Abstract
Raloxifene was approved in 2007 by the FDA for the chemoprevention of breast cancer in postmenopausal women at high risk for invasive breast cancer. Approval was based in part on the improved safety profile for raloxifene relative to the standard treatment of tamoxifen. However, recent studies have demonstrated the ability of raloxifene to form reactive intermediates and act as a mechanism-based inhibitor of cytochrome P450 3A4 (CYP3A4) by forming adducts with the apoprotein. However, previous studies could not differentiate between dehydrogenation to a di-quinone methide and the more common oxygenation pathway to an arene oxide, as the most likely intermediate to inactivate CYP3A4. In the current work, 18O-incorporation studies were utilized to carefully elucidate CYP3A4-mediated oxygenation versus dehydrogenation of raloxifene. These studies established that 3′-hydroxyraloxifene is produced exclusively via CYP3A4-mediated oxygenation and provide convincing evidence for the mechanism of CYP3A4-mediated dehydrogenation of raloxifene to a reactive di-quinone methide, while excluding the alternative arene oxide pathway. Furthermore, it was demonstrated that 7-hydroxyraloxifene, which was previously believed to be a typical O2-derived metabolite of CYP3A4, is in fact produced by a highly unusual hydrolysis pathway from a putative ester, formed by the conjugation of raloxifene di-quinone methide with a carboxylic acid moiety of CYP3A4, or other proteins in the reconstituted system. These findings not only confirm CYP3A4-mediated dehydrogenation of raloxifene to a reactive di-quinone methide, but also suggest a novel route of raloxifene toxicity.
Breast cancer is the second most common form of cancer in women and second most common cause of cancer mortality in the United States (1). Tamoxifen, the prototypical SERM, has been the mainstay treatment for hormone-dependent breast cancer (2, 3), and more recently used as a chemopreventive agent in women at risk of developing breast cancer (4). Despite the effectiveness of tamoxifen in the treatment of breast cancer, its use has been linked to an increased risk of endometrial cancer (5-8) through formation of DNA adducts (9-11). It has been proposed that toxicity of tamoxifen is caused by the dehydrogenation of 4-hydroxytamoxifen (the active metabolite of tamoxifen) to reactive intermediates, such as a quinone methide (12-14) which forms DNA and protein adducts.
As a result of tamoxifen’s potential side effects, several second generation SERMs have been developed to reduce potential toxicities. One such SERM, raloxifene, was originally used clinically for the treatment and prevention of osteoporosis in postmenopausal women (15, 16). Due to recent studies and the clinical trial for chemoprevention of breast cancer (STAR trial: Study of Tamoxifen and Raloxifene) that have shown raloxifene to be as effective as tamoxifen in reducing breast cancer, with a reduced risk of endometrial cancer and blood clots (17-19), the FDA approved raloxifene for the chemoprevention of breast cancer in 2007. However, as with tamoxifen, recent work has shown that the metabolism of raloxifene via cytochrome P450 3A4 (CYP3A4) can generate several reactive quinone species (20-22). Furthermore, raloxifene has been shown to be a mechanism-based inactivator of CYP3A4, forming adducts with the apoprotein (21, 23-25). Although the inactivating species has not been explicitly identified, it is theorized that dehydrogenation of raloxifene to a di-quinone methide is responsible for the inactivation of CYP3A4 (20-22, 26). The efficient excretion of raloxifene by presystemic intestinal glucuronidation decreases the potential for abnormally high concentrations that could be toxic (27). Thus, it appears that this SERM may be substantially safer than tamoxifen, or other first-generation SERMs. In fact, even though raloxifene reactive intermediates bind extensively to microsomal proteins, it has been characterized as “a non-hepatotoxic drug” in a recent comparison of drugs that bind extensively to microsomal proteins, to agents that do not bind extensively (28). In addition, an analogue of the new SERM, arzoxifene, with a fluorine substituted for the hydroxyl group at the critical 4′-position that must have a hydroxyl group to be dehydrogenated to a di-quinone methide, was not metabolized to an electrophilic intermediate (29). To facilitate the development of less toxic SERMs, it is critical to fully elucidate the mechanisms of CYP3A4-mediated metabolism of raloxifene and identify the inactivating specie(s).
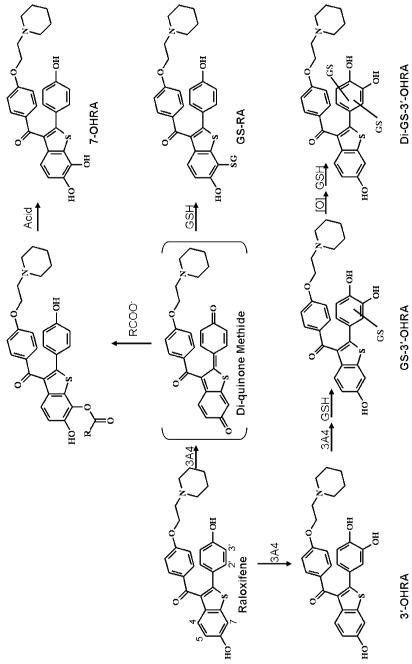
Recent studies have identified several oxygenated raloxifene metabolites and several GSH adducts (20, 21). Despite the high quality of these reports, due to the complexity of raloxifene metabolism, they were unable to fully characterize CYP3A4-mediated oxygenation versus dehydrogenation of raloxifene. Specifically, the oxygenated metabolites and GSH adducts could have been produced from either the epoxide or di-quinone methide intermediates (21). In this study, we utilized 18O-incorporation studies to determine that 3′-hydroxyraloxifene (3′-OHRA) was formed directly via P450-mediated oxygenation. In contrast, it was determined that 7-hydroxyraloxifene (7-OHRA) was not formed via CYP3A-mediated oxygenation. Rather, 7-OHRA was formed exclusively through dehydrogenation of raloxifene to a di-quinone methide intermediate. In a novel mechanism, this reactive di-quinone methide conjugates with a carboxylic acid moiety of CYP3A4 or another protein, and is subsequently released by acid-catalyzed hydrolysis of the ester to form 7-OHRA.
Material and methods
Materials
Raloxifene, NADPH, reduced GSH, silver oxide, H218O and propionic acid were purchased from Sigma-Aldrich (St. Louis, MO). 18O2 was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). 7-OHRA and 3′-OHRA synthesized standards (20) were generous gifts from Dr. Judy L. Bolton (University of Illinois, Chicago). The purity of the standards was confirmed with 1H NMR. All other chemicals for synthesis or analysis were of analytical grade or equivalent and obtained at the highest grade commercially available.
