Abstract
The α-helix is a ubiquitous structural element in proteins, and a number of studies have addressed the mechanism of helix formation and melting in simple peptides. However, fundamental issues remain to be resolved, particularly the temperature (T) dependence of the rate. In this work we report application of a novel kHz repetition rate solid-state tunable NIR (pump) and deep UV Raman (probe) laser system to study the dynamics of helix unfolding in Ac-GSPEA3KA4KA4-CO-D-Arg-CONH2, a peptide designed for helix stabilization in aqueous solution. Its T-dependent UV resonance Raman (UVRR) spectra, excited at 197 nm for optimal enhancement of amide vibrations, were decomposed into variable contributions from helix and coil spectra. The helix fractions derived from the UVRR spectra and from far UV CD spectra were coincident at low T, but deviated increasingly at high T, the UVRR curve giving higher helix content. This difference is consistent with the greater sensitivity of UVRR spectra to local conformation than CD. After a laser-induced T-jump the UVRR-determined helix fractions defined mono-exponential decays, with time-constants of ~120 ns, independent of the final T (Tf =18 to 61°C), provided the initial T (Ti) was held constant (60C). However, there was also a prompt loss of helicity, whose amplitude increased with increasing Tf, thereby defining an initial enthalpic phase, distinct from the subsequent entropic phase. These phases are attributed to disruption of H-bonds followed by reorientation of peptide links, as the chain is extended. When Ti was raised in parallel with Tf (10° C T-jumps), the prompt phase merged into an accelerating slow phase, an effect attributable to the shifting distribution of initial helix lengths. Even greater acceleration with rising Ti has been reported in T-jump experiments monitored by IR and fluorescence spectroscopies. This difference is attributable to the longer range character of these probes, whose responses are therefore more strongly weighted toward the H-bond-breaking enthalpic process.
Introduction
The formation of α-helical segments in proteins is a fundamental aspect of the protein folding problem. With the advent of laser-induced T-jump methods, it has become possible to measure sub-microsecond folding and unfolding rates of small α-helical peptides1–10, which are amenable to theoretical analysis and modeling11–19. The T jump perturbs the helix/coil equilibrium, and relaxation to the new equilibrium position can be monitored by optical techniques, including fluorescence2, 12, 20–23, infrared1, 4–6, 9, 24–29 and Raman10, 30–34 spectroscopies. Raman spectroscopy is advantageous because the entire vibrational fingerprint region can be monitored, whereas IR spectroscopy is limited largely to the amide I′ (carbonyl stretching) region in D2O, because of the strong IR absorption of water. Fluorescence monitors the local environment of a fluorophore, which responds indirectly to the peptide conformation. Raman excitation in the deep ultraviolet region produces enhancement of peptide backbone vibrations, via resonance with the amide π-π* electronic transitions35–38. We have found 197 nm excitation to be optimal for amide UVRR (ultraviolet resonance Raman) signals39. An additional advantage of the UVRR experiment is that the probe laser spot is small, permitting large T-jumps to be achieved by focusing the pump laser.
We have recently implemented a high-repetition (1 kHz) rate T-jump/UVRR apparatus of novel design32, and here report its application to helix-coil transition monitoring. We adopt the techniques pioneered by Lednev et al.10, who reported the first T-jump/UVRR study of a helical peptide.
Alanine-based peptides of ~20 residues, containing a few charged residues to maintain solubility, have significant helical content in aqueous solution, especially if they have effective end-capping groups. The end-caps form H-bonds with amide groups near the helix termini that have no interhelical H-bond partners40–43. T-jump studies have been carried out on a number of these peptides. The relaxation rates are consistently in the 100–400 ns range,1–10, 20, 22, 24–26, 30, 31,34 setting the time scale for helix nucleation and elongation models11–19, 44. However, there is discord about the temperature coefficient, some studies finding strong3, 4, 8, 29 and others finding weak dependence10 on temperature of the relaxation rate. Since the activation enthalpy of helix formation is of fundamental interest, this disagreement is a matter for concern.
Here we use T-jump Raman to study a peptide (GSP; Ac-GSPEA3KA4KA4-CO-D-Arg-CONH2), that is closely related to the peptide previously studied by Gai and coworkers8 using T-jump IR. The peptide differs only in the removal of a Tyr residue, which interferes with the Raman spectrum. To determine how changes near the helical terminus can affect the kinetics of helix formation, we also studied a non-acetylated version of the peptide. We expected that removal of the N-terminal acetyl group would slightly destabilize the helix.
Our results suggest that part of the disagreement stems from different ways of doing the T-jump experiment, since both initial and final temperature can be varied. We find, at least in the case of the peptide selected for this study, Ac-GSPEA3KA4KA4-CO-D-Arg-CONH2, that the temperature coefficient of the relaxation is zero, provided that the initial temperature is held constant, and the magnitude of the T-jump is varied. However, there is a prompt drop in helicity, whose amplitude increases as the final temperature is raised. When the T-jump is held constant, the relaxation accelerates as both initial and final temperatures are raised, but the prompt response is minimal. These results provide evidence for a two-stage mechanism for helix unfolding: an enthalpic stage, in which inter-residues are broken, and an entropic stage, in which the peptide links reorient as the chain becomes extended. The steeper temperature dependence reported for T-jump transients monitored by IR or fluorescence spectroscopy is attributed to greater weighting of the enthalpic stage, due to the longer range sensitivity of these probes.
Methods
Materials
Synthesis and purification of the Ac-GSP peptide, Ac-GSPEAAAKAAAAKAAAA-CO-D-Arg-CONH2 and its non-acetylated derivative are described elsewhere8.
For static and T-jump/UVRR experiments, ~ 3 mg/ml peptide was dissolved in 50 mM phosphate buffer at pH 7.4, with 0.2 M sodium perchlorate added as a Raman intensity standard. About 4 ml of the peptide solution was circulated through a wire-guided free-flowing cell to minimize decomposition and background scattering. Temperature control of the sample was achieved with a water bath (RTE-100, Neslab), which regulates the temperature of the sample cell and reservoir. UVRR spectra were collected with 10 minutes accumulation time. Temperature-controlled N2 gas was purged around the sample to maintain constant temperature and to displace atmospheric O2 (thereby avoiding interference from the O2 Raman band at 1550 cm−1).
