes (SSH) related to Al resistance in common bean genotype ‘Quimbaya’
| Clone ID | GenBank acc. no. | Annotation | Identity (%) | E-value | Up-/down-regulated | Putative function |
|---|---|---|---|---|---|---|
| 0_153 | DQ072165 | VD anion-selective channel* | 97 | 1E-154 | up | Anion transport |
| 0_156 | M64246 | ATP synthase (F1 alpha) | 99 | 0E + 00 | up | ATP production through proton gradient |
| 0_178 | DQ340249 | Cytochrome P450 monooxygenase | 92 | 0E + 00 | up | Detoxification of toxic compounds |
| 4_86 | L12157 | NADP-Isocitrate dehydrogenase | 88 | 2E-138 | down | Conversion of isocitrate to 2-oxoglutarate in the TCA cycle. |
* VD = voltage-dependent.
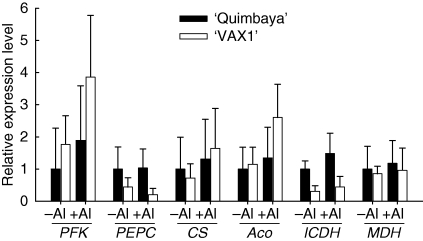
In addition candidate genes were selected based on the physiological mechanisms of Al resistance in bean, which is a sustained release of citrate from the root tips (Fig. 1). The expression of two groups of genes was closely investigated using qRT-PCR. The first group included genes encoding the enzymes involved in citrate metabolism, while the second group consisted of genes encoding ion transporters. Aluminium treatment did not significantly alter the expression of genes coding for enzymes which play a role in citrate synthesis and degradation (Fig. 3). Besides, there were few or no genotypic differences in Al-induced changes in gene expression. However, constitutively lower expression of genes coding for phosphoenolpyruvate carboxylase (PEPC) and isocitrate dehydrogenase (ICDH) was observed in ‘VAX 1’ than in ‘Quimbaya’. The expression pattern did not change when the plants were treated for 8 or 24 h (data not shown).
Fig. 3.
Expression of genes encoding enzymes involved in citrate metabolism in common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) grown in nutrient solution treated with or without 20 µm Al for 4 h. Total RNA was extracted from root tips. Quantitative RT-PCR was performed using the β-tubulin gene as internal standard and untreated plants of the Al-resistant genotype ‘Quimbaya’ as calibrator. Relative gene expression was calculated from three biological and two technical replicates; standard errors are shown.
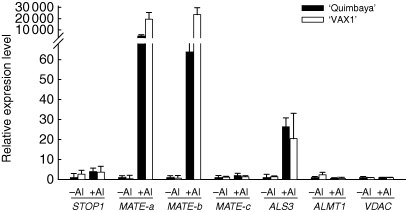
The release of organic acid anions is mediated by membrane-localized anion transporters, whose expressions were known to be regulated by the STOP1 transcription factor. In beans, short-term Al treatment (4 h) slightly increased the expression of STOP1 (Fig. 4) but the expression levels of MATE-a, MATE-b and ALS3 were greatly enhanced by Al treatment in both bean genotypes. Yet, Al did not affect the transcript abundance of MATE-c, ALMT1 and VDAC (voltage-dependent anion-selective channel; Fig. 4).
Fig. 4.
Expression of genes regulating/encoding ion transporters in the common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) grown in nutrient solution treated with or without 20 µm Al for 4 h. Total RNA was extracted from root tips. Quantitative RT-PCR was performed using the β-tubulin gene as internal standard and untreated plants of the Al-resistant genotype ‘Quimbaya’ as calibrator. Relative gene expression was calculated from three biological and two technical replicates; standard errors are shown.