Instrumentation
LC/MS was conducted using a Thermo LCQ Advantage MAX mass spectrometer, coupled with an LC system consisting of a Finnigan Surveyor LC pump and Surveyor Autosampler (Thermo Fisher Scientific, Waltham, MA). Electrospray ionization (ESI) with positive ionization was utilized. The source temperature was set to 250°C, ionization voltage to 5 kV, capillary voltage 45 V, and sheath gas (N2) flow rate of 50 units. Parameters for MS/MS by CID with helium gas were as follows: activation amplitude at 35.0%, activation Q at 0.250, activation time at 30 ms and isolation width of 2 amu. Chromatography was conducted using a Phenomenex Gemini 3μ C6-Phenyl (150 × 2.00 mm) reverse-phase column (Phenomenex Inc., Torrance, CA). The mobile phase consisted of solvent A: acetonitrile and solvent B: 10% methanol containing 0.4% formic acid (v/v/v). For raloxifene analysis, the mobile phase was linear from 5 to 20% solvent A over 40 min, increasing to 100% solvent A over 10 min, at a flow rate of 0.2 ml/min. Identification of raloxifene metabolites was based on product-ion spectra obtained from CID of the [M+H]+ ions to their product-ions previously described by Yu et al.(20) and Chen et al. (21) LCQ Zoom scans (minimum of 10 scans) of the [M+H]+ or [M+2H]2+ ion for each metabolite was utilized to quantitate isotope peak distribution. For testosterone analysis, the mobile phase was linear from 5 to 70% solvent A over 35 min, increasing to 95% solvent A over 5 min, with a flow rate of 0.2 ml/min. Identification of testosterone metabolites was based on detection of the hydroxylated metabolites at [M+H]+ of 305 m/z, and by comparison of retention times to standards. For raloxifene/carboxylic acid conjugate analysis, the mobile phase was linear from 5 to 40% solvent A over 20 min, increasing to 95% solvent A over 10 min, at a flow rate of 0.2 ml/min.
Preparation of reconstituted CYP3A4 and oxidoreductase
A CYP3A4 construct containing C-terminal polyhistidine tag cloned into the pSE380 vector was a generous gift from Dr. James R. Halpert (University of California, San Diego, CA), and the rat cytochrome P450-oxidoreductase (POR) construct pOR262 was a generous gift from Dr. Charles B. Kasper (University of Wisconsin, Madison, WI). CYP3A4 was expressed in Escherichia coli DH5-α cells (Invitrogen, Carlsbad, CA) and purified as described previously (30). Rat POR was expressed in JM-109 cells and purified as described previously (31).
Incubation of raloxifene, 7-hydroxyraloxifene, and 3′-hydroxyraloxifene with recombinant CYP3A4
The reconstituted system contained 50 pmol of purified CYP3A4, 100 pmol of recombinant POR, 100 pmol of cytochrome b5 (Invitrogen, Carlsbad, CA), 0.04% (w/v) sodium cholate, and 20 μg of lipid mix (equal weights of DOPC, DLPC and DLPS). The mixture was gently shaken at room temperature for 10 min. To the system was added potassium phosphate buffer (50 mM, pH 7.4), GSH (4 mM), MgCl2 (15 mM), and substrate (50 μM) in a final volume of 500 μl. The mixture was preincubated at 37°C for 5 min, and the reaction was initiated by the addition of 2 mM of NAPDH. The reaction was allowed to proceed for 30 min at 37°C and then was terminated by one of two methods. Method 1: addition of 60 μl of trichloroacetic acid (TCA). The mixture was vortexed, followed by centrifugation at 21000 × g for 15 min to remove the protein. Raloxifene and metabolites were extracted using C-18 Sep-Pak cartridges (Waters, Taunton, MA). The resulting eluate was concentrated to dryness by evaporation under nitrogen and reconstituted in 10% acetonitrile (v/v) for analysis via LC/MS. Method 2: addition of 500 μl of cold methanol. The mixture was vortexed, followed by centrifugation at 21000 × g for 15 min to remove the protein. The supernatant was concentrated to dryness by evaporation with nitrogen for analysis via LC/MS.
To investigate the effects of GSH on raloxifene metabolism, raloxifene was incubated with a reconstituted system as described above, with or without GSH. The mixture was preincubated at 37°C for 5 min, and the reaction was initiated by the addition of 2 mM of NADPH. The reaction was allowed to proceed for 10 min at 37°C and then terminated by the addition of 100 μl of 60% TCA (v/v), containing 11β-hydroxytestosterone as an internal standard. The samples were then extracted using method 1. All incubations were performed 5 times (i.e. 5 separate reconstituted systems).
18O-incorporation studies
To measure the incorporation of oxygen from water, incubations of CYP3A4 and raloxifene were conducted as described above, with the substitution of 250 μl of H218O (97%). Reactions were terminated by method 1. To account for fractional incorporation and to determine maximal incorporation from H218O, prior to termination, a 25 μl aliquot from each incubation was removed and added to 2 mg of 2-chloronicotinolyl chloride (Alfa Aesar, Ward Hill, MA) to form the acid product. To this, anhydrous acetonitrile (100 μl) was added, and the resulting mixture was analyzed by direct infusion into the LCQ. The ratio of 16O to 18O incorporated into the acid product was calculated using Brauman’s Least Square method (32, 33). Briefly, the contribution of each isotope peak for a compound was determined by primary Zoom scans of unlabeled standard. The ion intensities of the 18O labeled compounds were corrected for the ion overlap due to normal isotopic species, by subtracting the percent contribution of each isotope from the compound of interest. This resulted in a series of simultaneous equations, which were solved by Brauman’s method to calculate the percent of molecules containing 18O versus the percent of molecules that contained 16O.
To measure the incorporation of oxygen from molecular oxygen, incubations were conducted in an air-tight four-flask manifold apparatus that was constructed so that it could be evacuated, flushed with ultrapure nitrogen gas, and repressurized with 18O2 introduced. The incubations were conducted as described above, with the following modifications. The potassium phosphate buffer was purged with nitrogen prior to addition to the reconstituted system. The manifold was then evacuated and flushed with ultrapure nitrogen three times to remove any atmospheric oxygen. 18O2 (97%) was then introduced to the system. The incubations (one per flask) were preincubated at 37°C for 5 min, and the reactions were initiated by the addition of 2 mM of NAPDH via syringe through air-tight septa. Reactions were terminated by method 1. To account for fractional incorporation and determine maximal incorporation from 18O2, 200 μM of testosterone replaced raloxifene in one of the incubations. The testosterone incubation was analyzed via LC/MS, and the ratio of 16O to 18O incorporated into 6β-hydroxytestosterone was calculated using Brauman’s Least Square method.
Chemical oxidation studies
To chemically form SG-raloxifene, the procedure from Yu et al. (20) was utilized. Briefly, raloxifene (5 mg) was dissolved in 5 ml of anhydrous acetonitrile and heated to 60°C. Silver oxide (0.5 g) was added and stirred for 10 s. The resulting slurry was immediately filtered into a secondary solution consisting of 10 ml of 50 mM potassium phosphate buffer (pH 7.4) containing 0.5 mM GSH. The secondary solution was divided into one portion, which was concentrated under nitrogen gas for analysis via LC/MS, and a second portion to which TCA was added to a final concentration of 12% (v/v) and concentrated under nitrogen gas for analysis via LC/MS.
To form raloxifene/carboxylic acid conjugate, raloxifene (5 mg) and 1 μl of glacial acetic acid or propionic acid was added to 4 ml of anhydrous acetonitrile and heated to 60°C. Silver oxide (0.5 g) was subsequently added and stirred for 10 s. The resulting slurry was immediately filtered into a secondary solution consisting of 5 ml of 50 mM potassium phosphate buffer (pH 8.0). The secondary solution was divided in half, one portion was concentrated under nitrogen gas for analysis via LC/MS, and TCA was added to a second portion to produce a final concentration of 12% (v/v), and concentrated under nitrogen gas for analysis via LC/MS.
18O-labeled propionic acid was synthesized by incubating 5 μl of 16O-propionic acid in 100 μl of H218O (97%, pH 1 with HCl). The oxygen exchange proceeded for 3 days at 65°C. The solution was concentrated to dryness under nitrogen and reconstituted in 10 μl anhydrous acetonitrile. Incorporation of 18O was 90%, measured by direct infusion into the MS. The chemical oxidation of raloxifene was repeated, with the exception that 5 μl of the 18O-labeled propionic acid solution was added to the anhydrous acetonitrile solution. The products were analyzed via LC/MS.