CD measurements
Far-UV circular dichroism spectra were measured with an AVIV 62DS spectropolarimeter equipped with a temperature controlled cuvette holder, using a 1 mm quartz cell. The 20 °C CD spectra were averages of three repeated scans with 1 nm/sec spacing from 260 to 190 nm. The temperature profiles were recorded at 222 nm from 0 to 98 °C with 2 °C/sec steps and 1 min equilibration time. A reverse scan, from 98 to 0° C, was also carried out to verify reversibility. The peptide concentration was ~0.20 mM. The data were converted to mean residue ellipticity [θ] in deg.cm2/dmol using the equation [θ] = (θobs/10lc)/r, where θobs is the ellipticity measured in millidegrees, l is the optical path length (cm), c is the concentration of the peptide (M), and r is the number of residues.
Static and T-jump/time-resolved UV resonance Raman spectroscopy
The experimental set-up and the spectral acquisition scheme has been described elsewhere32, 45, 46. Nanosecond T-jumps were induced using IR laser pulses at 1.9 μm (10 ns, 0.4–0.8 mJ/pulse, 1 kHz) obtained from a potassium titanyl arsenate (KTA) crystal-based optical parametric oscillator (OPO) operating in an intracavity configuration of a diode pumped Nd-YLF laser (Photonics International Inc.). T-jump measurements were made at 6 °C initial temperature with varied ΔT and constant ΔT of ~ 10 °C with varied initial and final temperatures.
Raman spectra were excited using 197 nm pluses ( 1 μJ/pulse, 20 ns, 1 kHz) obtained from the fourth harmonic output of a Ti:sapphire laser (Photonics international) pumped by the second harmonic (12 W) of a Q-switched Nd:YLF laser (Photonics international, GM-50-527). Pump (IR) and probe (UV) beams were directed collinearly using a dichroic mirror (transmits IR and reflects UV), and focused onto the sample stream with a quartz lens. Raman scattered light was collected at 135° with a pair of fused quartz lenses, f-matched to a 1.26 m spectrograph (Spex 1269), which was equipped with a holographic grating (3600 groove mm−1) and intensified photodiode-array detector (Roper Scientific). The timing between pump and probe pulses was adjusted with a computer-controlled pulse generator (DG 535, Stanford Research Systems). The data acquisition sequence subtracts averages of paired readings, pump-probe minus probe-pump, in order to cancel effects of slowly changing conditions45. Static UVRR spectra at different temperatures were obtained by equilibrating the sample for ~10 mins at each measured temperature.
The spectra were calibrated using the standard Raman spectrum of acetone. Spectral analyses were carried out using GRAMS AI (Themo Electron Corp., formerly Galactic Industries), Solver module in MS Excel, and Matlab (Mathworks, Natick, MA) platforms.
Results
Circular dichroism
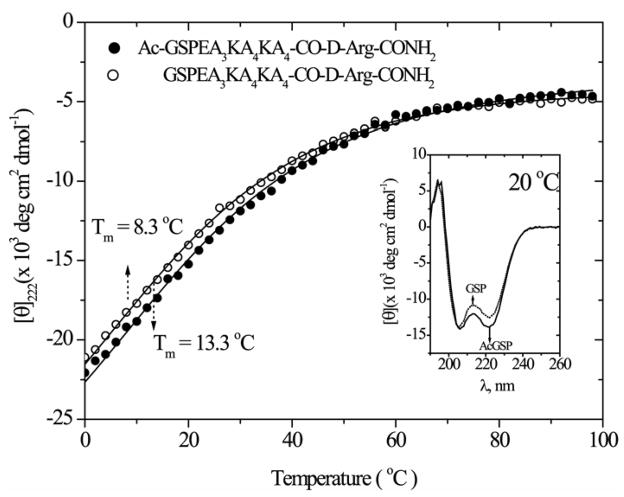
The far-UV CD spectrum of the Ac-GSP and GSP peptides show typical α-helical features when measured at 20°C (Figure 1 inset). As the temperature rises, the characteristic 222 nm trough steadily diminishes. As expected, the GSP peptide was less stable than the N-terminally modified Ac-GSP peptide. To allow comparison with earlier studies of monomeric helical peptides we used a two-state fit47, which gave a melting temperature of Tm = 13°C for Ac-GSP, and a lower value 8 °C for GSP. Thermodynamic parameters for Ac-GSP were ΔH = 9.3 kcal mol−1, ΔS = 33 cal K−1mol−1, and ΔCp = 0 cal K−1mol−1 and similar values were obtained for GSP. However, we note that this is a rough approximation for a helix-coil transition, and that application of a two-state treatment to an intrinsically multi-state process results in lower computed values of the enthalpy change than the true values.
Figure 1.
CD temperature profile for aqueous Ac-GSP and GSP peptide (200 μM, 50 mM sodium phosphate buffer, pH 7.4 contain 0.1 M sodium perchlorate) measured using the molar ellipticity at 222 nm (θ222). The continuous line corresponding to fitting of the two-state model using the equation47, θ222 = (θH − θC)/( 1+Keq ) + θC; θH and θC are the baseline ellipticities for helix and coil; the equilibrium contant, Keq = exp (−ΔG/RT) is related to enthalpy, entropy, and heat capacity changes via ΔG = ΔH + ΔCp ( T −Tm ) − T[ΔS + ΔCp ln (T/Tm)]. The inset shows the far UV CD spectrum measured at 20 °C.