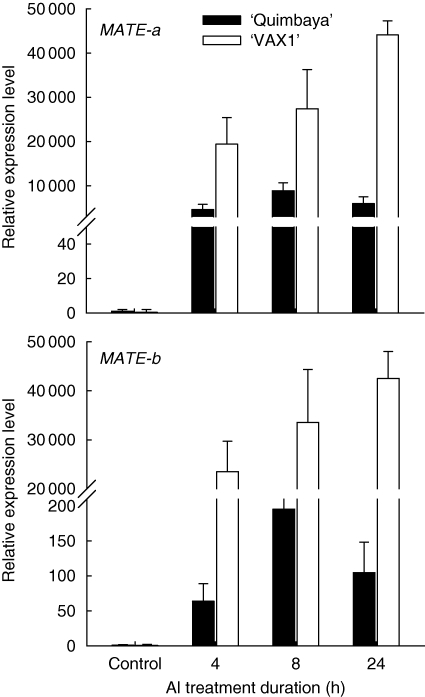
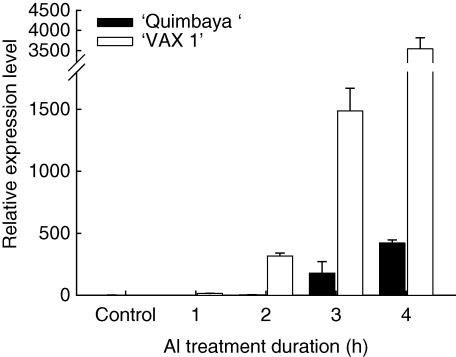
Upon extended Al treatment duration, the expression of MATE-a and MATE-b continued to increase in the Al-sensitive genotype ‘VAX 1’ but not in the Al-resistant genotype ‘Quimbaya’ (Fig. 5). In order to find out the exact timing of MATE-gene induction, root samples were collected at 1-h intervals during the first 4 h of Al treatment. Enhanced expression of the MATE-a gene was observed as early as 2 h Al treatment in ‘VAX 1’, but in ‘Quimbaya’ the expression was delayed until 3 h (Fig. 6). The expression levels continued to increase with time in both genotypes. Similar to Fig. 5, the expression level was much higher in ‘VAX 1’ than in ‘Quimbaya’. The earlier induction of MATE genes in this genotype is in agreement with the earlier induction of citrate exudation in ‘VAX 1’ (Fig. 1B).
Fig. 5.
Expression of two MATE genes under extended duration of Al treatment in the common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) grown in nutrient solution treated with or without 20 µm Al for up to 24 h. Total RNA was extracted from root tips. Quantitative RT-PCR was performed using the β-tubulin gene as internal standard and untreated plants of the Al-resistant genotype ‘Quimbaya’ as calibrator. Relative gene expression was calculated from three biological and two technical replicates; standard errors are shown. Control plants were not treated with Al.
Fig. 6.
Expression of MATE-a genes under short-term Al treatment in the common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) grown in nutrient solution treated with or without 20 µm Al for up to 4 h. Total RNA was extracted from root tips. Quantitative RT-PCR was performed using the β-tubulin gene as internal standard and untreated plants of the Al-resistant genotype ‘Quimbaya’ as calibrator. Relative gene expression was calculated from three biological and two technical replicates; standard errors are shown. Control plants were not treated with Al.
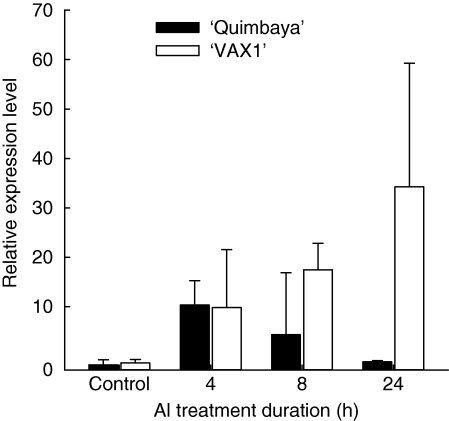
Finally, the expression of a typically stress-induced gene coding the ACC-oxidase (1-aminocyclopropane-1-carboxylic acid oxidase) which was differentially expressed according to the SSH study (Table S1 in Supplementary data) was determined (Fig. 7). Early Al stress (4 h) resulted in a >10-fold increase in the expression of ACC-oxidase gene in both bean genotypes. In a subsequent analysis, enhanced gene expression could be found in both genotypes as early as 1 h after Al treatment (data not shown). But with Al treatment duration, it decreased to the original expression level in ‘Quimbaya’ while it continued to increase in ‘VAX 1’. This stress response is similar to the response in Al-induced inhibition of root growth (Fig. 1A) and thus might be used as an internal marker of Al stress.
Fig. 7.
Gene expression of ACC-oxidase in the common bean genotypes ‘Quimbaya’ (Al-resistant) and ‘VAX 1’ (Al-sensitive) grown in nutrient solution treated with or without 20 µm Al for up to 24 h. Total RNA was extracted from root tips. Quantitative RT-PCR was performed using the β-tubulin gene as internal standard and untreated plants of the Al-resistant genotype ‘Quimbaya’ as calibrator. Relative gene expression was calculated from three biological and two technical replicates; standard errors are shown. Control plants were not treated with Al.