Results
Incubation of raloxifene and hydroxyraloxifenes with recombinant CYP3A4
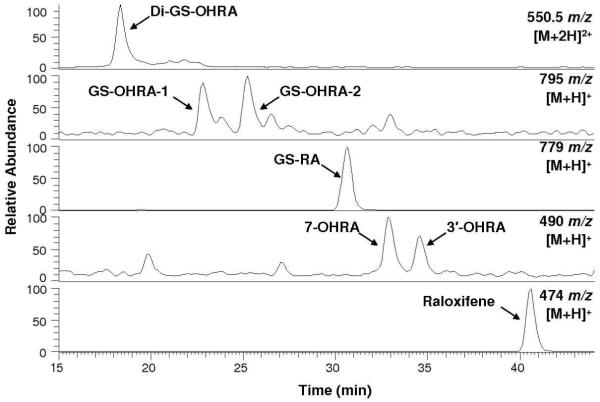
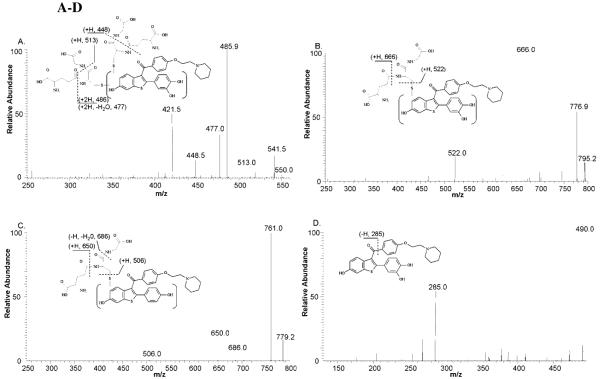
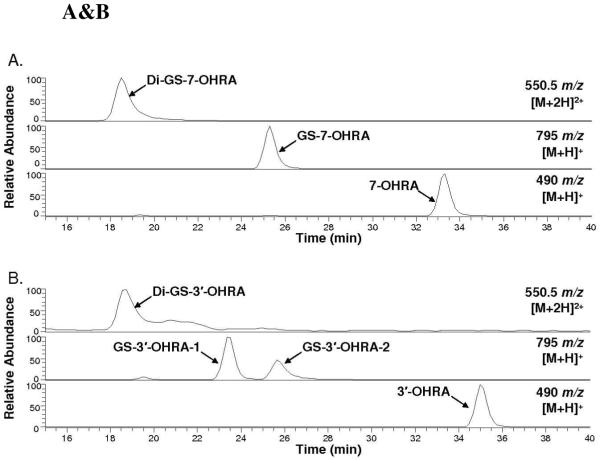
Incubations of raloxifene with recombinant CYP3A4, terminated by method 1, were analyzed by LC/MS/MS to identify raloxifene metabolites (Figure 1). We identified a di-GS-hydroxyraloxifene (di-GS-OHRA) metabolite, based on the detection of a strong [M+2H]2+ peak at 550.5 m/z at 18.3 min (a much weaker [M+H]+ peak at 1101 m/z was detected also at 18.3 min). MS/MS analysis of the diprotonated molecule at 550.5 m/z produced doubly-charged fragment ions at 486 and 421.5 m/z, corresponding to losses of one and two pyroglutamates moieties from GSH, respectively (Figure 2A). The doubly-charged fragment ions at 513 and 448.5 m/z corresponded to a loss of one glycine, and to the concurrent loss of glycine and pyroglutamate. The doubly charged fragment ions at 541.5 and 477 m/z corresponded to a loss of water, and the concurrent loss of water and pyroglutamate. Two GS-hydroxyraloxifene metabolites (assigned GS-OHRA-1 and GS-OHRA-2) were identified based on the detection of two weak [M+H]+ peaks at 795 m/z at 22.8 and 25.3 min (Figure 1). MS/MS analysis of the molecules at 795 m/z produced fragment ions at 522, 666, and 777 m/z, corresponding to cleavage of the thioester, loss of pyroglutamate, and loss of water, respectively (Figure 2B). There were no differences between the MS/MS spectra of GS-OHRA-1 and GS-OHRA-2. One mono-GS-raloxifene (GS-RA) metabolite was identified based on the detection of a strong [M+H]+ peak at 779 m/z at 30.7 min (Figure 1). MS/MS analysis of the molecule at 779 m/z produced strong fragment ions at 650 and 761 m/z, corresponding to losses of pyroglutamate and water, respectively (Figure 2C). Weak fragment ions at 506 and 686 m/z corresponded to a cleavage adjacent to the thioester moiety leaving sulfur on the raloxifene, and to the loss of glycine with water. Two hydroxyraloxifene metabolites were identified based on the detection of two [M+H]+ peaks at 490 m/z at 32.9 and 34.6 min (Figure 1). MS/MS analysis of both molecules at 490 m/z produced only one strong fragment ion at 285 m/z, corresponding to the loss of 1-(2-phenoxy-ethyl)-piperidine (Figure 2D). Additional MSn analysis of these metabolites was unable to produce fragment ions that could differentiate 7-OHRA and 3′-OHRA. However, the assignment of 7-OHRA to the peak at 32.9 min and 3-OHRA to the peak at 34.6 min was confirmed by comparison of retention times to synthetic standards.
Figure 1.
Positive ESI LC/MS mass chromatograms of raloxifene metabolites and GSH conjugates from the incubation of raloxifene with CYP3A4. Di-GS-OHRA [M+2H]2+ ions were monitored at 550.5 m/z, GS-OHRA [M+H]+ ions were monitored at 795 m/z, GS-RA [M+H]+ ions were monitored at 779 m/z, OHRA [M+H]+ ions were monitored at 490 m/z, and raloxifene [M+H]+ ions were monitored at 474 m/z.
Figure 2 A-D.
Positive ESI product-ion mass spectra from CID of the raloxifene metabolites and GSH conjugates shown in Figure 1. Spectra were obtained from CID of (A) Di-GS-OHRA at 550.5 m/z, (B) GS-OHRA at 795 m/z, (C) GS-RA at 779 m/z, (D) OHRA 490 m/z. Mass spectra for GS-OHRA-1 and 2 were identical, therefore only a representative spectrum of GS-OHRA-1 is shown. Mass spectra for 7-OHRA and 3′-OHRA were identical, therefore only a representative spectrum of 7-OHRA is shown.
