Abstract
MicroRNA-mediated regulation of gene expression appears to be involved in a variety of cellular processes, including development, differentiation, proliferation, and apoptosis. Mir-146a is thought to be involved in the regulation of the innate immune response, and its expression is increased in tissues associated with chronic inflammation. Among the predicted gene targets for mir-146a, the chemokine CCL8/MCP-2 is a ligand for the CCR5 chemokine receptor and a potent inhibitor of CD4/CCR5-mediated HIV-1 entry and replication. In the present study, we have analyzed changes in the expression of mir-146a in primary human fetal microglial cells upon infection with HIV-1 and found increased expression of mir-146a. We further show that CCL8/MCP-2 is a target for mir-146a in HIV-1 infected microglia, as overexpression of mir-146a prevented HIV-induced secretion of MCP-2 chemokine. The clinical relevance of our findings was evaluated in HIV-encephalitis (HIVE) brain samples in which decreased levels of MCP-2 and increased levels of mir-146a were observed, suggesting a role for mir-146a in the maintenance of HIV-mediated chronic inflammation of the brain.—Rom, S., Rom, I., Passiatore, G., Pacifici, M., Radhakrishnan, S., Del Valle, L., Piña-Oviedo, S., Khalili, K., Eletto, D., Peruzzi, F. CCL8/MCP-2 is a target for mir-146a in HIV-1 infected human microglial cells.
Keywords: microRNA, chronic inflammation, brain
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that function by inhibiting translation of mRNAs (1, 2). They are endogenously encoded single-stranded RNAs of ∼22 nucleotides in length that inhibit protein translation predominantly through imperfect base-pairing with sequences, which are generally located in the 3′ untranslated region (UTR) of mRNA transcripts. Although the role of miRNAs in the control of proliferation, differentiation, and apoptosis has been previously recognized, the importance of these small noncoding RNAs in the immune system is just emerging (3,4,5). For instance, mir-150 controls B-cell differentiation by targeting the c-Myb transcription factor (6), and mir-155 regulates a variety of immune reactions, including production of cytokines by T and B cells and the germinal center B-cell response (7, 8). Expression profile of miRNAs in human monocytes has revealed that miR-146a/b, miR-132, and miR-155 responded to endotoxin treatment with elevated expression (9). Taganov et al. (9) also demonstrated that LPS-mediated induction of mir-146a is NF-κB dependent, which attenuates Toll-like receptor (TLR) signaling by targeting IL-1 receptor associated kinase (IRAK1) and TNF-associated receptor factor 6 (TRAF6). Expression of mir-146a is induced by the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) (10), a 6-transmembrane molecule that mimics cellular tumor necrosis factor receptor (TNFR) family members and contributes to the oncogenic potential of EBV. The primary mechanism by which LMP1 induces expression of mir-146a appears to be NF-κB dependent (10). miRNA-gene target prediction strategy suggested that mir-146a may be part of a negative feedback loop that plays a role in modulating interferon responses (10, 11).
Infection by the human immunodeficiency virus-1 (HIV-1) is often associated with a chronic inflammatory reaction of the brain [HIV-encephalitis (HIVE)], where macrophages/microglial cells are thought to be the major reservoir for the virus (12). Upon viral infection, microglial cells secrete a variety of cytokines and chemokines, including monocytes chemoattractant proteins, MCPs. Chemokine receptors also play a role in infectious diseases, and CCR5 is the primary coreceptor for macrophage tropic (R5) HIV-1 (13, 14). The chemokine MCP-2 shares ∼60% homology with MCP-1, and MCP-3 and has ∼30% identity with MIP-1α, MIP-1β, and RANTES (15,16,17). MCP-2 is an efficient ligand for CCR1, CCR2, and CCR5 and a potent inhibitor of CD4/CCR5-mediated HIV-1 entry and replication (18,19,20). Increased levels of MCP-2 have been previously observed in the supernatant of mixed brain cell cultures infected with HIV-1 (21).
In this study, we have investigated the expression levels of mir-146a in HIV-1-infected primary human fetal microglia and found that its expression increased during the course of infection. We have also identified the CCL8/MCP-2 mRNA as a direct target for mir-146a and have shown that overexpression of mir-146a prior to infection with HIV-1 prevents release of MCP-2 by infected cells. Evaluation of mir-146a and MCP-2 expression in HIVE brain samples also revealed an inverse correlation between levels of mir-146a and expression of its target gene MCP-2, with mir-146a being up-regulated in HIVE and MCP-2 down-regulated. Since MCP-2 chemokine is an efficient ligand for CCR5, our findings demonstrating that mir-146a mediates suppression of MCP-2 may indicate a role for the mir-146a/MCP-2 axis in the modulation of viral spread in the brain.
MATERIALS AND METHODS
Primary human fetal microglia
Fetal tissue specimens were obtained from the Temple University Center for Neurovirology tissue culture core facility in accordance with institutional, state, and U. S. National Institutes of Health (NIH) guidelines. The procedures involved in the preparation of microglial cells at the fetal developmental stage are based on the principle of tissue dissociation, followed by enzymatic digestion with or without a gradient to remove debris, and then purification of microglial cells by preferential adhesion (22). In brief, the tissue specimens obtained were carefully washed in HBSS without phenol red to remove any blood vessels and residual meninges. The tissues were dissociated gently to generate smaller pieces and incubated with TrypleExpress (Invitrogen, Carlsbad, CA, USA) for 30 min. At the end of this incubation time, the dissociating enzyme was inactivated by the addition of 10% FBS. Cold HBSS was added in the proportion of 5:1 (40 ml HBSS+10 ml tissue), and the tissue was gently triturated. After centrifugation at 500 g for 10 min, the tissue was washed another 2 times with HBSS. The tissue was finally resuspended and tritured in complete microglia medium [Dulbecco’s modified Eagle medium (DMEM), 10% FBS, 20 mM/ml l-glutamine, 1% penicillin/streptomycin/neomycin, and 1000 U/ml macrophage colony stimulating factor (MCSF)] using smaller pipettes and, ultimately, a fire-polished Pasteur pipette. The cell suspension was filtered through a 70-μm filter, and the cells were counted. Cells were seeded in T75 flasks at a concentration of 3.5–4 × 106 cells/ml in 25 ml of medium, and incubated undisturbed for 2 wk, with a single change of medium on d 7. Supernatant was collected on d 14, 21, and 28 from the original plating and centrifuged at 500 g for 10 min, and the pellet was resuspended in a small volume of medium. Microglial cells were counted, plated at the desired density, and cultured for another week in complete medium without MCSF. Cells were either maintained in culture for experiments or frozen. Microglial cultures were highly pure, consisting of >98% CD68+ cells.