DISCUSSION
Using the SSH method, a range of genes which are differentially expressed in response to Al stress were identified in common bean. Aluminium triggered the expression of genes related to plant-stress response, plant defence and signal transduction. For example, Al enhanced the expression of ACC-oxidase (Fig. 7). Similarly, Sun et al. (2007) reported increased gene expression and enzyme activity of ACC-oxidase leading to increased ethylene production in Lotus japonicus and Medicago truncatula under Al stress and suggested that ethylene is involved in the Al-induced inhibition of root elongation. However, the expression of ACC-oxidase and other differentially expressed stress-responsive genes observed in the present study are also triggered by other stress factors such as heat, cold, drought, waterlogging, or disease infection (Nie et al., 2002; Fekete et al., 2009; Zeller et al., 2009) and as such may not be specific for Al.
Genes that may be related to the citrate exudation-mediated Al resistance of bean include those encoding a VDAC and an NADP-specific ICDH. Several studies indicate that the release of organic acid anions is mediated by anion channels located in the plasma membrane (Kochian et al., 2004; Delhaize et al., 2007; and references therein). The VDAC is a family of eukaryotic pore-forming proteins, originally discovered in the outer membrane of mitochondria where it allows free permeability of low molecular-weight solutes (Colombini, 1979). It is found to be not only expressed in the mitochondria, but also in the plasma membrane (Lawen et al., 2005) and in peroxisomes (Arai et al., 2008). Thus, VDAC may mediate organic anion exudation in bean. However, its differential expression in response to Al treatment could not be verified by qRT-PCR.
Similar to the current observation, cytochrome P450 monooxygenase expression was found to be up-regulated in Al-resistant near isogenic wheat lines under Al stress by Guo et al. (2007) and Houde and Diallo (2008) and was implicated to play a role in Al resistance. Cytochrome P450s may serve as monooxygenases in the biosynthethis of lignin, defence compounds, hormones, pigments, fatty acids and signalling molecules and in the detoxification catalysing numerous endogenous and exogenous toxic compounds encountered in the environment (Schuler and Werck-Reichhart, 2003). Thus it may contribute to sustained root growth under Al stress conditions. In addition, mitochondrial F1-ATPase (alpha-subunit) was also up-regulated in bean root tips upon Al treatment. F1-ATPase is involved in the mitochondrial oxidative phosphorylation by which ATP is produced through a proton gradient. The energy stored in ATP could fuel the metabolic processes involved in Al resistance. Hamilton et al. (2001) hypothesized that the induction of V-ATPase and the F1F0-ATPase plays a role in Al resistance of wheat. The subunits of these enzymes were newly synthesized upon Al treatment and the proteins accumulate in an Al dose-dependent manner (Basu et al., 1994). In addition, accumulation of V-ATPase and F1F0-ATPase subunits segregated with the Al-resistant phenotype (Taylor et al., 1997). This indeed suggests that the up-regulated ATPase may play a yet unspecified role in Al resistance.
The best understood mechanism of Al resistance in plants is the release of organic acid anions such as citrate, malate and oxalate, which chelate Al and form non-toxic complexes (Ryan et al., 2001; Ma et al., 2001). Rangel et al. (2010) observed that Al-activated exudation of citrate plays a major role in Al resistance of common bean. Citrate exudation started after about 4 h of Al treatment despite the abundant citrate content in the root tissue (Fig. 1B and C). Moreover, the root growth of both Al-resistant and Al-sensitive bean genotypes was equally inhibited during this lag period, indicating that Al resistance in common bean is not constitutively expressed (Fig. 1A). This is in agreement with an earlier suggestion that Al resistance in bean is an inducible trait (Cumming et al., 1992). The lag phase between the beginning of Al treatment and the onset of citrate exudation shows that the induction process involves gene transcription and de novo synthesis of proteins which are necessary for citrate transport.