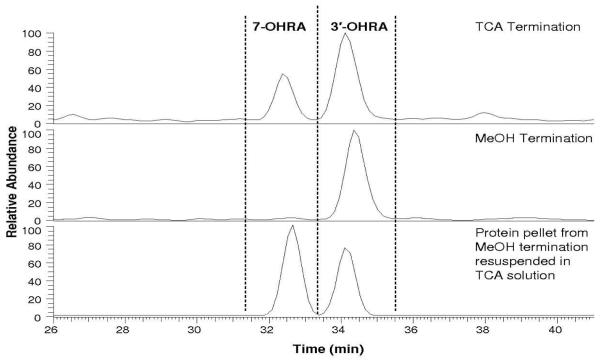
Incubations of raloxifene with CYP3A4, terminated by method 2, were analyzed via LC/MS/MS to identify raloxifene metabolites. Similar to the incubation terminated with acid, we were able to detect a single di-GS-OHRA, two GS-OHRA, and a single GS-RA metabolite. Retention times and MS/MS analysis of the metabolites were the same as described previously. However, in contrast to the acid-terminated incubations, only 3′-OHRA was detected in incubations terminated by methanol (Figure 3). Interestingly, when the protein pellets from the methanol-terminated incubations were resuspended in 50 mM potassium phosphate buffer (pH 7.4) and treated with acid, we were able to detect appreciable amounts of previously undetectable 7-OHRA. Lesser amounts of raloxifene and raloxifene metabolites, including 3′-OHRA, were also detected, but these molecules are believed to be adventitiously bound to the protein/lipid pellet and released during secondary acid treatment. 7-OHRA was the only raloxifene metabolite which required acid treatment to be liberated and detected.
Figure 3.
Positive ESI LC/MS mass chromatograms of hydroxyraloxifene metabolites (490 m/z) from incubations of raloxifene with CYP3A4. The top chromatogram shows that both 7-OHRA and 3′-OHRA are detected after the incubation was terminated with TCA. The middle chromatogram shows that only 3′-OHRA was detected after the incubation was terminated with methanol. In the bottom chromatogram, the protein pellet from the methanol termination was resuspended in a TCA solution, and both 3′-OHRA and 7-OHRA were recovered from the protein pellet.
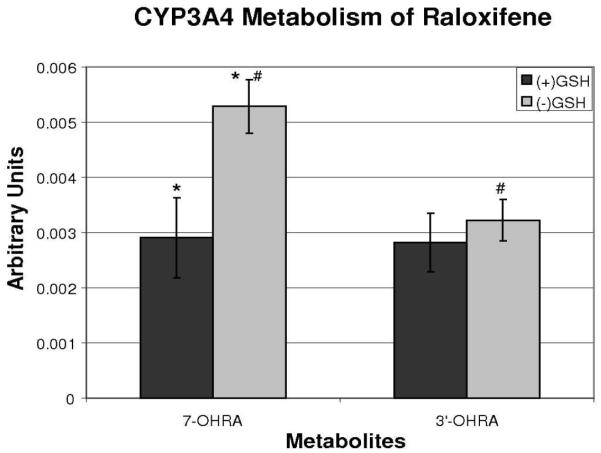
To investigate the role of GSH in the metabolism of raloxifene, incubations were conducted in the presence and absence of GSH. These incubations were terminated by method 1, and the relative amounts of hydroxylated metabolites quantified by use of an internal standard. The presence of GSH had no significant effect on the production of 3′-OHRA (Figure 4). However, the production of 7-OHRA was significantly decreased (55%) in the presence of GSH, compared to production of 7-OHRA in the absence of GSH.
Figure 4.
The effects of glutathione on CYP3A4-mediated production of 7-hydroxyraloxifene and 3′-hydroxyraloxifene, with incubation terminated by addition of TCA. The relative amounts of metabolites were estimated with an internal standard and concentration is expressed in arbitrary units (AU). (* and #) indicates significant difference (p < 0.05, n = 5).
Incubations of 7-OHRA and 3′-OHRA standards with recombinant CYP3A4, terminated by method 1, were analyzed by LC/MS and LC/MS/MS to identify metabolites (Figure 5 A&B). Similar to previous studies with rat and human liver microsomes by Yu et al. (20), the metabolism of 7-OHRA produced one mono-GSH conjugate (GS-7-OHRA), identified by detection of [M+H]+ peak at 795 m/z at 25.3 min and one di-GSH conjugate (di-GS-7-OHRA) identified by detection of [M+2H]2+ peak with 550.5 m/z at 18.5 min (Figure 5A). CYP3A4-mediated metabolism 3′-OHRA produced two mono-GSH conjugates (GS-3′-OHRA-1 and GS-3′-OHRA-2), identified by detection of two [M+H]+ peaks at 795 m/z at 23.3 and 25.6 min, and one di-GSH conjugate (di-GS-3′-OHRA) identified by detection of a [M+2H]2+ peak at 550.5 m/z at 18.7 min (Figure 5B). MS/MS analysis of the GSH conjugates produced similar spectra to that of the GSH conjugates produced from CYP3A4-mediated metabolism of raloxifene (data not shown). Based on identical retention times, GS-OHRA-1 eluting at 22.8 min in Figure 1 was determined to be hydroxylated at 3′ position. However, the hydroxylation site of GS-OHRA-2, eluting at 25.3 min in Figure 1, could not be assigned unequivocally. Similarly, the hydroxylation site of the di-GS-OHRA metabolite in Figure 1 could not be assigned.
Figure 5 A&B.
Positive ESI LC/MS mass chromatograms of the GSH conjugates from incubation of CYP3A4 with (A) 7-OHRA standard and (B) 3′-OHRA standard. All incubations were terminated by addition of TCA. Di-GS-OHRA [M+2H]2+ ions were monitored at 550.5 m/z, and GS-OHRA [M+H]+ ions were monitored at 795 m/z.
18O incorporation studies
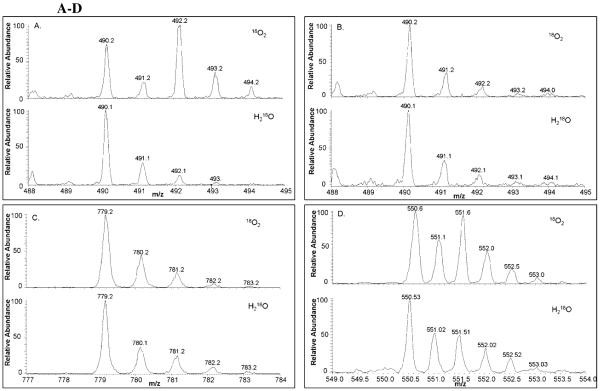
To determine the source of oxygen for the hydroxylated metabolites, raloxifene was incubated with CYP3A4 in the presence of 18O2. The incubations were terminated by method 1 and analyzed via LC/MS utilizing Zoom scans to quantitate isotopic abundance. Maximal possible 18O incorporation in the raloxifene metabolites was determined to be 45%, by measuring the 18O incorporated into the positive control, 6β-hydroxytestosterone. By inspection of the 3′-OHRA mass spectrum, it was apparent from the increase of the +2 isotope peak (492 m/z) that 3′-OHRA efficiently incorporated 18O (Figure 6A). Using Brauman’s Least Square method and accounting for fractional incorporation, greater than 99% of 3′-OHRA incorporated oxygen from O2 (i.e. P450-mediated oxygenation). By inspection of the 7-OHRA and GS-RA mass spectra, the absence of an increase in the +2 isotope peaks (492 and 781 m/z respectively), indicate that 7-OHRA and GS-RA did not incorporate 18O (Figure 6 B&C). Using Brauman’s Least Square method no appreciable amount of oxygen in 7-OHRA or GS-RA originated from O2. It is apparent from the increase of the +2 isotope peak (551.5 m/z, due to diprotonation) that di-GS-OHRA incorporated 18O (Figure 6D). Using Brauman’s Least Square method and accounting for fractional incorporation, greater than 99% of di-GS-OHRA incorporated oxygen from O2. Due to the minimal amounts of GS-OHRA metabolites formed, we were unable to measure the amount of 18O incorporation by Brauman’s Least Square method. However, by visual inspection, the GS-OHRA metabolites appeared to incorporate an appreciable amount of 18O.