Two different preparations of microglia were utilized in the present study, and all of the experiments were performed using both microglia preparations, each in triplicate.
Preparation of JRFL HIV-1 virus stock
U937 (Human leukemic monocyte lymphoma) cell line was used to make a stock of HIV, strain JRFL. Approximately 5 × 106 cells were infected with HIV (4 μg p24) in 5 ml of serum-free OptiMEM for 5 h. At the end of adsorption, cells were pelleted, virus inoculum was removed, and cells were resuspended in RPMI containing 10% FBS. One-third of the cell culture medium was changed every 2–3 d until a majority of the cells exhibited cytopathic effect in the form of syncitia (usually 10 d postinfection). After 10 d, cells were pelleted, and supernatant was divided into 1-ml aliquots and stored at −80°C. The concentration of p24 in the new stock was determined with an enzyme-linked immunosorbent assay (ELISA) kit (PerkinElmer, Waltham, MA, USA), according to the manufacturer’s protocol.
Quantitative RT-PCR
RNA was extracted using the miRVana miRNA extraction kit (Ambion, Austin, TX, USA). Real-time RT-PCR was performed using the mirVana qRT-PCR miRNA detection kit (Ambion) per the manufacturer’s protocol, as described previously (23). PCR primer pairs for reverse transcription and detection of mature miRs were purchased from Ambion (hsa-mir-146a and control U6). In general, quantitative real-time RT-PCR (qRT-PCR) on primary human fetal microglia was performed on 2 independent experiments using 25 ng of template. For each sample, qRT-PCR was performed in quadruplicate. Each sample was normalized for the corresponding values of U6 control. Relative quantification of the selected miRNAs was calculated using the Roche analysis software (based on the 2nd derivative maximum; Roche, Indianapolis, IN, USA).
qRT-PCR to detect levels of mir-146a in brain samples was performed essentially as described above. RNA was extracted from frozen brain tissue samples of 4 control brains, 3 HIVE brains, and 2 HIV brains without brain pathology, using the miRVana miRNA extraction kit, and qRT-PCR was performed using the mirVana qRT-PCR miRNA detection kit following the manufacturer’s instructions. qRT-PCR was performed in quadruplicate using 25 ng of template. Analysis of results was performed as described above.
miRNA functional analysis
The sequence of the hsa-mir-146a stem loop was retrieved using miRBase database no. MI0000477: CCGAUGUGUAUCCUCAGCUUUGAGAACUGAAUUCCAUGGGUUGUGUCAGUGUCAGACCUCUGAAAUUCAGUUCUUCAGCUGGGAUAUCUCUGUCAUCGU.
For cloning into BlockIt (Invitrogen), the following oligos were used: forward, 5′-TGCTGTGAGAACTGAATTCCATGGGTTGTTTTGGCCACTGACAACCCATGATTCAGTTCTCA, and reverse, CCTGTGAGAACTGAATCATGGGTTGTCAGTGGCCAAAACAACCCATGGAATTCAGTTCTCAC. The oligos were annealed and cloned as double strands into the expression vector following the manufacturer’s protocols.
For the cloning of the perfect match sequence into PsiCheck-2 vector (Promega, Madison, WI, USA), the PCR primers were based on the mature sequence of hsa-miR-146a but in the antisense orientation. The primers were as follows: forward, 5′-CCGCTCGAGAACCCATGGAATTCAGTTCTCAAGATCTGCGGCCGCTAAACTAT-3′; and reverse, 5′-ATAGTTTAGCGGCGCAGATCTTGAGAACTGAATTCCATGGGTTCTCGAGCGG-3′. The MCP-2 3′UTR sequence was PCR amplified from genomic DNA from human microglial cells and cloned downstream to the Renilla sequence in the PsiCheck2 reporter vector. The primers were as follows: forward, 5′-CCGCTCGAGAGTCAGAGCTTGAAG, and reverse: 5′-ATAAGAATGCGGCCGCCATAGTTATATACAGTAACATC, containing XhoI and NotI restriction sites, respectively (underscored). The MCP-2 3′UTR mutated in the mir-146a seeding sequence was generated by site directed mutagenesis (Stratagene, La Jolla, CA, USA) using the psiCHECK2/MCP-2 3′UTR as a template. The oligonucleotides for the mutagenesis were as follows: forward, 5′-ATTCTTTGGCTAAGTCAGGAGC (mutated bases in the mir-146a seeding sequence are underscored). The reverse primer was complementary to the forward.
Dual luciferase assay
For miR target validation, 293T cells were plated at a concentration of 8 × 104 cells/well in a 12-well plate. The next day, a total amount of 0.6 μg/well of DNA was transfected using Fugene 6 (Invitrogen) at the DNA:Fugene 6 ratio of 1:3. pcDNA3 was added to keep the total amount of DNA constant. Samples were harvested 48 h post-transfection and subjected to the dual luciferase assay system (Promega) following the manufacturer’s instructions using Femtomaster FB12 chemiluminometer (Zylex Corp., Coeur D Alene, ID, USA). Relative units represent the ratio between Renilla values and the luciferase internal control. The experiments were performed in duplicate and repeated ≥3 times.