In the present study, the expression of genes encoding organic anion transporters was examined. Among the candidate genes tested, MATE-a (GenBank accession no. CV535133) and MATE-b (GenBank accession no. CV534527) were strongly expressed upon Al treatment in bean. Both ESTs (denoted as MATE-a and MATE-b) of common bean have high sequence similarity to previously characterized MATE genes of Lupinus albus (GenBank accession no. AY631874) and Glycine max (GenBank accession nos EU591739 and EU591741). Nucleotide sequences of MATE-a and MATE-b have no significant similarity and also they do not belong to the same contig assembly of ESTs in the TIGR database. Whether they are two different genes or just different sequence regions of the same gene will be clarified in the ongoing study through full-length cDNA sequencing.
The MATE proteins are a large family of membrane transport-proteins which have 58 members (paralogues) known just in the arabidopsis genome (Hvorup et al., 2003). The arabidopsis FRD3 gene which is important for iron transport in the xylem as ferric citrate is also a MATE protein (Durrett et al., 2007; Rogers et al., 2009). The role of a MATE protein for Al resistance was first observed in sorghum (Magalhaes et al., 2007) and barley (Furukawa et al., 2007) almost simultaneously, later in wheat (Ryan et al., 2009) and further characterized in arabidopsis (Liu et al., 2009). The MATE protein was described as an Al-activated citrate transporter which is responsible for Al resistance of both sorghum and barley (Furukawa et al., 2007; Magalhaes et al., 2007). In sorghum SbMATE was expressed only in the root tips of the Al-resistant genotype in an Al-inducible way. Similarly, barley HvMATE was constitutively expressed mainly in the root apices and correlated with Al-activated citrate exudation and Al resistance in a set of barley cultivars. In contrast, the MATE gene of bean is highly expressed in both resistant and sensitive genotypes used in the present study (Figs 4–6). This result corroborates the observation that citrate exudation was induced by Al in both Al-resistant and -sensitive genotypes (Fig. 1B). Regardless of the ample amount of citrate in the root tissue (Fig. 1B), exudation started only after about 4 h of Al treatment, the time lag which is required for activation of MATE-gene transcription, translation and formation of the functioning protein (Fig. 6). Similarly, Al enhanced the expression of a citrate transporter gene in soybean after 4 h (Z. M. Yang, Jilin University, China, pers. comm.). After the MATE protein is in place, citrate exudation progressed and resulted in a reduction in the citrate content of the root tissue (Fig. 1B and C). As a result of citrate exudation, both bean genotypes transiently recovered from the stress which equally affected both of them in the early hours of Al treatment (Fig. 1A). The remarkable difference between the two bean genotypes was observed in their capacity to replenish the tissue citrate reserve and to sustain citrate exudation in order to protect the growing root tip. The Al-resistant genotype ‘Quimbaya’ was able to restore the citrate pool in the root tissue and to continue to release citrate, whereas the Al-sensitive genotype ‘VAX 1’ was unable to restore the internal citrate pool and failed to further release citrate after the short recovery period (Fig. 1B and C). These observations underline that sustained synthesis of citrate as well as constant expression and activity of a citrate transporter are vital for Al resistance in common bean.
Although the role of organic acid anion exudation for Al resistance and the importance of organic acid anion transporters are currently well defined, the significance of organic acid metabolism and accumulation in the root tissue are still not well understood (Ryan et al., 2001; Horst et al., 2007). In plant species, where organic acid anion release started directly after Al treatment, no correlations were observed between internal organic acid concentrations and efflux. For example, Al-sensitive and Al-resistant wheat genotypes did not differ in root concentrations of malate, although the Al-resistant genotypes released up to 10-fold more malate than the Al-sensitive genotypes (Delhaize et al., 1993). Similarly, contrasting maize genotypes did not differ in tissue citrate content and Al equally increased citrate accumulation in the root tissue of both genotypes but significant citrate exudation was only observed in the Al-resistant genotype (Pellet et al., 1995). In contrast, in soybean (pattern II plant) the Al-enhanced internal accumulation of citrate contributed to the enhanced citrate exudation (Silva et al., 2001). Reports on the role of enzymes involved in the organic acid metabolism for Al-induced organic acid anion efflux are also diverse. In wheat, Al-induced malate exudation occurred without significant changes to the activities of PEPC or malate dehydrogenase (NAD-MDH). Moreover, the activities of these enzymes were not significantly different between genotypes (Ryan et al., 1995). In contrast, an increased citrate synthase (CS) activity was reported in P. vulgaris (Mugai et al., 2000) and Cassia tara (Yang et al., 2004) after Al treatment. Similarly, Al treatment enhanced the gene expression as well as enzyme activity of mitochondrial CS in soybean (Z. M. Yang, Jilin University, China, pers. comm.). Furthermore, over-expression of enzymes involved in organic acid metabolism has been proven to be effective in enhancing exudation of organic acid anions leading to Al resistance in transgenic plants of arabidopsis (Koyama et al., 2000), alfalfa (Tesfaye et al., 2001) and canola (Anoop et al., 2003).