Figure 6 A-D.
Positive ESI Zoom scan mass spectra of raloxifene metabolites and GSH conjugates from the incubation of raloxifene with CYP3A4 in the presence of 18O2 or H218O. Maximal possible 18O incorporation in the raloxifene metabolites was determined to be 45% from 18O2 and 50% from H218O by measuring the 18O incorporated into the controls, 6β-hydroxytestosterone and 2-chloronicotinolyl acid, respectively. The isotope patterns are shown for (A) 3′-OHRA, (B) 7-OHRA, (C) GS-RA, and (D) di-GS-OHRA.
To determine if any of the hydroxylated raloxifene metabolites were produced from hydration of a reactive intermediate, raloxifene was incubated with CYP3A4 in the presence of H218O. The incubations were terminated by method 1 and analyzed via LC/MS with Zoom scan to quantitate the isotope peaks distribution. Maximal possible 18O incorporation into the raloxifene metabolites was determined to be 50%, by measuring the 18O incorporated into 2-chloronicotinolyl acid. By visual inspection, due to the lack of an increase in their +2 isotope peaks, it was concluded that 3′-OHRA, 7-OHRA, and GS-RA did not incorporate 18O from water (Figure 6 A-C). Using Brauman’s Least Square method and accounting for fractional incorporation, no appreciable amount of oxygen was found to originate from H2O in these metabolites. Careful inspection of the di-GS-OHRA mass spectra, showed a small, but appreciable, increase in the +2 isotope peak (Figure 6 D). Using Brauman’s Least Square method and accounting for fractional incorporation, it was determined that roughly 20% of di-GS-OHRA incorporated oxygen from H2O. Due to the minute amounts of GS-OHRA metabolites formed, we were unable to accurately measure the amount of 18O incorporation from H218O. However, the GS-OHRA metabolites did not appear to incorporate an appreciable amount of 18O from water.
Chemical oxidation
Silver oxide was used to chemically oxidize raloxifene to a di-quinone methide. The raloxifene di-quinone methide is very unstable, with a half-life estimated to be less than 1 s in phosphate buffer (pH 7.4) at 5°C (20). To assay for this unstable electrophilic di-quinone methide, it was added to a secondary solution containing GSH to form stable GSH conjugates. The resulting mixture was separated into two portions; one was immediately analyzed, while TCA was added to the second portion prior to analysis by LC/MS/MS. The raloxifene di-quinone methide formed a single GSH adduct, which was identified as GS-RA by comparison of its retention time and MS/MS spectrum to the GS-RA metabolite produced from CYP3A4-mediated metabolism of raloxifene (data not shown). No hydroxyraloxifene peaks were detected and the addition of TCA had no effect on the product profile, demonstrating that the di-quinone methide did not conjugate with the carboxyl groups on GSH.
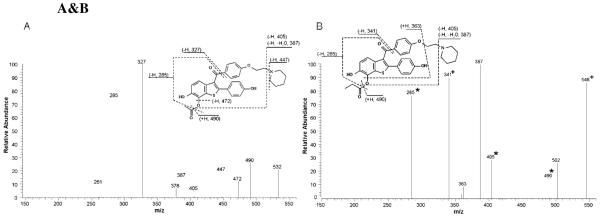
To determine if raloxifene di-quinone methide could form conjugates with compounds containing carboxyl groups, the electrophilic di-quinone methide was formed in the presence of either acetic acid or propionic acid. The resulting mixtures were separated into two portions, one was immediately analyzed, and the other acidified with TCA prior to analysis via LC/MS/MS. Analysis of the non-TCA-treated samples resulted in the detection of an ion consistent with the putative molecular weight of raloxifene conjugated to acetic acid, [M+H]+ = 532 m/z, and propionic acid, [M+H]+ = 546 m/z. MS/MS analyses of the 532 m/z peak produced fragment ions at 285, 327, 387, 405, 447, 472, and 490 m/z (Figure 7A). The fragment ions at 327 and 285 m/z corresponded to the loss of the 1-(2-phenoxy-ethyl)-piperidine moiety, and concurrent cleavage of the ester, leaving an oxygen. The fragment ion at 405 and 387 m/z corresponded to the loss of piperidine and the cleavage of the ester, leaving oxygen, and concurrent loss of water. The fragment ion at 472 m/z corresponded to the loss of the acetic acid moiety. The fragment ion at 490 m/z corresponded to the cleavage of the ester, leaving oxygen. MS/MS analyses of the 546 m/z peak produced fragment ions similar to that of the 532 m/z peak, including at 285, 387, 405, and 490 m/z, which corresponded to the same fragment pattern as discussed above (Figure 7B). However, the 546 m/z peak also had fragment ions at 341 and 363 m/z. The fragment ion at 341 m/z corresponded to the loss of the 1-(2-phenoxy-ethyl)-piperidine moiety. The fragment ion at 363 m/z corresponded to the concurrent loss of 1-ethyl-piperidine and propionic acid. LC/MS/MS analysis of the TCA-treated samples detected a single hydroxyraloxifene peak, [M+H]+ at 490 m/z, with a fragment ion at 285 m/z. This hydroxyraloxifene product was identified as 7-OHRA by comparison to the synthetic standard.
Figure 7A&B.
Positive ESI product-ion mass spectra from CID of the (A) raloxifene/acetic acid conjugate, [M+H]+ = 532 m/z, and (B) raloxifene/propionic acid conjugate, [M+H]+ = 546 m/z, produced by adding chemically synthesized raloxifene di-quinone methide in the presence of the carboxylic acids. (*) Indicates fragment ions that increased by 2 amu and (+) indicates fragment ions that increased by 4 amu when 18O-labeled propionic acid was used to produce the adduct, [M+H]+ = 550 m/z.
The raloxifene di-quinone methide was trapped by incubation with 18O-labeled propionic acid (incorporation of two oxygen atoms), and analyzed by LC/MS. A peak with the putative molecular weight of raloxifene conjugated to 18O-labeled propionic acid, [M+H]+ = 550 m/z eluted at the same retention time as the adduct formed with unlabeled propionic acid (546 m/z, raloxifene conjugated to 16O-propionic acid, data not shown). MS/MS analysis of the labeled adduct produced fragment ions (identified in Figure 7B) that were highly similar to the unlabeled adduct, but with mass shifts of 2 or 4 amu, consistent with the addition of 1 or 2 18O atoms. Major fragment ions of the adduct peak with a mass of 550 m/z showed mass shifts of 285→287 m/z (+1 18O), 405→407 m/z (+1 18O), 490→492 m/z (+1 18O) and 341→345 m/z (+2 18O). These results confirmed the identity of the di-quinone methide, produced by chemical oxidation of raloxifene, and its ability to be trapped with carboxylic acids to form ester adducts.