Immunohistochemistry
Six archival samples of formalin-fixed, paraffin-embedded brain tissue from patients with HIVE and 6 samples from age-matched control patients who died of nonneurological conditions and had no confirmed brain pathology were collected from the NIH-funded HIV National Tissue Consortium at the Manhattan Brain Bank (Mount Sinai Medical School, New York, NY, USA). In all samples, the frontal lobe was selected for analysis since it constitutes the most frequently affected region in HIVE. For immunohistochemistry, 4-μm-thick sections were cut using a microtome and placed on charged glass slides. Immunohistochemistry was performed according to the manufacturer’s instructions (Vector Laboratories, Burlingame, CA, USA). Our protocol includes deparaffination, rehydration in alcohol up to water, antigen retrieval in citrate buffer at pH 6.0, endogenous peroxidase quenching, and blocking in normal horse serum for 2 h at room temperature. The primary antibody against MCP-2 was a goat polyclonal (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and was incubated overnight in a humidified chamber. After rinsing with PBS, secondary biotinylated antibody was incubated for 1 h (1:200 dilution), followed by avidin-biotin peroxidase complex incubation (ABC Elite kit, Vector Laboratories) and developing with diaminobenzidine (DAB, Boehringer Ingelheim, Ingelheim, Germany), counterstained with hematoxylin, and coverslipped.
In situ hybridization
For in situ hybridization, we utilized miRCURY 3′-DIG-labeled LNA-miRNAs (mir-146a and mir-128a; Exiqon, Woburn, MA, USA) detection probes. A miRCURY detection probe for the brain-enriched mir-128a and scrambled miRNA were used as positive and negative controls, respectively. Samples from 4 frozen brains of patients with HIVE and 4 brains of age-matched control patients who died of nonneurological diseases were collected from the pathology archives of the Manhattan Brain Bank at Mount Sinai Medical School. Sections of 10 μm thickness from the frontal lobe were cut from using a cryostat and mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA, USA). Sections were fixed in 70% alcohol for 2 min. Slides were washed 3 times with PBS, acetylated for 10 min, and rinsed again in PBS. The probe (20 ng DIG-mir-146a or DIG-mir-128a) was diluted in hybridization solution (50% deionized formamide; 0.3 M NaCl; 20 mM Tris-HCl, pH 8.0; 5 mM EDTA; 10 mM NaPO4, pH 8.0; 10% dextran sulfate; 1× Denhardt’s; and 0.5 mg/ml yeast RNA). The hybridization mixture was heated at 65°C for 5 min to linearize the probe and chilled on ice. Fifty to 100 μl of hybridization solution was added to each slide; sections were covered with RNase-free plastic coverslips and incubated overnight at a temperature 20–22°C below the temperature of melting (Tm) of the miRCURY LNA detection probe in a chamber humidified with 50% formamide and 1× SSC (in soaked KimWipes; Kimberly-Clark, Irving, TX, USA). Slides were washed with 5× SSC at room temperature and 2 times for 30 min each at a temperature 20–22°C below Tm of the miRCURY LNA detection probe in a solution containing 50% formamide, 0.1% Tween-20 and 1× SSC. Slides were then washed for 15 min in 0.2× SSC at room temperature, and an additional 15 min in PBS at room temperature. For antibody detection, the protocol was the following: slides were incubated for 1 h at room temperature in blocking solution (0.5% blocking powder, 10% heat-inactivated goat serum, 0.1% Tween-20, and 1× PBS). Slides were incubated for 3 h at room temperature in blocking solution preincubated for 1 h with AP-conjugated anti-DIG Fab fragment (1:1500 in blocking solution; Roche). Slides were washed 2 times for 30 min in 0.1% Tween-20 in PBS followed by 2 times for 20 min in PBS. For detection, slides were incubated with 250 μl of BM Purple AP Substrate (Roche) on slides overnight in the dark at room temperature. The reaction was stopped by placing the slides into stop solution (PBS, pH 5.5; and 1 mM EDTA). Slides were mounted in a water-based medium (VectaShield; Vector Laboratories).
ELISA assay
ELISA to detect CCL8/MCP-2 in the medium of HIV-1-infected cells was performed following standard procedures. In brief, 96-well plates were coated with supernatant from HIV-1-infected and control cells and left at 4°C overnight. Wells were washed with PBST 6 times and blocked with 10% milk in PBST for 2 h at 37°C. Anti-MCP-2 (C-17; Santa Cruz Biotechnology) was added to the wells at a dilution of 1:200 in PBST and left overnight at 4°C. Next day, wells were washed 6 times with PBST and incubated with anti-goat HRP secondary antibody for 2 h at 37°C. Wells were washed 6 times with PBST and incubated with 150 μl of 1× TMB substrate solution (eBioscience Inc, San Diego, CA, USA). Samples were analyzed using an ELISA microplate reader at 630 nm. As a positive control, wells were coated with CCL8/MCP-2 peptide (C-17; Santa Cruz Biotechnology) at concentrations of 0.002, 0.2, 0.6, 1, and 2 μg.
Statistics
Results were analyzed by an unpaired, 2- or 1-sided Student’s t test. Values of P ≤ 0.05 were considered statistically significant.
RESULTS
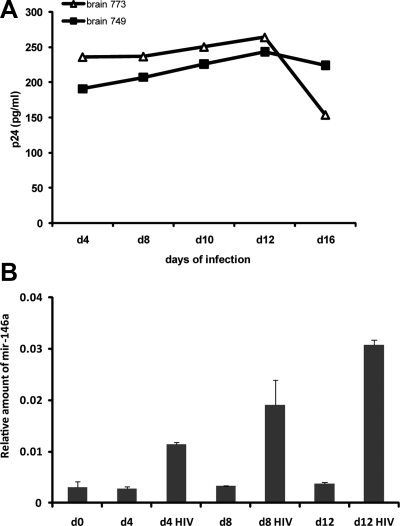
Two different preparations of primary human fetal microglial cells were infected with HIV-1, and levels of p24 were determined by ELISA at 4, 8, 10, 12, and 16 d postinfection as an indicator of viral replication (Fig. 1A). Next, we performed qRT-PCR to determine changes in the expression levels of mir-146a in HIV-1-infected primary human fetal microglial cells compared to uninfected cells. RNA samples from HIV-infected and control cells were collected at 0, 4-, 8-, and 12-d time points. qRT-PCR was performed for each sample in quadruplicate, and relative quantification of mir-146a, normalized for U6 levels, was calculated by the Roche 480 LyteCycler software. Results in Fig. 1B represent the average expression of mir-146a in the 2 experimental samples and controls. Quantitatively, an increase in mir-146a levels of ∼4-, 6-, and 8-fold was observed in HIV-1-infected microglia at 4-, 8-, and 12-d time points, respectively, when compared to controls. In addition, we monitored cell survival on HIV-1 infection during the course of the experiment, and no statistically significant difference in the number of viable cells was observed between HIV-1-infected and control cultures. Trypan blue exclusion test performed at each time point during infection demonstrated an average of 83 ± 2 and 84 ± 5% live cells in control and HIV-1-infected cells, respectively. These results show HIV-1 mediated induction of mir-146a expression in primary human fetal microglial cells.