UVRR spectra
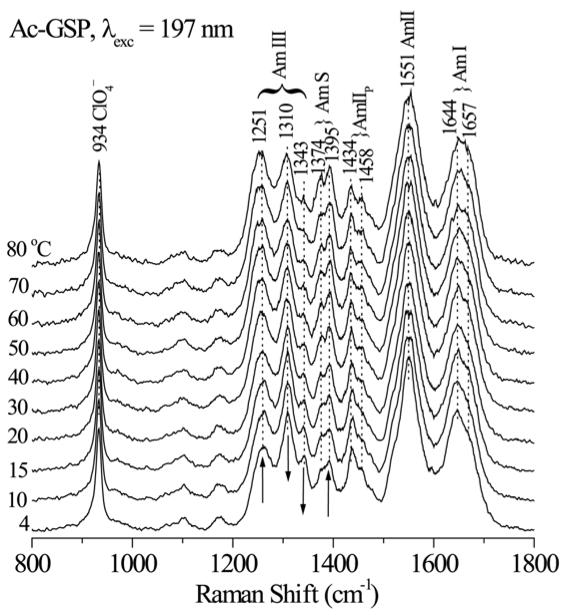
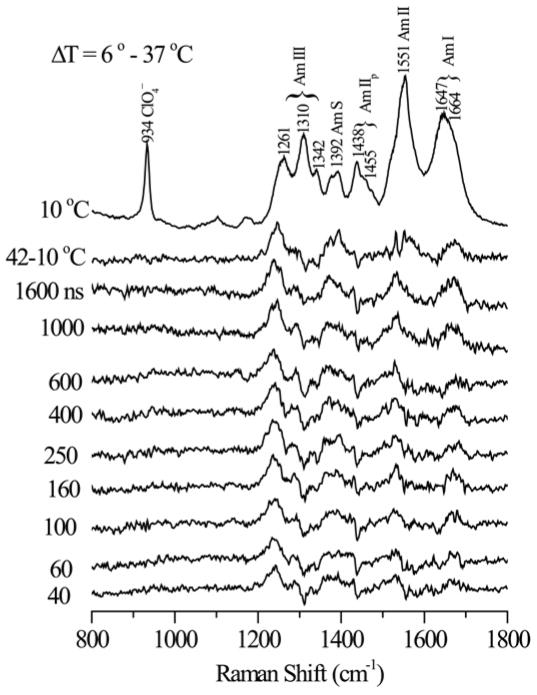
197 nm - excited UVRR spectra of Ac-GSP peptide (Figure 2) contain strongly enhanced amide bands (see labels) at expected positions for a mixture of α-helical and coil conformations35–39. As the temperature increases, coil contributions grow at the expense of helix contributions (arrows). To model the temperature dependence, we made the simple assumption of two component spectra, contributing in proportion to the helix and coil fractions. It is important to note that this assumption does not require that the peptide be in a two-state conformational equilibrium, but only that each residue in the peptide have only two states, one being helical and the other being an ensemble of other conformers with similar spectral characteristics. At a given temperature the peptide would be expected to adopt a large ensemble of different conformations with various helical lengths and positions within the peptide. A least squares algorithm was used to compute the basis spectra for the helical and non-helical states, using initial estimates of the helix fraction from the CD analysis and from the intensity of the Am S band (see below) and then iterating to find the fractions that give the smallest residuals across the entire spectrum, with no negative peaks.
Figure 2.
Raman spectra of aqueous Ac-GSP peptide (3 mg/ml, 50 mM sodium phosphate buffer, pH 7.4 contain 0.1 M sodium perchlorate) excited at 197 nm (1.2 mW) at the indicated temperatures. Band positions and assignments are marked at the top.
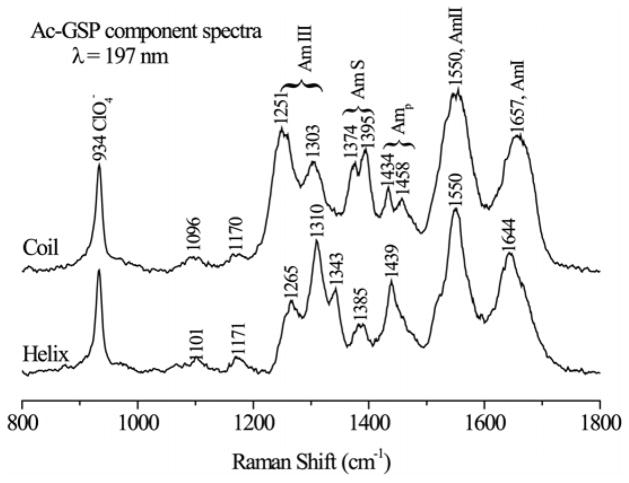
The computed basis spectra (Figure 3) have the expected helix and coil features, and agree well with canonical secondary structure spectra extracted from a set of structurally characterized proteins39, 48. Amide I (C=O stretching) shows a well-established upshift, from 1647 in the helix spectrum to 1656 cm−1 in the coil spectrum, which is attributable to the intra-helical H-bonds being stronger than the solvent H-bonds of the coil conformation39, 48. AmII (out-of-phase combination of C-N stretching and N-H bending) shows little dependence on conformation. AmIII (in-phase C-N stretching and N-H bending, plus other coordinates) has multiple components, with distinct patterns for different peptide conformations39, 48. Helix bands at 1265, 1310 and 1343 cm−1 are replaced by coil bands at 1251 and 1303 cm−1(‘coil’ in this context simply means melted -helix; there is considerable evidence that melting leads to local conformations which are similar to the polyproline II (PPII) conformation with some contribution from β-strand confromations)49–60. AmS is mainly CαH bending in character61; there are two components, at 1377 and 1395 cm−1 in the coil spectrum. At the α-helix conformation the CαH bending coordinate is dispersed among several modes, and there is no dominant AmS mode35, 62. Consequently AmS is a marker for non-helix structure. Nevertheless, we see remnant intensity at 1385 cm−1 in the helix spectrum indicating that the low-temperature component contains some population of non-helical conformations.
Figure 3.
Helix and coil UVRR basis spectra obtained from linear least squares fit of the data in Fig. 2 (see text).
Finally, we note the contribution from AmIIP, at ~1450 cm−1, from the single proline residue, near the start of the sequence. Because the X-Pro peptide link has no NH group, the AmIIP frequency is greatly shifted from the AmII frequency of the remaining amides, and its per-residue intensity is also higher63. The frequency is sensitive to the H-bonding status of the X-Pro carbonyl63, and the two coil AmIIP components suggest alternative conformations with stronger (1458 cm−1) or weaker (1434 cm−1) H-bonding. This conformational heterogeneity is quenched in the low-temperature spectrum, which shows a single component, at 1439 cm−1.