Discussion
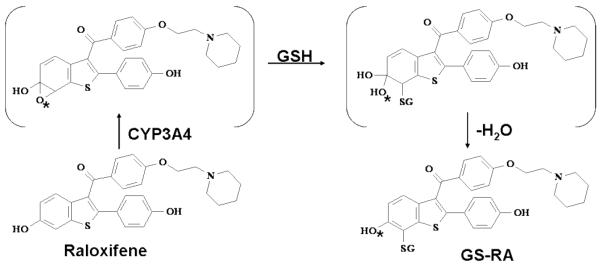
Raloxifene is a second generation SERM used for the treatment of osteoporosis in postmenopausal women (15, 16), and for the chemoprevention of breast cancer. However, P450-mediated metabolism of raloxifene produces reactive o-quinones and multiple GSH adducts (20, 21). Raloxifene is also a mechanism-based inactivator of CYP3A4, forming at least two adducts with the apoprotein (21, 23-25). The current study also demonstrates that CYP3A4 dehydrogenates raloxifene to form a highly reactive di-quinone methide. Due to the instability of this electrophile, it can only be detected by trapping it as its mono-GSH adduct (GS-RA). The origins of this adduct could arise from either dehydrogenation or via the formation of an epoxide intermediate, followed by subsequent loss of water (21). In this study, we utilized recombinant CYP3A4, 18O incorporation studies and chemically-oxidized raloxifene to elucidate the mechanisms of CYP3A4-mediated metabolism of raloxifene.
Raloxifene incubated with reconstituted CYP3A4 and terminated with TCA, produced two hydroxyraloxifene metabolites and several GSH adducts (Figure 1). The hydroxyraloxifene metabolites were identified as 7-OHRA and 3′-OHRA. One mono-GSH adduct, GS-RA, was identified by LC/MS/MS analysis. Previous studies identified up to three mono-GSH raloxifene adducts (20, 21). However, those studies used rat and human liver microsomes which contained a multitude of P450 enzymes, which could potentially produce additional mono-GSH adducts. It was possible that multiple mono-GSH adducts co-eluted in our analysis, but careful examination of the products using several highly varied HPLC conditions did not indicate the presence of additional adducts (data not shown). Two peaks corresponding to GS-OHRA metabolites and one peak corresponding to di-GS-OHRA were produced by CYP3A4 and detected by LC/MS/MS. Inclusion of the 7-OHRA and 3′-OHRA standards in the CYP3A4 incubation confirmed that the GS-OHRA-1 adduct was hydroxylated at the 3′-position. The site of hydroxylation on the remaining GS-OHRA and di-GS-OHRA adducts could not be unequivocally identified.
To establish the mechanism of CYP3A4-mediated oxygenation versus dehydrogenation of raloxifene, 18O incorporation studies were utilized to track the source of oxygen in the raloxifene metabolites. Data indicated that 3′-OHRA obtained greater than 99% of its 3′ oxygen from molecular oxygen (i.e. through P450-mediated oxygenation). Surprisingly, 7-OHRA did not incorporate its oxygen atom from either molecular oxygen or water. Therefore, 7-OHRA was not formed through CYP3A4-mediated oxygenation or through hydration of the di-quinone methide. GS-RA did not incorporate oxygen from either molecular oxygen or water, and these results would be expected if the GS-RA adduct was formed from the oxygenated/hydrated di-quinone methide intermediate. Alternatively, if GS-RA was formed via CYP3A4-mediated epoxidation of the C6-C7 double bond to an arene oxide intermediate, the nucleophilic attack at C7 by GSH would result in the formation of a gem diol (21) (Scheme 1). Subsequent dehydration of the unstable gem diol could produce the GS-RA metabolite. However, because there is an equal statistical probability that either of the gem diol oxygens would be lost from dehydration, if the arene oxide was formed at the C6-C7 position, one would expect 50% of the 6-hydroxyl oxygen to contain 18O incorporated from 18O2. There was no detectable 18O incorporated into the GS-RA adduct. Therefore, these results strongly suggest that raloxifene di-quinone methide was directly produced by CYP3A4 and it was the precursor of GS-RA.
Scheme 1.
Formation of raloxifene adducts with GSH. The mono-glutathione-raloxifene adduct at the C7 carbon is shown to be produced through CYP3A4-mediated epoxidation at the C6-C7 position, followed by gem diol intermediate formation at the C6 position. *O indicates 18O from molecular oxygen, where 50% of the mono-glutathione adduct would theoretically incorporate 18O from molecular oxygen through formation of the C6-C7 epoxide. Conversely, epoxidation at the C4-C5 position (not shown), followed by epoxide hydration and aromatization through water loss would produce the C5-C6 catechol. Oxidation of the catechol would form the ortho-quinone at C5-C6, which could trap GSH to form an adduct that would theoretically fail to incorporate 18O from molecular oxygen. However, the adduct would have an additional hydroxyl group, most likely at C5, and the adduct that was detected did not.
Additional experiments investigated the origins of 7-OHRA. If 7-OHRA production required P450 turnover, but did not obtain its oxygen atom from either O2 or H2O, what was the source of the oxygen? Incubations in the presence of catalase or superoxide dismutase had no effect on 7-OHRA production, and incubations of raloxifene in the presence of lipid hydroperoxides and H2O2 failed to produce 7-OHRA (data not shown). Interestingly, it was noted that when incubations of raloxifene with CYP3A4 were terminated with methanol, all of the raloxifene metabolites and adducts were observed, with the exception of 7-OHRA. Only when the incubations were terminated with TCA, or other acids such as perchloric acid (data not shown), was 7-OHRA formed. Furthermore, when acid was added to the protein pellet resulting from methanol quenching and extraction, we could recover appreciable amounts of 7-OHRA. GSH has previously been shown to decrease, but not prevent, the irreversible inhibition of CYP3A4 via raloxifene (21). Our work demonstrated that while GSH had no effect on 3′-OHRA production, the presence of GSH significantly reduced 7-OHRA production. Together, these results suggest that the reactive intermediate that inactivates CYP3A4, is also the reactive intermediate required for 7-OHRA production. Previous work by Yukinaga et al. (25) showed that raloxifene formed an adduct with the OH group of tyrosine (Tyr) 75 on the CYP3A4 apoprotein. However, the resulting ether bond would be extremely difficult to hydrolyze with any aqueous acidic condition. Therefore, we theorized that the raloxifene di-quinone methide reacted with carboxyl groups on CYP3A4 or other proteins in the reconstituted system (i.e. oxidoreductase and b5), to produce an ester, which could be hydrolyzed by acid to release 7-OHRA. A similar unusual mechanism was observed in the P450-mediated metabolism of benzo[a]pyrene (BaP) to its diol epoxide, which covalently modified the Asp47 carboxylic side chain of human hemoglobin to form an ester (34, 35). Subsequent hydrolysis of the ester formed the alcohol, BaP 7,8,9,10-tetrahydrotetrol.
Due to the reactivity and short half-life of the di-quinone methide (<1 s) (20), it was unlikely that it would travel far from the CYP3A4 active site, suggesting CYP3A4 was most likely site of ester formation. However, the previously reported raloxifene/CYP3A4 adducts on Cys239, which is adjacent to an exit channel (36), or Tyr75 on the solvent-exposed surface of CYP3A4 (observed from the crystal structure), demonstrated that the di-quinone methide was stable enough to travel from the active site and alkylate these remote nucleophilic residues. Furthermore, when raloxifene was linked to biotin and then metabolized with rat liver microsomal incubations, the raloxifene congener still formed covalent adducts with microsomal proteins that were not P450s (22). Thus, the di-quinone methide must have been stable enough to exist outside the active-site long enough to diffuse and react with nucleophiles on a protein that was different from the original bioactivation P450 enzyme. Therefore, we cannot exclude a mechanism where the di-quinone methide leaves the active site and forms an ester with carboxyl groups on surrounding proteins in the reconstituted system (i.e. oxidoreductase and b5).