Figure 1.
Increased levels of mir-146a in HIV-1-infected microglial cells. A) ELISA to detect p24 in medium collected at indicated time points from 2 different preparations of HIV-1-infected primary human fetal microglia (brains 7739 and 7490). B) qRT-PCR to detect levels of mature mir-146a in HIV-1-infected and control microglial cells at 0, 4, 8, and 12 d postinfection. qRT-PCR was performed on 25 ng of template, each in quadruplicate. Each sample was normalized for corresponding values of U6 control. Relative amount of mir-146a was calculated by the Roche LightCycler software. Graph represents average values obtained from microglial cell cultures isolated from brain samples 7739 and 7490.
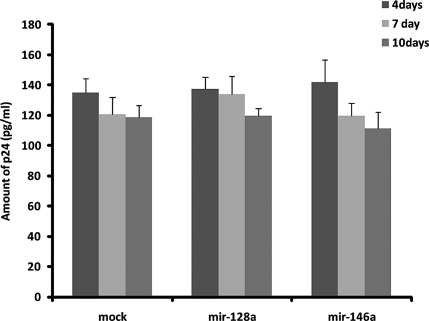
Initially, we hypothesized that mir-146a up-regulation by HIV-1 might function to promote the latency of the virus in microglial cells, which may involve a decrease in viral replication. Accordingly, we cloned a genomic sequence corresponding to the human mir-146a into the pBlockIt miR expression plasmid to generate Blo/146a (see Materials ad Methods). Expression of mature mir-146a in 293T cells was verified by qRT-PCR at 24 and 48 h post-transfection (data not shown). To test a possible role of mir-146a in viral replication, microglial cells were transfected with control empty vector, a nonrelevant miRNA (mir-128a) or mir-146a, 72 h prior to infection with HIV-1, or with mock control medium. HIV-1 p24 protein levels were determined at 4, 7, or 10 d following the infection by p24-specific ELISA. Results in Fig. 2 show no statistically significant differences between the samples, suggesting that the activity of mir-146a does not interfere with viral replication in this cellular system.
Figure 2.
mir-146a does not affect viral replication. ELISA to determine amounts of p24 protein in medium of HIV-1-infected human fetal microglia. Cells were transfected with mir-146a or nonrelevant miRNA, mir-128a, 72 h prior to infection with HIV-1 or control mock (see Materials and Methods). Supernatant was collected after 4, 7, and 10 d of infection. Each time point was performed in triplicate; whole experiment was repeated twice.
Evaluation of CCL8/MCP-2 protein levels during HIV-1 infection of human fetal primary microglial cells
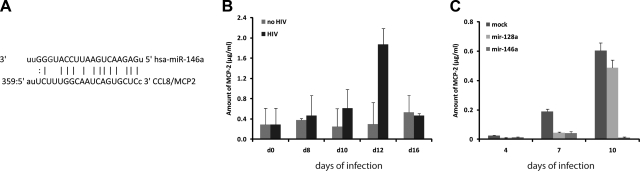
Since mir-146a appears to play a role in the innate immune response (24), we next asked whether it could regulate expression of cytokines and chemokines that are secreted by HIV-1-infected cells. We analyzed predicted gene targets for mir-146a and found that CCL8/MCP-2 mRNA has a putative binding site for mir-146a in its 3′UTR. Figure 3A shows the sequence alignment between mir-146a and MCP-2 mRNA according to miRNApath database (http://lgmb.fmrp.usp.br/mirnapath/tools.php).
Figure 3.
mir-146a inhibits secretion of CCL8/MCP-2 by HIV-1-infected microglial cells. A) Diagram showing putative mir-146a binding sites in the 3′UTR-MCP-2 mRNA sequence. B) Graph of a representative ELISA showing amount (μg/ml) of secreted CCL8/MCP-2 chemokine in medium of HIV-1-infected cells and control mock-infected cells. Experiment was repeated 3 times, each in duplicate. C) ELISA to monitor amount of secreted MCP-2 on supernatant of primary human fetal microglial cells transfected with mir-146a, a nonrelevant miRNA (mir-128a), or control empty vector (mock), followed by infection with HIV-1. Experiment was repeated 3 times, each in quadruplicate.
Primary human fetal microglial cells were infected with HIV-1, and supernatants from HIV-infected and control cells were collected every second day starting from d 2 for up to d 16 following infection. CCL8/MCP-2 levels in the medium of HIV-1-infected cells were detected by ELISA following standard procedures. As a positive control, wells were coated with CCL8/MCP-2 peptide at the concentrations ranging from 0.002 to 2 μg. Results in Fig. 3B show a gradual increase in the amount of MCP-2 chemokine secreted by microglial cells during the first 12 d after infection. Interestingly, a reduction in the release of MCP-2 occurred between d 12 and 16, which may reflect the inhibitory action of HIV-induced mir-146a.
In the next series of experiments, we determined the effect of mir-146a on the expression of CCL8/MCP-2 by measuring levels of MCP-2 protein in the medium of microglial cells overexpressing mir-146a or controls after HIV-1 infection (Fig. 3C). As a negative control, we utilized a plasmid containing a miRNA (mir-128a) that is not predicted to target CCL8/MCP-2 3′UTR sequence. Primary human fetal microglial cells were transfected with BlockIt/146a or BlockIt/128a following standard protocols. Seventy-two hours post-transfection, cells were infected with HIV-1. Untransfected, but HIV-1 infected, cells were used as a negative control (mock). MCP-2 ELISA was performed on supernatants obtained at 0, 4, 7, and 10 d after infection of cells with HIV-1. Untransfected, HIV-1-infected microglial cells release CCL8/MCP-2 in the supernatant with a maximal amount secreted on d 10 postinfection. Similar results were obtained when the cells were transfected with control miRNA (mir-128a) prior to infection with HIV-1. Notably, secretion of MCP-2 was almost completely suppressed in cells overexpressing mir-146a.