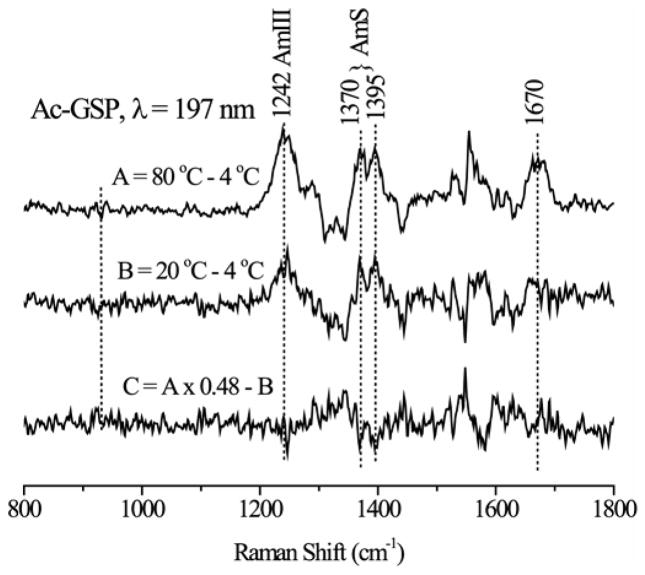
Our least squares procedure assumes that that the two basis spectra themselves are temperature independent. If there were a large temperature dependence, the two-component fitting procedure would fail. A small temperature dependence would still allow two basis spectra to be computed, but the bands would be broadened. Our computed spectra show narrow bands, with widths similar to amide bands in Raman spectra of other aqueous short peptides and polymers. The bands are no broader for the coil than the helix, even though the coil structure must have multiple conformations, whose distribution would shift with temperature. To check the spectral response to temperature, we subtracted the Ac-GSP spectrum at 4°C from the 80°C spectrum, producing the expected coil-helix difference spectrum (Figure 4). The same spectrum, with the expected reduced amplitude, was found in the 20 – 4°C difference spectrum. When these two difference spectra were subtracted, with appropriate scale factor, the residual was essentially at baseline. No sigmoidal features were seen, as would have been expected had the amide bands shifted between 20°C and 80°C.
Figure 4.
Temperature difference UVRR spectrum, and the double difference spectrum, showing negligible band shifts with temperature.
In their earlier helix-coil UVRR study, Lednev et al.10 assumed that the coil spectrum would vary with temperature, and modeled the temperature dependence with the difference UVRR spectrum between penta-alanine and tri-alanine, thus using the middle of the penta-alanine peptide as a surrogate for the coil structure. They found appreciable temperature coefficients for the amide band frequencies, and used these to correct their coil basis spectrum as a function of temperature. It is possible that the temperature insensitivity of our component spectra reflect accidental compensation between an intrinsic temperature dependence of the spectrum, reflecting anharmonic effects, and a shifting distribution of conformations with slightly different band positions.
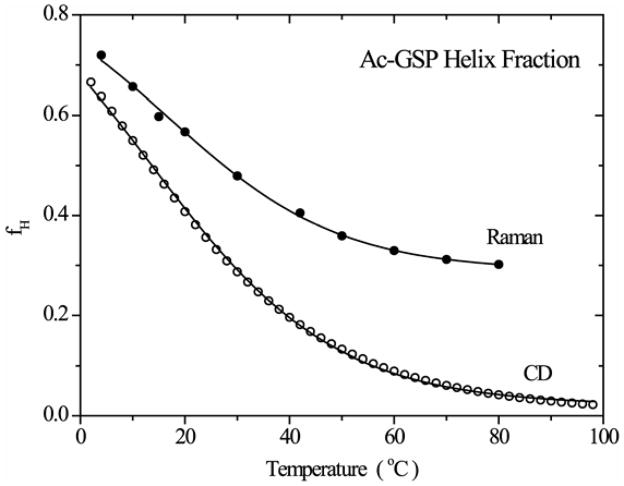
Figure 5 plots the temperature profile of the helix fraction obtained from our UVRR fitting procedure, and also from the two-state modeling of the CD ellipticity. The two curves approach convergence at low temperature, but deviate strongly at higher temperature, with the Raman data yielding higher helicity. This phenomenon has been noted before64, and is attributed to the different physical mechanisms underlying the two spectroscopies. The Raman bands are sensitive to the local conformation of the peptide bonds, whereas the CD ellipticity depends on delocalized electronic interactions along the ordered peptide chain65. Consequently CD is more sensitive to the length and degree of order of the helix. The divergence of the two curves indicates that as the temperature rises long helices are melted. The divergence between CD and IR at high temperature indicates that in the unfolded ensemble the average residue samples the alpha-helical phi/psi angles about one third of the time.
Figure 5.
Fractional helix content (fH) as a function of temperature for aqueous Ac-GSP peptide obtained by the two-state fitting of the CD (open circle) and UVRR (solid circle) spectra (see text)
T-jump/UVRR
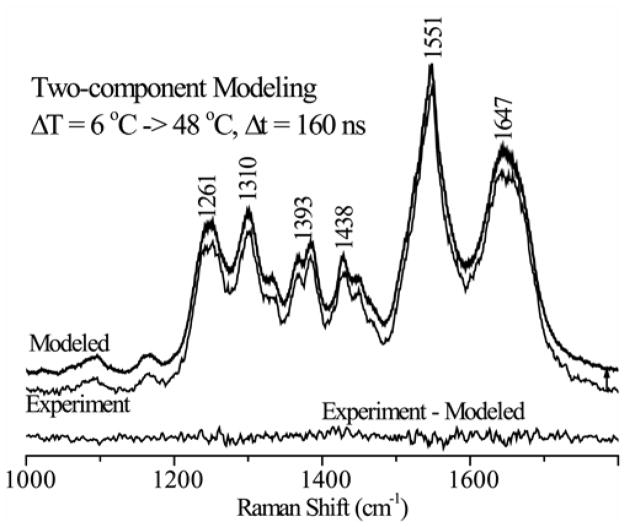
Figure 6 shows pump-probe difference spectra recorded at various delays following a 6–37°C temperature jump. The spectra evolve on the sub-microsecond time scale toward a response closely resembling the static difference spectrum for a similar temperature interval. The parent UVRR spectra were fit to our helix and coil basis spectra via least squares, to obtain the helix fractions. Figure 7 illustrates the quality of data fitting. Additional T-jumps were carried out, all starting at 6°C, and ending at temperatures between 18 and 61°C.
Figure 6.
Difference UVRR spectra (pump/probe - pobe) of aqueous Ac-GSP peptide (3 mg/ml, 50 mM sodium phosphate buffer, pH 7.4 contain 0.1 M sodium perchlorate) at the indicated delays following a 6 ° – 37 °C T-jump. [λT-jump= 1.9 μm, 0.6 mJ/pulse, 1 kHz; λUVRR= 197 nm, 1.2 μJ/pulse, 1 kHz]. The probe-only spectrum and a static 42 ° – 10 °C difference spectra are shown at the top.
Figure 7.
Sample data showing the two-state modeling of the experimentally measured T-jump/UVRR spectra using basis spectra corresponding to helix and coil components (Fig. 3). Thin and thick line spectra correspond to experimental and modeled (vertically displaced for clarity) spectra respectively. The residual shown at the bottom reflects the accuracy of the modeling.