To test the theory of ester formation, silver oxide was used to chemically produce raloxifene diquinone methide, which was trapped with GSH to produce GS-RA, verifying that the di-quinone methide was formed by oxidation of raloxifene. Acetic acid and propionic acid were employed instead of GSH as trapping agents to mimic protein carboxylic acid moieties and used to trap the di-quinone methide. LC/MS analysis of these products detected molecules with [M+H]+ = 532 m/z and 546 m/z, the precise [M+H]+ ions predicted for raloxifene conjugated with acetic acid and propionic acid, respectively. Furthermore, when TCA was added to these products, 7-OHRA was recovered; strongly suggesting that 7-OHRA was not produced directly via CYP3A4-mediated oxygenation. Rather, CYP3A4 dehydrogenates raloxifene to a reactive di-quinone methide, which was subsequently trapped by a carboxylic acid functional side chain on CYP3A4 or nearby protein, forming an ester. 7-OHRA was released only by the hydrolysis of the ester under acidic conditions, and therefore was not present during the incubation.
Altogether, the results of this study change our current understanding of CYP3A4-mediated metabolism of raloxifene (Scheme 2). This study demonstrated that 3′-OHRA appears to be the only hydroxylated metabolite produced via CYP3A4-mediated oxygenation. Furthermore, lack of 18O incorporation from molecular oxygen demonstrates that GS-RA must be produced from CYP3A4-mediated dehydrogenation of raloxifene to a di-quinone methide intermediate, not an arene oxide intermediate (Scheme 1). Unfortunately, due to the lack of GS-RA standards, we could not quantify the production of GS-RA. However, due to the relatively abundant production of 7-OHRA and GS-RA, dehydrogenation appears to be a major pathway of raloxifene metabolism.
Scheme 2.
Proposed pathway of raloxifene oxidation by CYP3A4. RCOO− = carboxylic acid residue on CYP3A4 or alternative protein, GSH = glutathione.
In contrast, 7-OHRA was not a “normal” metabolite of CYP3A4. Rather, we propose a highly unusual pathway, in which raloxifene di-quinone methide conjugates with carboxyl groups of CYP3A4 or nearby proteins to form an ester bond. Interestingly, this ester bond only forms on the benzothiophene moiety and not the phenol moiety. Acidic conditions are required to hydrolyze this ester bond to release 7-OHRA. Therefore, if 7-OHRA is not produced during the incubation of raloxifene with CYP3A4, the secondary metabolites (i.e. GS-OHRA and di-GS-OHRA) are most likely formed by additional metabolism of 3′-OHRA. In support of this rationale, greater than 99% of the di-GS-OHRA metabolite incorporated oxygen from molecular oxygen, suggesting it is efficiently formed from 3′-OHRA. Unfortunately, due to the lower levels of GS-OHRA metabolites, we could not determine the source of oxygen in these metabolites. However, there remains the possibility that GS-RA is hydroxylated by CYP3A4 to form the GS-OHRA and di-GS-OHRA products. Additional study is required to determine the fate of GS-RA, and the source of the secondary metabolites.
In summary, CYP3A4 oxygenates raloxifene to 3′-OHRA and dehydrogenates raloxifene to a reactive di-quinone methide. The reactive species can be trapped by GSH in the form of thioether conjugates, form adducts with Cys239 and Tyr75 of CYP3A4, or in a novel mechanism, conjugate with carboxyl groups, forming ester conjugates with CYP3A4 or nearby proteins. Furthermore, these results suggest that the benzothiophene moiety, and not the phenol moiety, of raloxifene is the structural feature responsible for formation of the reactive species. These findings support the rational design of new SERMs without the benzothiophene structure to reduce bioactivation liabilities. Other putative heterocyclic functional groups that could be “structural alerts” include the benzofuran, indole, and benzimidazole moieties.
Acknowledgements
The authors would like to thank Dr. Judy Bolton, University of Illinois, Chicago, for providing 3′-hydroxyraloxifene and 7-hydroxyraloxifene for these studies.
Abbreviations
- SERM
selective estrogen receptor modulator
- GSH
glutathione
- LC/MS
liquid chromatography-mass spectrometry
- ESI
electrospray ionization
- CID
collision-induced dissociation
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DLPC
1,2-dilauryl-sn-glycero-3-phophocholine
- DLPS
1,2-diacyl-sn-glycero-phospho-L-serine
Footnotes
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences [Grant GM074249]
References
- (1).Horner MJ, R. L, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK. SEER Cancer Statistics Review, 1975-2006. National Cancer Institute; Bethesda, MD: 2009. [Google Scholar]
- (2).Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- (3).Hortobagyi GN. Treatment of breast cancer. N Engl J Med. 1998;339:974–984. doi: 10.1056/NEJM199810013391407. [DOI] [PubMed] [Google Scholar]
- (4).Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- (5).Magriples U, Naftolin F, Schwartz PE, Carcangiu ML. High-grade endometrial carcinoma in tamoxifen-treated breast cancer patients. J Clin Oncol. 1993;11:485–490. doi: 10.1200/JCO.1993.11.3.485. [DOI] [PubMed] [Google Scholar]
- (6).Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: Findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–537. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- (7).Curtis RE, Boice JD, Jr., Shriner DA, Hankey BF, Fraumeni JF., Jr. Second cancers after adjuvant tamoxifen therapy for breast cancer. J Natl Cancer Inst. 1996;88:832–834. doi: 10.1093/jnci/88.12.832. [DOI] [PubMed] [Google Scholar]
- (8).Swerdlow AJ, Jones ME. Tamoxifen treatment for breast cancer and risk of endometrial cancer: A case-control study. J Natl Cancer Inst. 2005;97:375–384. doi: 10.1093/jnci/dji057. [DOI] [PubMed] [Google Scholar]
- (9).Han XL, Liehr JG. Induction of covalent DNA adducts in rodents by tamoxifen. Cancer Res. 1992;52:1360–1363. [PubMed] [Google Scholar]
- (10).Randerath K, Moorthy B, Mabon N, Sriram P. Tamoxifen: Evidence by 32P-postlabeling and use of metabolic inhibitors for two distinct pathways leading to mouse hepatic DNA adduct formation and identification of 4-hydroxytamoxifen as a proximate metabolite. Carcinogenesis. 1994;15:2087–2094. doi: 10.1093/carcin/15.10.2087. [DOI] [PubMed] [Google Scholar]
- (11).Kim SY, Suzuki N, Laxmi YR, Shibutani S. Genotoxic mechanism of tamoxifen in developing endometrial cancer. Drug Metab Rev. 2004;36:199–218. doi: 10.1081/dmr-120033997. [DOI] [PubMed] [Google Scholar]
- (12).Marques MM, Beland FA. Identification of tamoxifen-DNA adducts formed by 4-hydroxytamoxifen quinone methide. Carcinogenesis. 1997;18:1949–1954. doi: 10.1093/carcin/18.10.1949. [DOI] [PubMed] [Google Scholar]