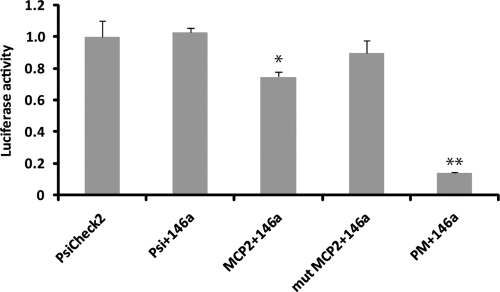
To test the possibility that expression of MCP-2 was post-transcriptionally regulated by mir-146a, we performed a functional assay using MCP-2 reporter system (23). The reporter construct contained the full-length MCP-2 3′UTR fused to the Renilla gene in the PsiCheck2 vector, which also contained the firefly luciferase gene for normalization.
For Renilla luciferase assays, the reporter vectors were coexpressed together with mir-146a in 293T cells and assayed 48 h post-transfection. As shown in Fig. 4, the ratio Renilla/luciferase (R/L) in the control vectors (PsiCheck2 empty vector, Psicheck2 3′UTR MCP-2, and Psicheck2 mut 3′UTR MCP-2) was scaled to 1 (100%). No significant difference between the ratios was observed when PsiCheck2 was cotransfected with mir-146a (psi+146a). Instead, coexpression of 3′UTR MCP-2 with mir-146a (MCP-2+146a) resulted in a reproducible 25% reduction of R/L values (P<0.05). The control reporter plasmid carrying the MCP-2 3′UTR mutated in the mir-146a seeding site was not affected by the addition of mir-146a (mutMCP-2+146a), confirming the specificity of the mir-146a activity. As a positive control, the R/L ratio relative to the reporter plasmid carrying the mir-146a perfect match sequence (PM) resulted in a strong (∼90%) inhibition, confirming that mir-146a is overexpressed and functional in this cell system (P<0.001).
Figure 4.
Inhibition of MCP-2 3′UTR by mir-146a. Luciferase assay on 293T cells transfected with empty vector alone (PsiCheck2), or cotransfected with mir-146a and MCP-2 3′UTR, with mir-146a and the 3′UTR-mutated sequence (mut MCP-2), or with the control mir-146a perfect matching sequence (PM). Luciferase and Renilla values were determined 48 h post-transfection. Relative units represent ratio between Renilla values and luciferase internal control. Experiment was repeated twice, each in triplicate. *P < 0.05; **P < 0.001.
Evidence that MCP-2 inhibits viral replication in microglial cells
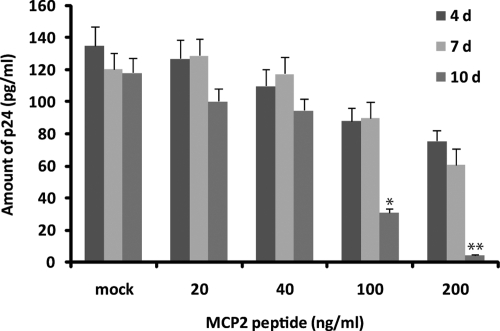
CCL8/MCP-2 chemokine is known to bind CCR5 receptor with high affinity and to inhibit HIV-1 replication (18, 19). We tested this possibility in our cellular system, and results are shown in Fig. 5. Human fetal microglial cells were treated with increasing concentrations of MCP-2 peptide (20, 40, 100, and 200 ng/ml) prior to infection with HIV-1. Amounts of p24 protein were determined by ELISA on the supernatant collected at d 4, 7, and 10 after infection. Note that production of p24 decreased with increasing concentration of MCP-2 protein confirming the important role of MCP-2 in viral replication (18, 19).
Figure 5.
Effects of MCP-2 on HIV-1 infection. ELISA showing amounts of p24 in medium of HIV-1-infected microglial cells in absence or presence of MCP-2 peptide at 4, 7, and 10 d postinfection. Experiment was repeated twice, each in duplicate. *P < 0.05; **P < 0.001.
Relevance to HIVE
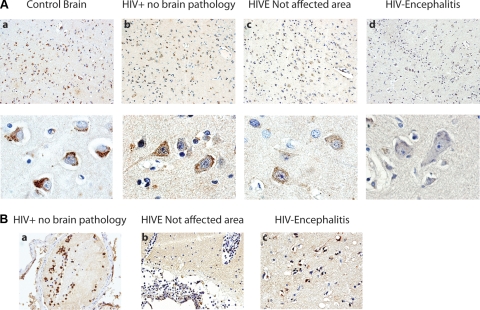
Our results indicate a possible role for mir-146a in modulating expression of MCP-2 during the course of HIV infection in cultured microglial cells. The next series of experiments was aimed at investigating the effect of such modulation in clinical samples of HIVE. Immunohistochemistry for MCP-2 was performed in cortical sections from archival brains of 3 patients with HIVE and compared to sections from 3 control patients and 3 HIV+ patients with no brain pathology. Figure 6Aa shows representative images from the control brain, where a robust immunolabeling was found in virtually all neurons, compared with the HIV+ no-pathology brain, in which the expression of MCP-2 was slightly reduced (Fig. 6Ab), and the HIVE brain, where MCP-2 was significantly reduced (Fig. 6Ac, d). Even within the same HIVE section, the expression of MCP-2 was dramatically reduced from nonaffected areas (Fig. 6Ac), to areas of severe perivascular inflammatory cuffs and microglial nodules, in which the expression of MCP-2 was reduced to a couple of weak speckles (Fig. 6Ad).
Figure 6.