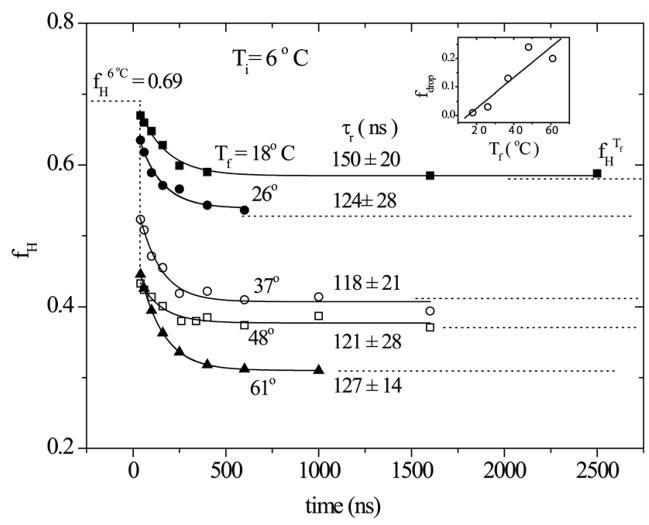
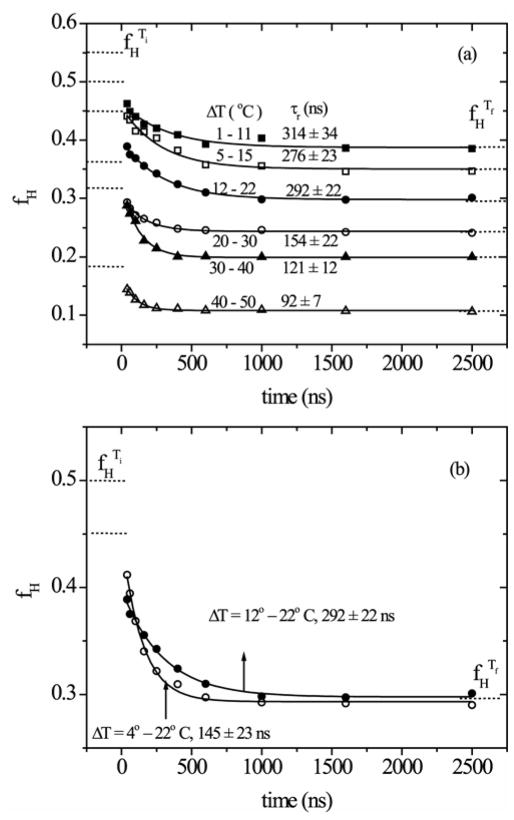
The decay curves for the computed helix fractions are shown in Figure 8. The data are adequately fit by mono-exponential decays. The time constants are all close to 120 ns. Moreover, the curves level off at helix fractions that correspond to the equilibrium fractions at the final temperatures (indicated as fHTf (- - -) in Figure 8), establishing that there is no unaccounted slower phase. However, there is a substantial drop from the equilibrium helix fraction at 6 °C to that of our first time point, 40 ns, and this prompt response increases with increasing final temperature (Figure 8 inset).
Figure 8.
T-jump induced time-resolved helical content change (ΔfH) obtained from two-state spectral modeling of the UVRR spectra for aqueous Ac-GSP peptide at the indicated Tf following T-jumps from 6 °C. The continuous line corresponds to a single exponential fit. Dotted lines indicate the equilibrium helix fractions at 6°C and at the final temperature following the T-jump. The inset plots the magnitude of the initial drop in helicity as a function of Tf.
The rate constant corresponding to the measured relaxation is essentially independent of temperature (Figure 8), a result that might appear to differ markedly from previous IR and fluorescence studies, which found large temperature dependencies. A large part of the discrepancy lies in the different time resolutions of the experiments (40 ns in the present study vs. < 10 ns in previous IR and fluorescence studies), and the fact that the IR measurements show complex kinetics that can only be fit with stretched exponentials or multiple exponentials. At the lowest temperature, we see very little missing amplitude in the prompt phase, and the rate constant we measure in the second phase is in good agreement with IR studies of a related peptide. However, with increasing temperature the IR studies show an increasingly rapid process that ultimately exceeds the resolution of our measurements. Consistent with this expectation we find missing amplitude in the prompt phase that increases with increasing final temperature. Additionally we hypothesized that the differing behavior might result, in part, from differing experimental protocols; since the IR and fluorescence studies varied both initial (Ti) and final (Tf) temperatures, with constant T-jumps (ΔT).
To test this idea further, we turned to the GSP peptide, which had a slightly lower helical potential Ac-GSP, allowing us to modulate the fraction of helix at a given temperature. The GSP peptide showed a relaxation time of 145 ns for a T-jump from 4 °C to 22 °C (Fig. 9), which is similar to the value observed for the Ac-GSP for similar initial and final temperatures (Fig. 8). However, we found that the rate of relaxation varied markedly with the initial temperature, with the relaxation time increasing to 292 ns when the initial temperature was 12 °C. These results are consistent with previous IR studies, which showed an increase in relaxation time with increasing size of the T-jump while holding Tf constant21. Further experiments showed that the relaxation rate increased in the T-jump/UVRR experiment when both Ti and Tf were raised (Figure 9a). Moreover the 10° C T-jumps showed that the prompt response remained nearly constant regardless of Ti.
Figure 9.
(a) As Figure 8, but with the initial temperature adjusted to produce 10°C T-jumps, for the GSP peptide. (b) Time course comparison for GSP peptide, with the same Tf, but different Ti.
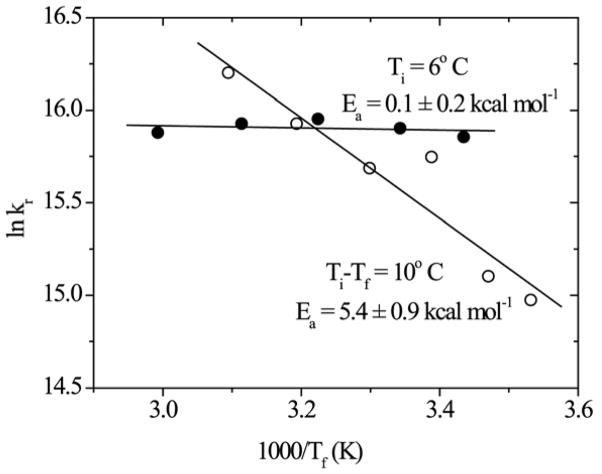
When the relaxation rate constants (lnk) are plotted against 1/T (Figure 10), the Arrhenius slopes give apparent activation energies of 0.1 kcal/mole for Ti = 6° C and 5.4 kcal/mole for variable Ti with ΔT = 10° C.