Rangel et al. (2010) studied the changes in activities of enzymes involved in citrate metabolism. Al treatment reduced the activity of ICDH leading to reduced internal citrate consumption and enhanced exudation. The citrate content in the root tissue is a function of citrate synthesis, exudation, degradation or consumption for other metabolic functions. Accordingly, continuous release of citrate while maintaining normal citrate concentration in root tissue requires enhanced synthesis and/or reduced degradation of citrate. Reduction in cytosolic NADP-isocitrate dehydrogenase activity resulted in citrate accumulation and subsequent release from mutant carrot cells which were able to grow on insoluble phosphate sources (Kihara et al., 2003). But according to Rangel et al. (2010), not only the reduction of NADP–ICDH but also maintaining the activities of CS and PEPC are important for sustained exudation of citrate in common bean. Failure of continuous citrate exudation in the Al-sensitive bean genotype ‘VAX 1’ was mainly attributed to the constitutively lower CS activity which was further inhibited by extended duration of Al treatment. In the current study, no significant change was observed in the expression of genes encoding enzymes involved in citrate metabolism (Fig. 3). Similarly, Kumari et al. (2008) who made a large-scale, transcriptomic analysis of root responses to Al, using a microarray representing about 93 % of the predicted genes in the genome of arabidopsis did not detect a significant increase in transcript abundance for any of the 52 genes of the TCA cycle present in the microarray, except for MDH. However this does not mean that there is no change in the activity of enzymes involved in the TCA cycle. Since Rangel et al. (2010) clearly demonstrate the changes in the enzyme activity of the above bean genotypes it can be concluded that the activities of these enzymes are regulated at the post-translational level.
The role of ATP-binding cassette transporter family proteins, ALS1 and ALS3 for Al resistance, was observed in arabidopsis (Larsen et al., 2005, 2007). Plant ATP-binding cassette transporters that have been functionally characterized so far were known to detoxify organic and inorganic compounds by sequestering in the vacuole (Schulz and Kolukisaoglu, 2006). Arabidopsis als1-1 and als3-1 mutants were hypersensitive to Al but the exact functions and substrates of ALS1 and ALS3 are not known. Whereas ALS1 is located in the tonoplast and the gene is expressed in root apices and the vascular system (Larsen et al., 2007), ALS3 is primarily located in the plasma membrane of leaf hydathode cells, the phloem and the root cortex (Larsen et al., 2005). The expression of ALS3 is induced by Al and was suggested to function in channelling accumulated Al away from Al-sensitive tissues in order to protect the growing root from Al toxicity. In agreement with Larsen et al. (2005) it was observed in the present study that Al treatment induced the expression of the ALS3 gene in both in Al-resistant and sensitive bean genotypes (Fig. 4). However, the suggested function of ALS3 could not be confirmed since the sensitive cultivar continued to accumulate Al in the root tissue (Rangel et al., 2010) regardless of ALS3 expression.
In conclusion, this study strongly suggests that in common bean a MATE gene is responsible for Al-induced citrate exudation. The expression of this gene is a prerequisite for Al resistance. However, sustained citrate release and genotypic Al resistance requires, in addition, the continuous synthesis and maintenance of a cytosolic citrate pool in the root apex.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Steve Beebe, Leader of the Bean Program of CIAT, for the supply of seed of the two common bean genotypes. This work was supported by the Federal Ministry of Economic Cooperation and Development (BMZ) and the German Technical Cooperation (GTZ) (05·7860·9–001·00) through a restricted core project, ‘Fighting drought and aluminium toxicity: Integrating functional genomics, phenotypic screening and participatory evaluation with women and small-scale farmers to develop stress-resistant common bean and Brachiaria for the tropics’ granted to the International Centre for Tropical Agriculture (CIAT).