Detection of MCP-2 protein in HIVE and control brains. A) Immunohistochemical analysis of MCP-2 in cortical sections of a control patient, an HIV+ patient with no brain pathology, and an HIVE patient revealed a gradual reduction in levels of MCP-2 from the control patient, in which MCP-2 in robustly labeled (a), to lesser expression in the HIV+ patient (b), to a significant reduction in HIVE (c, d). MCP-2 is also noticeably decreased within the same HIVE sample, from nonaffected areas (c) to areas of severe inflammation, where MCP-2 is almost undetectable (d). Original view: ×200 (top panels); ×1000 (bottom panels). B) As expected, MCP-2 is robustly expressed in leukocytes inside a subarachnoid blood vessel in an HIV+ patient with no brain pathology (a); in HIVE, leukocytes show a dramatic reduction in MCP-2 (b). Microglial nodules in HIVE demonstrate robust cytoplasmic expression of MCP-2 (c). Original view: ×400.
As expected, expression of MCP-2 was intense in leukocytes found inside subarachnoid blood vessels in the control patient (data not shown), and the HIV+ patient with no brain pathology (Fig. 6Ba). However, the levels of MCP-2 were significantly decreased in leukocytes from the HIVE patient (Fig. 6Bb). Although the general pattern of MCP-2 seemed to decrease in HIVE, it is interesting that parenchymal microglial nodules demonstrated a robust cytoplasmic immunolabeling for MCP-2 in HIVE (Fig. 6Bc).
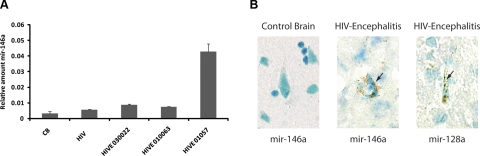
We have evaluated expression of mir-146a in HIVE and control brains by qRT-PCR. Archived frozen control brain and HIVE samples were obtained from the frontal lobe, as it represents the most frequently affected region of the brain, which results in the clinicopathological term of AIDS-dementia complex. RNA was extracted from brain tissues of three HIVE patients (HIVE 010063, 01057, and 030022), 4 control patients, and 2 HIV patients without encephalitis or any other pathology. Results in Fig. 7A show relative amounts of mir-146a in all the samples after normalization with levels of U6 RNA. Comparable levels of mir-146a were observed in the 4 control brains; their average is represented as CB in Fig. 7A. Levels of mir-146a were slightly higher in the HIV+ tissue without brain pathology. A 2-fold increase in mir-146a expression was detected in two HIVE samples, while the third (HIVE 010157) strikingly showed ∼12-fold increase in mir-146a expression.
Figure 7.
mir-146a is up-regulated in HIVE brain samples. A) qRT-PCR of mir-146a on RNA samples obtained from 4 HIVE patients (HIVE 030022, 010063, and 01057), 4 control brains (CB), and 2 HIV patients without brain pathology (HIV). qRT-PCR of mir-146a was performed in quadruplicate using 25 ng of RNA. B) Detection of hsa-mir-146a in HIVE 010063 and control CB 528B archived frozen samples. In situ hybridization was performed with miRCURY-LNA-DIG mir-146a (and control mir-128a) detection probes. Arrows indicate positive signal in cytoplasm of neurons. Original view: ×1000.
Finally, expression of mir-146a was monitored in the same samples used to monitor expression of mir-146a, 4 archived frozen HIVE brain samples and 4 control brain samples, by in situ hybridization. Samples were processed and subjected to hybridization with LNA miRNA probes (see Materials and Methods); representative results are shown in Fig. 7B. Images indicate undetectable levels of mir-146a in control non-HIV brain (Fig. 7B, CB 528B; left panel) and its abundance in the HIVE sample (HIVE 10157; middle panel), which was also stained with the neuronal mir-128a used as a positive control. Hybridization with a scrambled probe was negative in both control and HIVE samples (not shown). Altogether, these data support a correlation between mir-146a expression and down-regulation of MCP2 chemokine.
DISCUSSION
Similar to other neurodegenerative disorders, uncontrolled inflammation in the brain of HIV-1-infected individuals may account for the chronic progression of neuroAIDS (25). Therefore, understanding the mechanisms that trigger and/or sustain uncontrolled inflammation might be of critical importance for the development of new therapeutic modalities to prevent neuronal degeneration. Increasing evidence has accumulated that miRNAs function as key regulators in a wide variety of biological processes, including proliferation, differentiation, cell fate determination, apoptosis, and signal transduction, as well as organ development (2, 26,27,28,29,30,31,32). As a consequence, abnormal expression of miRNAs appears to be a common feature of many human diseases, ranging from muscular (26) and cardiovascular disorders (33, 34), to cancer (35, 36); developmental abnormalities; psychiatric disorders, such as schizophrenia (37); and most recently inflammatory diseases (38). For instance, expression of several miRNAs has been shown to be modulated (elevated or repressed) in activated T cells in vitro (39). In addition, mir-146a is thought to be involved in innate immunity by regulating, together with mir-155, the acute immune response after pathogen recognition by TLRs (9, 24, 40).
Here, we present evidence showing that expression of mir-146a is increased in primary human fetal microglia infected with HIV-1 (Fig. 1). This is in agreement with previous findings showing increased expression of mir-146a in T cells following proinflammatory stimuli (9), including viral infection (10, 41). Despite several studies demonstrating induction of mir-146a expression following inflammatory stimuli, there is still little information regarding its function (24). Because mir-146a expression levels increase during infection until at least d 12, we initially hypothesized that mir-146a could interfere with viral replication in the late stage of infection. However, mir-146a is not predicted to target any of the viral genes (42), and levels of p24 did not significantly change in cells that overexpress mir-146a (Fig. 2). Although we cannot exclude an indirect effect of mir-146a on viral proteins other than p24, we choose to study direct gene targets. Specifically, we have investigated the role of mir-146a in modulating the release of proinflammatory chemokines and found that MCP-2 is a target for mir-146a in HIV-1 infected primary human fetal microglia (Figs. 234). Activation of macrophages/microglia is thought to play a key role in development and progression of neuroAIDS (reviewed in ref. 43). Among the factors secreted by HIV-1-infected microglia/macrophages, MCP-1 is one of the most investigated chemokines and is thought to play a central role in the pathogenesis of HIV-1-associated dementia (44,45,46). Another member of the MCP family of chemokines, MCP-2, has been shown to inhibit CD4/CCR5-mediated HIV-1 entry and replication (18,19,20). Our data also show the inhibitory effect of MCP-2 on HIV-1 entry/replication, with the maximal effect at the highest concentration of MCP-2 at d 10 postinfection (Fig. 5). While we cannot explain the reason for the late effect of MCP-2, we could speculate that the multiplicity of infection (MOI) (47) may play a role; perhaps a highest MOI would result in earlier inhibition of p24 production by MCP-2. Nevertheless, our data are in line with the work of Yang et al. (19), which shows maximal inhibition of HIV-1 replication by MCP-2 at d 10 postinfection.