Figure 10.
Arrhenius plots of the T-jump relaxation rate constants at constant Ti (Ac-GSP peptide Fig. 8) and constant ΔT (GSP peptide, Fig. 9a).
Discussion
Table 1 lists the extant studies, in addition to the present one, which include the temperature dependence of the T-jump-induced relaxation of small α-helical peptides. The peptides are all based on the helix-forming poly-alanine sequence of Marqusee and Baldwin66, but have different patterns of interspersed charged residues (K or R), which insure solubility, and different capping groups at the N- and C-termini, which differentially stabilize the helix. Consequently the melting temperatures vary somewhat. Nevertheless, all the studies report relaxation time constants in the 100 – 400 ns range, when measured near the Tm temperature. Thus helix melting and formation occur on the sub-microsecond time frame, in the context of ~20-residue alanine-based peptides. However, the temperature coefficients of the observed relaxation rates vary widely.
Table 1.
Survey of helix-coil studies that included the temperature dependence of T-jump relaxations.
| Peptide | Monitor | Ti, °C | T, °C | Apparent Ea (kcal mol−1)⊥ | Ref. |
|---|---|---|---|---|---|
| Ac-WA3H+-(AAAR+A)3A | Fluorescence | varied | ~10.0 | 8.0 | [3] |
| A5(A3RA)3A | UVRR | 4.0 | varied | * | [10] |
| YGG(KA4)3COR | IR | varied | ~10.0 | 15.5 | [4] |
| AcYGSPEA3KA4KA4-DArg-CONH2 (Ac-Y-GSP peptide) | IR | varied | ~10.0 | 11.5 | [8] |
| [Rub2m]2+-CONH-AAAAA(AAARA)3A-CONH2 | IR | varied | ~10.0 | 13.5 | [19] |
| Ac-GSP peptide | UVRR | 6.0 | varied | 0.1 | Current Work |
| GSP peptide | varied | 10 | 5.4 | Current Work |
Obtained from the Arrhenius plots of the relaxation rate following the T-jumps.
A factor of 3 acceleration between Tf = 33 and 65° was decomposed via the helix/coil equilibrium constant into apparent Ea’s of 8.0 and −4.0 for unfolding and folding rates, assuming a 2-state kinetic model.
Part of the difference stems from different T-jump protocols, as noted above. The IR and fluorescence experiments employed relatively small T-jumps, less than 15°C, and varied both initial (Ti) and final (Tf) temperatures to sample the temperature dependence. In the UVRR studies it was possible to keep Ti constant and varied the magnitude of the T-jump in order to reach variable Tf. This is easier to do with UVRR than with IR or fluorescence monitoring, since the UVRR laser spot size is much smaller than the area of the IR or fluorescence probe beams. Consequently the pump laser can be focused more tightly in the UVRR experiment, producing larger T-jumps.
We find essentially no temperature dependence when Ti is held constant, but a substantial temperature dependence when both Ti and Tf are increased. This difference can be connected to the temperature dependence of the helix length distribution. The deviating CD and UVRR temperature profiles (Figure 5) indicate that the average helix length becomes shorter as the temperature rises. Thus relaxation in response to a constant T-jump accelerates as the distribution of structures shifts to shorter helices.
A key to interpreting these results is the drop in the UVRR-determined helicity within the ~40 ns time resolution of the experiment. There are two distinct kinetic phases. In the constant Ti (= 6°C) experiments, the amplitude of the fast phase is initially small, but increases steadily with increasing Tf, until it reaches 50 % of the total helicity change at Tf = 61°C. Thus the fast phase is thermally activated, whereas the slow phase is not. The two phases can be represented by
in which an ensemble of molecules with a temperature-dependent distributions of helix lengths is rapidly converted to a set of intermediate structures and more slowly to a set of final structures. The fast phase is enthalpic while the slow phase is entropic. At low temperatures the fast phase slows and merges with the slow phase; the enthalpic process is then rate-limiting. As the temperature increases the entropic process is rate-limiting, and the two phases separate. In the variable Ti experiment, however, the distribution of helix lengths shifts with each measurement, and the enthalpic process remains rate-limiting; the amplitude of the fast phase remains small and the slow phase accelerates.
We propose that the enthalpic process is disruption of helices via the breaking of inter-residue H-bonds, while the entropic process is the subsequent redistribution of conformations about the peptide bonds. Some bond rotation does accompany the H-bond breaking, as reflected in the UVRR change in the prompt kinetic phase, but even at the highest Tf, this prompt change represents only 50 % of the equilibrium change in helicity. At this stage many of the residues still have phi/psi angles close to the α-helical values, despite the loss of long-range order. Their subsequent reorientation to allow a more extended conformation of the chains is entropic in character, and becomes the slow step at high temperatures. Slow polymer dynamics is consistent with the concerted motions that would be required for chain extension.
In the constant ΔT experiment, Ti is allowed to rise in parallel with Tf. The prompt phase of the UVRR response is then of constant amplitude, while the slow phase accelerates. This behavior implies that the enthalpic phase remains largely rate-limiting as Ti is raised, so that UVRR relaxation now represents a combination of enthalpic and entropic phases. We know from the divergence of CD and UVRR temperature profiles that the consequence of raising Ti is to shift the distribution of helix lengths from longer to shorter helices. Thus there are fewer inter-residue H-bonds remaining to be broken, and helix refolding contributes increasingly to the overall rate of the first kinetic phase, which therefore slows down and merges with the entropic phase.
It is possible that the modest temperature coefficient observed by Lednev et al.,10 (factor of three from 33 to 65° C, vs no change in our experiment), despite carrying out their T-jump UVRR study at constant Ti, stems from a similar merging of the enthalpic and entropic phases. The AP peptide they studied may have had shorter average helices, since the N-terminal sequence contained only alanine, a less effective N-cap than our Ac-GSP sequence40–43. The less effective N-cap in our GSP may also have contributed to its temperature response.