LITERATURE CITED
- Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ. Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiology. 2003;132:2205–2217. doi: 10.1104/pp.103.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y, Hayashi M, Nishimura M. Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant and Cell Physiology. 2008;49:526–539. doi: 10.1093/pcp/pcn027. [DOI] [PubMed] [Google Scholar]
- Basu A, Basu U, Taylor GJ. Induction of microsomal membrane proteins in roots of an aluminum-resistant cultivar of Triticum aestivum L. under conditions of aluminum stress. Plant Physiology. 1994;104:1007–1013. doi: 10.1104/pp.104.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIAT. A planning document 1993–98 and an achieving document 1978–92. 1992. Constraints to and opportunities for improving bean production. Cali, Colombia. [Google Scholar]
- Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature. 1979;279:643–645. doi: 10.1038/279643a0. [DOI] [PubMed] [Google Scholar]
- Cumming JR, Cumming AB, Taylor GJ. Patterns of root respiration associated with the induction of aluminium tolerance in Phaseolus vulgaris L. Journal of Experimental Botany. 1992;43:1075–1081. [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiology. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Ryan PR. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Letters. 2007;581:2255–2262. doi: 10.1016/j.febslet.2007.03.057. [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiology. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete C, Fung RWM, Szabó Z, et al. Up-regulated transcripts in a compatible powdery mildew-grapevine interaction. Plant Physiology and Biochemistry. 2009;47:732–738. doi: 10.1016/j.plaphy.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, et al. An aluminum-activated citrate transporter in barley. Plant and Cell Physiology. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- Guo P, Bai G, Carver B, Li R, Bernardo A, Baum M. Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum tolerance under aluminum stress. Molecular Genetics and Genomics. 2007;277:1–12. doi: 10.1007/s00438-006-0169-x. [DOI] [PubMed] [Google Scholar]
- Hamilton CA, Good AG, Taylor GJ. Induction of vacuolar ATPase and mitochondrial ATP synthase by aluminum in an aluminum-resistant cultivar of wheat. Plant Physiology. 2001;125:2068–2077. doi: 10.1104/pp.125.4.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst WJ, Kollmeier M, Schmohl N, et al. Significance of the root apoplast for aluminium toxicity and resistance of maize. In: Sattelmacher B, Horst WJ, editors. The apoplast of higher plants: compartment of storage, transport and reactions: the significance of the apoplast for mineral nutrition of higher plants. Dordrecht: Springer; 2007. pp. 49–66. [Google Scholar]
- Houde M, Diallo AO. Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics. 2008;9:400. doi: 10.1186/1471-2164-9-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier J. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. European Journal of Biochemistry. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- Kihara T, Ohno T, Koyama H, Sawafuji T, Hara T. Characterization of NADP-isocitrate dehydrogenase expression in a carrot mutant cell line with enhanced citrate excretion. Plant and Soil. 2003;248:145–153. [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant and Cell Physiology. 2000;41:1030–1037. doi: 10.1093/pcp/pcd029. [DOI] [PubMed] [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK. Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Molecular Genetics and Genomics. 2008;279:339–357. doi: 10.1007/s00438-007-0316-z. [DOI] [PubMed] [Google Scholar]
- Larsen P, Cancel J, Rounds M, Ochoa V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta. 2007;225:1447–1458. doi: 10.1007/s00425-006-0452-4. [DOI] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. The Plant Journal. 2005;41:353–363. doi: 10.1111/j.1365-313X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- Lawen A, Ly JD, Lane DJR, Zarschler K, Messina A, Pinto VD. Voltage-dependent anion-selective channel 1 (VDAC1): a mitochondrial protein, rediscovered as a novel enzyme in the plasma membrane. The International Journal of Biochemistry & Cell Biology. 2005;37:277–282. doi: 10.1016/j.biocel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiology. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. The Plant Journal. 2009;57:389–399. doi: 10.1111/j.1365-313X.2008.03696.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma JF, Ryan PR, Delhaize E. Aluminium tolerance in plants and the complexing role of organic acids. Trends in Plant Science. 2001;6:273–278. doi: 10.1016/s1360-1385(01)01961-6. [DOI] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimaraes CT, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nature Genetics. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- Manrique G, Rao IM, Beebe S. Identification of aluminum resistant common bean genotypes using a hydroponic screening method. 2006 Paper presented at the 18th World Congress of Soil Science, Philadelphia, USA, July 9–15. [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. [Google Scholar]