According to our model, acute infection of microglia with HIV-1 would trigger the production and secretion of MCP-2 chemokine (Fig. 3B). Interestingly, the rapid decline of MCP-2 levels after the peak at 12 d is coincident with the up-regulation of mir-146a expression. Transfection of mir-146a prior to infection with HIV-1 prevented the expression of MCP-2 (Fig. 3C) in microglial cells, further emphasizing a function for this particular miRNA as a negative regulator of MCP-2 chemokine. When tested in a miRNA functional assay, the 3′UTR of MCP-2 mRNA was inhibited by >20% by the action of mir-146a (Fig. 4), and a mutation in the mir-146a seeding sequence of the MCP-2 3′UTR abolished the inhibitory activity of mir-146a. Perhaps the up-regulation of mir-146a at a certain point during infection might be a feedback mechanism to rapidly decrease the otherwise deleterious effects of an overproduction of MCP-2. This mechanism may work during an acute inflammatory response, where expression of mir-146a is negatively regulated later during infection. One such mechanism can involve the activity of NF-κB, which is up-regulated during viral infection and is an inducer of mir-146a promoter (9). Indeed, NF-κB was found to be activated in microglia and macrophages during HIV-1-encephalitis (48), and we found increased expression of mir-146a in HIVE brain samples (Fig. 7), emphasizing a role for the NF-κB/mir-146a pathway in HIVE. Lukiw et al. (49) have provided evidence for a NF-κB-sensitive mir-146a-mediated inflammatory pathway in Alzheimer’s disease and in stressed human brain cells.
There are a limited number of reports in which expression of MCP-2 in normal or diseased brains has been investigated. McManus et al. (50) found prominent immunoreactivity for MCP-2 in the center of multiple sclerosis (MS) lesions, with reactivity diminishing at the lesion edges. In our study, immunohistochemical analysis of HIVE and control brain samples revealed a substantial presence of MCP-2 in the control brain, which was strongly attenuated in HIVE, particularly in areas of inflammation (Fig. 6). Although interesting, we can only speculate on the reason why MCP-2 immunolabeling was stronger in microglial nodules of HIVE brain tissues. This could be a result of either the presence of cells that actively produce MCP-2 or it could be due to the presence of factors that increase the stability of this chemokine.
We have also noticed an intense immunolabeling for MCP-2 in neurons of normal brain. Although there are few studies supporting either the presence or the function of this chemokine in neurons, the MCP-2 main receptor, CCR2, is expressed in neurons (51, 52). Alternatively, as also proposed by Coughlan et al. (51), neurons themselves may produce chemokines, which may affect cellular function within the CNS.
In summary, we have identified a mechanism implicated in the negative regulation of inflammation in HIV-1 infected macrophages/microglia. It involves HIV-mediated increased expression of mir-146a, which, in turn, inhibits the expression of the proinflammatory chemokine MCP-2. Our data in cultured cells and in clinical samples support a role for mir-146 in the dynamic modulation of inflammatory processes in neuronal cells.
Acknowledgments
The authors thank Martyn White for editorial assistance. The work was supported by NIH grants MH071162 and MH079751, awarded to F.P. The parts of this work related to HIV-1 infection and immunohistochemistry were partially supported by grant P01 NS30916, awarded to L.D.V., S.A., and K.K. The authors thank Susan Morgello (Director of the Manhattan Brain Bank at Mt. Sinai School of Medicine, New York, NY, USA) for providing HIVE clinical samples.
References
- Gebauer F, Hentze M W. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Bagasra O, Prilliman K R. RNA interference: the molecular immune system. J Mol Histol. 2004;35:545–553. doi: 10.1007/s10735-004-2192-8. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado D P, Galler G, Thai T H, Patterson H C, Wang J, Rajewsky N, Bender T P, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren M V, Couttet P, Soond D R, van Dongen S, Grocock R J, Das P P, Miska E A, Vetrie D, Okkenhaug K, Enright A J, Dougan G, Turner M, Bradley A. Requirement of bic/ microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T H, Calado D P, Casola S, Ansel K M, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok J L, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Taganov K D, Boldin M P, Chang K J, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J E, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer B C, Flemington E K. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Pina-Oviedo S. HIV disorders of the brain: pathology and pathogenesis. Front Biosci. 2006;11:718–732. doi: 10.2741/1830. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Decock B, Conings R, Lenaerts J P, Billiau A, Van Damme J. Identification of the monocyte chemotactic protein from human osteosarcoma cells and monocytes: detection of a novel N-terminally processed form. Biochem Biophys Res Commun. 1990;167:904–909. doi: 10.1016/0006-291x(90)90609-q. [DOI] [PubMed] [Google Scholar]
- Proost P, Wuyts A, Van Damme J. Human monocyte chemotactic proteins-2 and -3: structural and functional comparison with MCP-1. J Leukoc Biol. 1996;59:67–74. doi: 10.1002/jlb.59.1.67. [DOI] [PubMed] [Google Scholar]
- Van Damme J, Proost P, Lenaerts J P, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, Howard O M, Turpin J A, Grimm M C, Ueda H, Gray P W, Raport C J, Oppenheim J J, Wang J M. Monocyte chemotactic protein-2 activates CCR5 and blocks CD4/CCR5-mediated HIV-1 entry/replication. J Biol Chem. 1998;273:4289–4292. doi: 10.1074/jbc.273.8.4289. [DOI] [PubMed] [Google Scholar]
- Yang O O, Garcia-Zepeda E A, Walker B D, Luster A D. Monocyte chemoattractant protein-2 (CC chemokine ligand 8) inhibits replication of human immunodeficiency virus type 1 via CC chemokine receptor 5. J Infect Dis. 2002;185:1174–1178. doi: 10.1086/339678. [DOI] [PubMed] [Google Scholar]