The differing experimental protocols for the UVRR and for the IR and fluorescence studies, constant Ti vs constant ΔT, do not entirely explain the differing temperature sensitivities. We observed only a factor of ~3 acceleration in the relaxation rate when Tf was raised from 10 to 50°C, with 10°C T-jumps, whereas the IR and fluorescence studies all found factors of ~10 acceleration over comparable ranges. This discrepancy is attributable to the differing physical interactions being monitored. The IR experiments monitor the peptide amide I band exclusively; its position in the IR spectrum is influenced by nearest-neighbor through bond coupling, and by non-nearest neighbor dipole-dipole coupling of C=O vibrators.60 The UVRR analysis covers the entire spectral region between 1200 and 1700 cm−1. While the amide I band is included in the analysis, its main determinants are amide III and S, which are more sensitive to the phi and psi angles around the individual peptide bonds. Thus the UVRR response is more local than the IR response, which may therefore emphasize longer helices, as does CD. The fluorescence measurements depend on quenching mechanisms, which are also long range. Thus, the cooperative unit that is being probed in the Raman experiments is smaller and more heterogeneous than in the IR and fluorescence measurements, leading to a smaller value of the apparent activation energy for the process.
Because of the longer range of the IR and fluorescence probes, they are more sensitive to the enthalpic phase of the melting, in which long range order is disrupted by the breaking of inter-residue H-bonds. The IR and fluorescence time-course are weighted toward the temperature-dependent phase, and consequently the apparent activation energy is higher.
There is also an unresolvable prompt response in the IR T-jump transients,2,4,5 but in this case a temperature-dependent spectral shift may be responsible. A marked temperature dependence of the amide I IR band has been documented67,68, and has been attributed to solvent H-bond changes. As described above, we find that the helix and coil UVRR spectra are invariant with temperature.
A new T-jump study from Asher’s lab34, using selectively isotope-labeled peptide (AdP) shows kinetic complexity that is unresolved in our experiment. Different relaxation rates are reported for the central and exterior peptide bonds, which respond differently to temperature. These interesting effects were attributed to involvement of local ‘defects’, including π-bulges and 310 helices in the melting process, induced via the presence hydrophilic residues. Such kinetic complexities are averaged in our experiment. Our averaged rates are nevertheless useful in revealing a dichotomy of enthalpic and entropic regimes.
Conclusions
When provided with good end-capping groups, a short alanine-based peptide gives a ~120 ns helix-coil T-jump/UVRR response, which is independent of final temperature, provided the initial temperature is held constant. However, an unresolvable fast phase is also seen, whose amplitude depends strongly on temperature. This experiment resolves an enthalpic fast phase of the unfolding mechanism from an entropic slow phase. We propose that the enthalpic phase involves breaking of inter-residue H-bonds, with partial reorientation of peptide bonds, while the entropic phase involves the further reorientation required for chain extension.
When the initial temperature is raised, in parallel with the final temperature, the fast phase of the T-jump/UVRR response is minimized, and merges with an accelerating slow phase. Thus the enthalpic process remains largely rate limiting, an effect attributable to the shift in the initial distribution of helix lengths from longer to shorter helices. Rates accelerate even more in the corresponding T-jump/IR and T-jump/fluorescence experiments, because the IR and fluorescence spectra are more responsive to long-range order than is the UVRR spectrum, and the signals are therefore weighted toward the H-bond-breaking enthalpic process.
Acknowledgments
This work was supported by NIH grant GM 25158 to TGS and by GM48130 to WFD.
References
- 1.Dyer RB, Gai F, Woodruff WH. Acc Chem Res. 1998;31:709. [Google Scholar]
- 2.Thompson PA, Eaton WA, Hofrichter J. Biochemistry. 1997;36:9200. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 3.Thompson PA, Munoz V, Jas GS, Henry ER, Eaton WA, Hofrichter J. J Phys Chem B. 2000;104:378. [Google Scholar]
- 4.Huang CY, Klemke JW, Getahun Z, DeGrado WF, Gai F. J Am Chem Soc. 2001;123:9235. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]
- 5.Huang CY, Getahun Z, Wang T, DeGrado WF, Gai F. J Am Chem Soc. 2001;123:12111. doi: 10.1021/ja016631q. [DOI] [PubMed] [Google Scholar]
- 6.Huang CY, Getahun Z, Zhu YJ, Klemke JW, DeGrado WF, Gai F. Proc Natl Acad Sci U S A. 2002;99:2788. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, Du DG, Gai F. Chem Phys Lett. 2003;370:842. [Google Scholar]
- 8.Wang T, Zhu YJ, Getahun Z, Du DG, Huang CY, DeGrado WF, Gai F. J Phys Chem B. 2004;108:15301. doi: 10.1021/jp037272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams S, Causgrove TP, Gilmanshin R, Fang KS, Callender RH, Woodruff WH, Dyer RB. Biochemistry. 1996;35:691. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 10.Lednev IK, Karnoup AS, Sparrow MC, Asher SA. J Am Chem Soc. 1999;121:4076. doi: 10.1021/ja003381p. [DOI] [PubMed] [Google Scholar]
- 11.Brooks CL. J Phys Chem. 1996;100:2546. [Google Scholar]
- 12.Hagen SJ, Eaton WA. J Mol Biol. 2000;301:1019. doi: 10.1006/jmbi.2000.3969. [DOI] [PubMed] [Google Scholar]
- 13.Doig AJ. Biophys Chem. 2002;101:281. doi: 10.1016/s0301-4622(02)00170-9. [DOI] [PubMed] [Google Scholar]
- 14.Scheraga HA, Vile JA, Ripoll DR. Biophys Chem. 2002;101:255. doi: 10.1016/s0301-4622(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 15.Werner JH, Dyer RB, Fesinmeyer RM, Andersen NH. J Phys Chem B. 2002;106:487. [Google Scholar]
- 16.Doshi U, Munoz V. Chem Phys. 2004;307:129. [Google Scholar]
- 17.Doshi UR, Munoz V. J Phys Chem B. 2004;108:8497. [Google Scholar]
- 18.Gnanakaran S, Garcia AE. Prot Str Funct Bioinf. 2005;59:773. doi: 10.1002/prot.20439. [DOI] [PubMed] [Google Scholar]