- (13).Beland FA, McDaniel LP, Marques MM. Comparison of the DNA adducts formed by tamoxifen and 4-hydroxytamoxifen in vivo. Carcinogenesis. 1999;20:471–477. doi: 10.1093/carcin/20.3.471. [DOI] [PubMed] [Google Scholar]
- (14).Bolton JL, Yu L, Thatcher GR. Quinoids formed from estrogens and antiestrogens. Methods Enzymol. 2004;378:110–123. doi: 10.1016/S0076-6879(04)78006-4. [DOI] [PubMed] [Google Scholar]
- (15).Jones CD, Jevnikar MG, Pike AJ, Peters MK, Black LJ, Thompson AR, Falcone JF, Clemens JA. Antiestrogens. 2. Structure-activity studies in a series of 3-aroyl-2-arylbenzo[b]thiophene derivatives leading to [6-hydroxy-2-(4-hydroxyphenyl)benzo[b]thien-3-yl] [4-[2-(1-piperidinyl)ethoxy]-phenyl]methanone hydrochloride ( LY156758), a remarkably effective estrogen antagonist with only minimal intrinsic estrogenicity. J Med Chem. 1984;27:1057–1066. doi: 10.1021/jm00374a021. [DOI] [PubMed] [Google Scholar]
- (16).Clemett D, Spencer CM. Raloxifene: A review of its use in postmenopausal osteoporosis. Drugs. 2000;60:379–411. doi: 10.2165/00003495-200060020-00013. [DOI] [PubMed] [Google Scholar]
- (17).DeMichele A, Troxel AB, Berlin JA, Weber AL, Bunin GR, Turzo E, Schinnar R, Burgh D, Berlin M, Rubin SC, Rebbeck TR, Strom BL. Impact of raloxifene or tamoxifen use on endometrial cancer risk: A population-based case-control study. J Clin Oncol. 2008;26:4151–4159. doi: 10.1200/JCO.2007.14.0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vogel VG. The NSABP study of tamoxifen and raloxifene (STAR) trial. Expert Rev Anticancer Ther. 2009;9:51–60. doi: 10.1586/14737140.9.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wickerham DL, Costantino JP, Vogel VG, Cronin WM, Cecchini RS, Ford LG, Wolmark N. The use of tamoxifen and raloxifene for the prevention of breast cancer. Recent Results Cancer Res. 2009;181:113–119. doi: 10.1007/978-3-540-69297-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yu L, Liu H, Li W, Zhang F, Luckie C, van Breemen RB, Thatcher GR, Bolton JL. Oxidation of raloxifene to quinoids: Potential toxic pathways via a diquinone methide and o-quinones. Chem Res Toxicol. 2004;17:879–888. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- (21).Chen Q, Ngui JS, Doss GA, Wang RW, Cai X, DiNinno FP, Blizzard TA, Hammond ML, Stearns RA, Evans DC, Baillie TA, Tang W. Cytochrome P450 3A4-mediated bioactivation of raloxifene: Irreversible enzyme inhibition and thiol adduct formation. Chem Res Toxicol. 2002;15:907–914. doi: 10.1021/tx0200109. [DOI] [PubMed] [Google Scholar]
- (22).Liu J, Li Q, Yang X, van Breemen RB, Bolton JL, Thatcher GR. Analysis of protein covalent modification by xenobiotics using a covert oxidatively activated tag: Raloxifene proof-of-principle study. Chem Res Toxicol. 2005;18:1485–1496. doi: 10.1021/tx0501738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Baer BR, Wienkers LC, Rock DA. Time-dependent inactivation of P450 3A4 by raloxifene: Identification of Cys239 as the site of apoprotein alkylation. Chem Res Toxicol. 2007;20:954–964. doi: 10.1021/tx700037e. [DOI] [PubMed] [Google Scholar]
- (24).Pearson JT, Wahlstrom JL, Dickmann LJ, Kumar S, Halpert JR, Wienkers LC, Foti RS, Rock DA. Differential time-dependent inactivation of P450 3A4 and P450 3A5 by raloxifene: A key role for C239 in quenching reactive intermediates. Chem Res Toxicol. 2007;20:1778–1786. doi: 10.1021/tx700207u. [DOI] [PubMed] [Google Scholar]
- (25).Yukinaga H, Takami T, Shioyama SH, Tozuka Z, Masumoto H, Okazaki O, Sudo K. Identification of cytochrome P450 3A4 modification site with reactive metabolite using linear ion trap-Fourier transform mass spectrometry. Chem Res Toxicol. 2007;20:1373–1378. doi: 10.1021/tx700165q. [DOI] [PubMed] [Google Scholar]
- (26).Liu H, Qin Z, Thatcher GR, Bolton JL. Uterine peroxidase-catalyzed formation of diquinone methides from the selective estrogen receptor modulators raloxifene and desmethylated arzoxifene. Chem Res Toxicol. 2007;20:1676–1684. doi: 10.1021/tx7001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kemp DC, Fan PW, Stevens JC. Characterization of raloxifene glucuronidation in vitro: contribution of intestinal metabolism to presystemic clearance. Drug Metab Dispos. 2002;30:694–700. doi: 10.1124/dmd.30.6.694. [DOI] [PubMed] [Google Scholar]
- (28).Obach RS, Kalgutkar AS, Soglia JR, Zhao SX. Can in vitro metabolism-dependent covalent binding data in liver microsomes distinguish hepatotoxic from nonhepatotoxic drugs? An analysis of 18 drugs with consideration of intrinsic clearance and daily dose. Chem Res Toxicol. 2008;21:1814–1822. doi: 10.1021/tx800161s. [DOI] [PubMed] [Google Scholar]
- (29).Liu H, Liu J, van Breemen RB, Thatcher GR, Bolton JL. Bioactivation of the selective estrogen receptor modulator desmethylated arzoxifene to quinoids: 4′-fluoro substitution prevents quinoid formation. Chem Res Toxicol. 2005;18:162–173. doi: 10.1021/tx049776u. [DOI] [PubMed] [Google Scholar]
- (30).Domanski TL, Liu J, Harlow GR, Halpert JR. Analysis of four residues within substrate recognition site 4 of human cytochrome P450 3A4: Role in steroid hydroxylase activity and alpha-naphthoflavone stimulation. Arch Biochem Biophys. 1998;350:223–232. doi: 10.1006/abbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- (31).Shen AL, Porter TD, Wilson TE, Kasper CB. Structural analysis of the FMN binding domain of NADPH-cytochrome P-450 oxidoreductase by site-directed mutagenesis. J Biol Chem. 1989;264:7584–7589. [PubMed] [Google Scholar]
- (32).Brauman JI. Least squares analysis and simplification of multi-isotope mass spectra. Anal Chem. 1966;38:607–610. [Google Scholar]
- (33).Korzekwa K, Howald WN, Trager WF. The use of Brauman’s least squares approach for the quantification of deuterated chlorophenols. EnvironBiomed Environ Mass Spectrom. 1990;19:211–217. doi: 10.1002/bms.1200190402. [DOI] [PubMed] [Google Scholar]
- (34).Day BW, Skipper PL, Zaia J, Tannenbaum SR. Benzo[a]pyrene anti-diol epoxide covalently modifies human serum albumin carboxylate side chains and imidazole side chain of histidine(146) J Am Chem Soc. 1991;113:8505–8509. [Google Scholar]
- (35).Skipper PL, Naylor S, Gan LS, Day BW, Pastorelli R, Tannenbaum SR. Origin of tetrahydrotetrols derived from human hemoglobin adducts of benzo[a]pyrene. Chem Res Toxicol. 1989;2:280–281. doi: 10.1021/tx00011a002. [DOI] [PubMed] [Google Scholar]
- (36).Fishelovitch D, Shaik S, Wolfson HJ, Nussinov R. Theoretical characterization of substrate access/exit channels in the human cytochrome P450 3A4 enzyme: involvement of phenylalanine residues in the gating mechanism. J Phys Chem B. 2009;113:13018–13025. doi: 10.1021/jp810386z. [DOI] [PMC free article] [PubMed] [Google Scholar]