- Mugai EN, Agong SG, Matsumoto H. Aluminium tolerance mechanisms in Phaseolus vulgaris L.: citrate synthase activity and TTC reduction are well correlated with citrate secretion. Soil Science and Plant Nutrition. 2000;46:939–950. [Google Scholar]
- Nie X, Singh RP, Tai GCC. Molecular characterization and expression analysis of 1-aminocyclopropane-1-carboxylate oxidase homologs from potato under abiotic and biotic stresses. Genome. 2002;45:905–913. doi: 10.1139/g02-062. [DOI] [PubMed] [Google Scholar]
- Pellet DM, Grunes DL, Kochian LV. Organic acid exudation as an aluminum-tolerance mechanism in maize (Zea mays L.) Planta. 1995;196:788–795. [Google Scholar]
- Rangel AF, Mobin M, Rao IM, Horst WJ. Proton toxicity interferes with the screening of common bean (Phaseolus vulgaris L.) genotypes for aluminium resistance in nutrient solution. Journal of Plant Nutrition and Soil Science. 2005;168:607–616. [Google Scholar]
- Rangel AF, Rao IM, Horst WJ. Spatial aluminium sensitivity of root apices of two common bean (Phaseolus vulgaris L.) genotypes with contrasting aluminium resistance. Journal of Experimental Botany. 2007;58:3895–3904. doi: 10.1093/jxb/erm241. [DOI] [PubMed] [Google Scholar]
- Rangel AF, Rao IM, Braun H-P, Horst WJ. Aluminum resistance in common bean (Phaseolus vulgaris) involves induction and maintenance of citrate exudation from root apices. Physiologia Plantarum. 2010;138:176–190. doi: 10.1111/j.1399-3054.2009.01303.x. [DOI] [PubMed] [Google Scholar]
- Rao IM. Role of physiology in improving crop adaptation to abiotic stresses in the tropics: the case of common bean and tropical forages. In: Pessarakli M, editor. Handbook of plant and crop physiology. New York, NY: Marcel Dekker; 2001. pp. 583–613. [Google Scholar]
- Rogers EE, Wu X, Stacey G, Nguyen HT. Two MATE proteins play a role in iron efficiency in soybean. Journal of Plant Physiology. 2009;166:1453–1459. doi: 10.1016/j.jplph.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ryan PR, DiTomaso JM, Kochian LV. Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. Journal of Experimental Botany. 1993;44:437–446. [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ. Characterisation of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta. 1995;196:103–110. [Google Scholar]
- Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:527–560. doi: 10.1146/annurev.arplant.52.1.527. [DOI] [PubMed] [Google Scholar]
- Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiology. 2009;149:340–351. doi: 10.1104/pp.108.129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, et al. A wheat gene encoding an aluminum-activated malate transporter. The Plant Journal. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Annual Review of Plant Biology. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- Schulz B, Kolukisaoglu HÜ. Genomics of plant ABC transporters: the alphabet of photosynthetic life forms or just holes in membranes? Federation of European Biochemical Societies Letters. 2006;580:1010–1016. doi: 10.1016/j.febslet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Silva IR, Smyth TJ, Israel DW, Raper CD, Rufty TW. Magnesium ameliorates aluminum rhizotoxicity in soybean by increasing citric acid production and exudation by roots. Plant and Cell Physiology. 2001;42:546–554. doi: 10.1093/pcp/pce067. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Horst WJ. The distal part of the transition zone is the most aluminum-sensitive apical root zone of maize. Plant Physiology. 1998;116:155–163. [Google Scholar]
- Sun P, Tian QY, Zhao MG, et al. Aluminum-induced ethylene production is associated with inhibition of root elongation in Lotus japonicus L. Plant and Cell Physiology. 2007;48:1229–1235. doi: 10.1093/pcp/pcm077. [DOI] [PubMed] [Google Scholar]
- Taylor GJ, Basu A, Basu U, Slaski JJ, Zhang G, Good A. Al-induced, 51-kilodalton, membrane-bound proteins are associated with resistance to Al in a segregating population of wheat. Plant Physiology. 1997;114:363–372. doi: 10.1104/pp.114.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA. Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiology. 2001;127:1836–1844. [PMC free article] [PubMed] [Google Scholar]
- Von Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant and Soil. 1995;171:1–15. [Google Scholar]
- Yang ZM, Yang H, Wang J, Wang YS. Aluminum regulation of citrate metabolism for Al-induced citrate efflux in the roots of Cassia tora L. Plant Science. 2004;166:1589–1594. [Google Scholar]
- Zeller G, Henz SR, Widmer CK, et al. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. The Plant Journal. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.