- 19.Sorin EJ, Pande VS. Biophys J. 2005;88:2472. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballew RM, Sabelko J, Gruebele M. Proc Natl Acad Sci U S A. 1996;93:5759. doi: 10.1073/pnas.93.12.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snow CD, Qiu LL, Du DG, Gai F, Hagen SJ, Pande VS. Proc Natl Acad Sci U S A. 2004;101:4077. doi: 10.1073/pnas.0305260101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ervin J, Sabelko J, Gruebele M. J Photochem and Photobiol B-Biol. 2000;54:1. doi: 10.1016/s1011-1344(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Rigler R, Rabl CR, Jovin TM. Rev Sci Instrum. 1974;45:580. [Google Scholar]
- 24.Colley CS, Clark IP, Griffiths-Jones SR, George MW, Searle MS. Chem Comm. 2000:1493. [Google Scholar]
- 25.Decatur SM. Biopolymers. 2000;54:180. doi: 10.1002/1097-0282(200009)54:3<180::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Gilmanshin R, Williams S, Callender RH, Woodruff WH, Dyer RB. Proc Natl Acad Sci U S A. 1997;94:3709. doi: 10.1073/pnas.94.8.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulotta M, Rogatsky E, Callender RH, Dyer RB. Biophys J. 2003;84:1909. doi: 10.1016/S0006-3495(03)74999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinstadler D, Fabian H, Backmann J, Naumann D. Biochemistry. 1996;35:15822. doi: 10.1021/bi961810j. [DOI] [PubMed] [Google Scholar]
- 29.Huang CY, He S, DeGrado WF, McCafferty DG, Gai F. J Am Chem Soc. 2002;124:12674. doi: 10.1021/ja028084u. [DOI] [PubMed] [Google Scholar]
- 30.Huang CY, Balakrishnan G, Spiro TG. Biochemistry. 2005;44:15734. doi: 10.1021/bi051578u. [DOI] [PubMed] [Google Scholar]
- 31.JiJi RD, Balakrishnan G, Hu Y, Spiro TG. Biochemistry. 2006;45:34. doi: 10.1021/bi051507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan G, Hu Y, Spiro TG. Appl Spectrosc. 2006;60:347. doi: 10.1366/000370206776593799. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto K, Mizutani Y, Kitagawa T. Biophys J. 2000;79:485. doi: 10.1016/S0006-3495(00)76310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikhonin AV, Asher SA, Bykov SV, Murza A. J Phys Chem B. 2007;111:3280–3292. doi: 10.1021/jp0654009. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Purrello R, Jordan T, Spiro TG. J Am Chem Soc. 1991;113:6359. [Google Scholar]
- 36.Wang Y, Purrello R, Georgiou S, Spiro TG. J Am Chem Soc. 1991;113:6368. [Google Scholar]
- 37.Chen XG, Asher SA, Schweitzerstenner R, Mirkin NG, Krimm S. J Am Chem Soc. 1995;117:2884. [Google Scholar]
- 38.Asher SA, Chi ZH, Li PS. J Raman Spectrosc. 1998;29:927. [Google Scholar]
- 39.Huang CY, Balakrishnan G, Spiro TG. J Raman Spectrosc. 2006;37:277. [Google Scholar]
- 40.Richardson JS, Richardson DC. Science. 1988;240:1648. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 41.Regan L. Proc Natl Acad Sci U S A. 1993;90:10907. doi: 10.1073/pnas.90.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aurora R, Rose GD. Protein Sci. 1998;7:21. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doig AJ, Baldwin RL. Protein Sci. 1995;4:1325. doi: 10.1002/pro.5560040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobs DJ, Wood GG. Biopolymers. 2004;75:1. doi: 10.1002/bip.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao XJ, Tengroth C, Chen RP, Simpson WR, Spiro TG. J Raman Spectrosc. 1999;30:773. [Google Scholar]
- 46.Balakrishnan G, Hu Y, Nielsen SB, Spiro TG. Appl Spectrosc. 2005;59:776. doi: 10.1366/0003702054280702. [DOI] [PubMed] [Google Scholar]
- 47.Becktel WJ, Schellman JA. Biopolymers. 1987;26:1859. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 48.Chi ZH, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 49.Mikhonin AV, Myshakina NS, Bykov SV, Asher SA. J Am Chem Soc. 2005;127:7712. doi: 10.1021/ja044636s. [DOI] [PubMed] [Google Scholar]
- 50.Mikhonin AV, Asher SA. J Phys Chem B. 2005;109:3047. doi: 10.1021/jp0460442. [DOI] [PubMed] [Google Scholar]
- 51.Eker F, Griebenow K, Cao XL, Nafie LA, Schweitzer-Stenner R. Proc Natl Acad Sci U S A. 2004;101:10054. doi: 10.1073/pnas.0402623101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eker F, Griebenow K, Schweitzer-Stenner R. Biochemistry. 2004;43:6893. doi: 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- 53.Asher SA, Mikhonin AV, Bykov S. J Am Chem Soc. 2004;126:8433. doi: 10.1021/ja049518j. [DOI] [PubMed] [Google Scholar]
- 54.Schweitzer-Stenner R, Measey TJ. Proc Natl Acad Sci U S A. 2007;104:6649–6654. doi: 10.1073/pnas.0700006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweitzer-Stenner R, Measey T, Kakalis L, Jordan F, Pizzanelli S, Forte C, Griebenow K. Biochemistry. 2007;46:1587–1596. doi: 10.1021/bi062224l. [DOI] [PubMed] [Google Scholar]
- 56.Graf J, Nguyen PH, Stock G, Schwalbe H. J Am Chem Soc. 2007;129:1179–1189. doi: 10.1021/ja0660406. [DOI] [PubMed] [Google Scholar]
- 57.Shi ZS, Chen K, Liu ZG, Kallenbach NR. Chemical Reviews. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 58.Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA. Proc Natl Acad Sci U S A. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi ZS, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Proc Natl Acad Sci U S A. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torii H, Tasumi M. J Raman Spectrosc. 1998;29:81–86. [Google Scholar]
- 61.Krimm S, Bandekar J. Adv Protein Chem. 1986;38:181. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 62.Jordan T, Spiro TG. J Raman Spectrosc. 1994;25:537. [Google Scholar]
- 63.Jordan T, Mukerji I, Wang Y, Spiro TG. J Mol Struct. 1996;379:51. [Google Scholar]
- 64.Ozdemir A, Lednev IK, Asher SA. Biochemistry. 2002;41:1893. doi: 10.1021/bi010970e. [DOI] [PubMed] [Google Scholar]
- 65.Woody RW, Tinoco I. J Chem Phys. 1967;46:4927. [Google Scholar]
- 66.Marqusee S, Baldwin RL. Proc Natl Acad Sci U S A. 1987;84:8898. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manas ES, Getahun Z, Wright WW, DeGrado WF, Vanderkooi JM. J Am Chem Soc. 2000;122:9883. [Google Scholar]
- 68.Walsh STR, Cheng RP, Wright WW, Alonso DOV, Daggett V, Vanderkooi JM, DeGrado WF. Protein Sci. 2003;12:520. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]