- Gong X, Gong W, Kuhns D B, Ben-Baruch A, Howard O M, Wang J M. Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J Biol Chem. 1997;272:11682–11685. doi: 10.1074/jbc.272.18.11682. [DOI] [PubMed] [Google Scholar]
- Wang J, Gabuzda D. Reconstitution of human immunodeficiency virus-induced neurodegeneration using isolated populations of human neurons, astrocytes, and microglia and neuroprotection mediated by insulin-like growth factors. J Neurovirol. 2006;12:472–491. doi: 10.1080/13550280601039659. [DOI] [PubMed] [Google Scholar]
- Borgmann K, Gendelman H E, Ghorpade A. Isolation and HIV-1 infection of primary human microglia from fetal and adult tissue. Methods Mol Biol. 2005;304:49–70. doi: 10.1385/1-59259-907-9:049. [DOI] [PubMed] [Google Scholar]
- Eletto D, Russo G, Passiatore G, Del Valle L, Giordano A, Khalili K, Gualco E, Peruzzi F. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008;216:764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A E, Perry M M, Moschos S A, Larner-Svensson H M, Lindsay M A. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36:1211–1215. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- Gao H M, Hong J S. Why neurodegenerative diseases are progressive: uncontrolled inflammation drives disease progression. Trends Immunol. 2008;29:357–365. doi: 10.1016/j.it.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato A A, Lidov H G, Kang P B, North K N, Mitrani-Rosenbaum S, Flanigan K M, Neely L A, Whitney D, Beggs A H, Kohane I S, Kunkel L M. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon G J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Huppi K, Volfovsky N, Mackiewicz M, Runfola T, Jones T L, Martin S E, Stephens R, Caplen N J. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17:65–73. doi: 10.1016/j.semcancer.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X S, Liu C M, Liu D P, Liang C C. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- Kloosterman W P, Plasterk R H. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska E A, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert B L, Mak R H, Ferrando A A, Downing J R, Jacks T, Horvitz H R, Golub T R. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Miska E A. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang M L, Segnalini P, Gu Y, Dalton N D, Elia L, Latronico M V, Hoydal M, Autore C, Russo M A, Dorn G W, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson K L, Croce C M, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kong S W, Lu J, Bisping E, Zhang H, Allen P D, Golub T R, Pieske B, Pu W T. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- Calin G A, Croce C M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Croce C M, Calin G A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Hansen T, Olsen L, Lindow M, Jakobsen K D, Ullum H, Jonsson E, Andreassen O A, Djurovic S, Melle I, Agartz I, Hall H, Timm S, Wang A G, Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Wei T, Janson P C, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B S, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale S T, Sakaguchi S, Livesey F J, Fisher A G, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R M, Taganov K D, Boldin M P, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motsch N, Pfuhl T, Mrazek J, Barth S, Grasser F A. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari S K. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- D'Aversa T G, Eugenin E A, Berman J W. NeuroAIDS: contributions of the human immunodeficiency virus-1 proteins Tat and gp120 as well as CD40 to microglial activation. J Neurosci Res. 2005;81:436–446. doi: 10.1002/jnr.20486. [DOI] [PubMed] [Google Scholar]
- Bell M D, Taub D D, Perry V H. Overriding the brain’s intrinsic resistance to leukocyte recruitment with intraparenchymal injections of recombinant chemokines. Neuroscience. 1996;74:283–292. doi: 10.1016/0306-4522(96)00083-8. [DOI] [PubMed] [Google Scholar]
- Gu L, Tseng S C, Rollins B J. Monocyte chemoattractant protein-1. Chem Immunol. 1999;72:7–29. doi: 10.1159/000058723. [DOI] [PubMed] [Google Scholar]
- Zink M C, Coleman G D, Mankowski J L, Adams R J, Tarwater P M, Fox K, Clements J E. Increased macrophage chemoattractant protein-1 in cerebrospinal fluid precedes and predicts simian immunodeficiency virus encephalitis. J Infect Dis. 2001;184:1015–1021. doi: 10.1086/323478. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jones M, Hingtgen C M, Bu G, Laribee N, Tanzi R E, Moir R D, Nath A, He J J. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000;6:1380–1387. doi: 10.1038/82199. [DOI] [PubMed] [Google Scholar]
- Dollard S C, James H J, Sharer L R, Epstein L G, Gelbard H A, Dewhurst S. Activation of nuclear factor κB in brains from children with HIV-1 encephalitis. Neuropathol Appl Neurobiol. 1995;21:518–528. doi: 10.1111/j.1365-2990.1995.tb01098.x. [DOI] [PubMed] [Google Scholar]
- Lukiw W J, Zhao Y, Cui J G. An NF-κB-sensitive microRNA-146a-mediated inflammatory circuit in Alzheimer’s disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C, Berman J W, Brett F M, Staunton H, Farrell M, Brosnan C F. MCP-1, MCP-2 and MCP-3 expression in multiple sclerosis lesions: an immunohistochemical and in situ hybridization study. J Neuroimmunol. 1998;86:20–29. doi: 10.1016/s0165-5728(98)00002-2. [DOI] [PubMed] [Google Scholar]
- Coughlan C M, McManus C M, Sharron M, Gao Z, Murphy D, Jaffer S, Choe W, Chen W, Hesselgesser J, Gaylord H, Kalyuzhny A, Lee V M, Wolf B, Doms R W, Kolson D L. Expression of multiple functional chemokine receptors and monocyte chemoattractant protein-1 in human neurons. Neuroscience. 2000;97:591–600. doi: 10.1016/s0306-4522(00)00024-5. [DOI] [PubMed] [Google Scholar]
- Van der Meer P, Ulrich A M, Gonzalez-Scarano F, Lavi E. Immunohistochemical analysis of CCR2, CCR3, CCR5, and CXCR4 in the human brain: potential mechanisms for HIV dementia. Exp Mol Pathol. 2000;69:192–201. doi: 10.1006/exmp.2000.2336. [DOI] [PubMed] [Google Scholar]